Abstract
Traditional virus-induced gene silencing (VIGS) is a powerful virus-based short interfering RNA-mediated RNA silencing technique for plant functional genomics. Besides short interfering RNAs, microRNAs (miRNAs) have also been shown to regulate gene expression by RNA silencing in various organisms. However, plant virus-based miRNA silencing has not been reported. In addition, a number of plant miRNAs have been identified or predicted, while their functions are largely unknown. Thus, there is an urgent need for the development of new technologies to study miRNA function. Here, we report that a modified cabbage leaf-curl geminivirus vector can be used to express artificial and endogenous miRNAs in plants. Using this viral miRNA expression system, we demonstrate that VIGS using artificial miRNAs, dubbed as “MIR VIGS,” was effective to silence the expression of endogenous genes, including PDS, Su, CLA1, and SGT1, in Nicotiana benthamiana. Silencing of SGT1 led to the loss of N-mediated resistance to Tobacco mosaic virus. Furthermore, using this viral miRNA expression system, we found that viral ectopic expression of endogenous miR156 and miR165 but not their mutants in N. benthamiana resulted in earlier abnormal developmental phenotypes, and expression of miR165 induced abnormal chlorotic spots on leaves. These results demonstrate that the cabbage leaf-curl geminivirus-based miRNA expression system can be utilized not only to specifically silence genes involved in general metabolism and defense but also to investigate the function of endogenous miRNAs in plants.
The explosive genome sequence data and increasing collections of a large number of ESTs demand simple and rapid reverse genetics tools to bridge the gap between sequence information and gene functions in plants. In recent years, RNA silencing has emerged as one of the most powerful techniques in plant functional genomics. It down-regulates gene expression at the posttranscriptional level through small RNAs, referred to as silencing RNAs, that act in a sequence-specific manner to target mRNA for degradation or to inhibit translation. Short interfering RNAs (siRNAs) and microRNAs (miRNAs) are two major classes of silencing RNAs (Llave et al., 2002; Reinhart et al., 2002; Chen, 2009). Techniques involving the use of both siRNA and miRNA have been developed to knock down gene expression in plants (Lu et al., 2003; Kusaba, 2004; Tenllado et al., 2004; Watson et al., 2005; Mansoor et al., 2006).
Traditional virus-induced gene silencing (VIGS) is a siRNA-mediated silencing approach and uses viral vectors carrying a fragment of a gene of interest to generate long double-stranded RNAs, which are then processed by Dicer to produce siRNAs for the silencing of the target gene. This traditional siRNA-mediated VIGS, dubbed as “SIR VIGS” hereafter, has been exploited as an effective and rapid gene “knockdown” technology and widely used to define gene functions (Robertson, 2004). This technique avoids stable plant transformation, and virus-induced silencing phenotypes can be readily observed within 3 to 4 weeks. However, SIR VIGS can generate a large number of different 21- to 24-nucleotide siRNAs, which may cause unintended nonspecific gene suppression (Jackson et al., 2003; Jackson and Linsley, 2004; Xu et al., 2006).
On the other hand, miRNAs are widely distributed noncoding small RNAs, usually 18 to 25 nucleotides long. They regulate gene expression and play an important role in plant development and response to biotic or abiotic stress (Lagos-Quintana et al., 2001, 2003; Carrington and Ambros, 2003; Bartel, 2004). In plants, miRNAs can inhibit gene expression by inducing target mRNA's degradation or translation repression (Bartel, 2009; Voinnet, 2009). Recently, artificial miRNA (amiRNA) technology has been developed using endogenous miRNA precursors to generate new target-specific miRNAs for gene silencing in animals and plants (Schwab et al., 2006; Qu et al., 2007; Khraiwesh et al., 2008; Ossowski et al., 2008; Warthmann et al., 2008; Zhao et al., 2008; Molnar et al., 2009). AmiRNA has been shown to efficiently and specifically silence both single and multiple target genes by well-designed precursor sequences (Schwab et al., 2006). Compared with siRNAs, miRNAs cause more accurate gene silencing, as deduced from genome-wide expression profiling (Schwab et al., 2005). Moreover, computational biology and high-throughput sequencing have predicted numerous putative miRNAs with unknown functions (Zhang et al., 2006; Sunkar and Jagadeeswaran, 2008). Therefore, there is a need to develop a toolbox for the induction of miRNA-mediated gene silencing and for functional analyses of miRNAs in plants.
Cabbage leaf curl virus (CaLCuV) is a member of the genus Begomovirus, family Geminiviridae. CaLCuV infects a broad range of plants, including cabbage (Brassica capitata), cauliflower (Brassica oleracea), Arabidopsis (Arabidopsis thaliana), and Nicotiana benthamiana, and replicates in the nucleus (Hill et al., 1998). Its genome consists of two approximately 2.6-kilonucleotide circular single-stranded DNA components, DNA-A and DNA-B. DNA-A possesses five genes, AR1, AL1, AL2, AL3, and AL4, encoding the coat protein, replication-associated protein, transcription activator, replication enhancer, and putative pathogenesis-related protein, respectively. DNA-B encodes two movement proteins, BR1 and BL1. The two genomic components share an almost identical common region of approximately 200 nucleotides, which includes the viral origin of replication and bidirectional promoters for both virion sense and complementary sense gene expression (Hill et al., 1998; Paximadis et al., 1999). The coat protein AR1 gene is not required for viral systemic infection, and an AR1 gene replacement CaLCuV-based VIGS vector has already been developed to trigger siRNA-mediated silencing in Arabidopsis (Turnage et al., 2002). Here, we report that a modified CaLCuV vector can be used not only to express amiRNAs to induce efficient VIGS using amiRNAs (MIR VIGS) but also to express endogenous miRNAs to study their biological functions.
RESULTS
Construction of a User-Friendly Infectious CaLCuV Vector
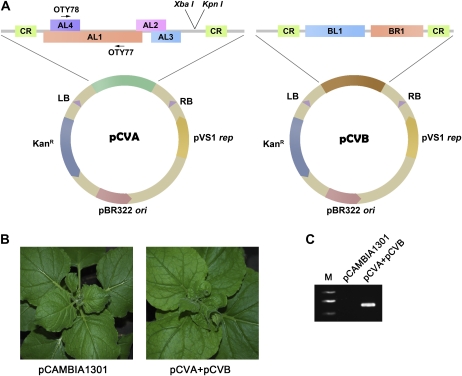
The currently available CaLCuV vector (Turnage et al., 2002) requires particle bombardment, a relatively tedious and less effective process, to introduce viral DNA into plants. In contrast, agroinfection is a simple and efficient approach to establish virus infection in plants (Grimsley et al., 1986; Vaghchhipawala and Mysore, 2008). In order to use this technique to infect plants with CaLCuV, we converted the currently available CaLCuV vector (Turnage et al., 2002) into T-DNA viral vectors by cloning the partially duplicated genome sequences of the A and B components of CaLCuV into pCAMBIA1301 (AF234297) to produce pCVA and pCVB, respectively (Fig. 1A). N. benthamiana plants infiltrated with Agrobacterium tumefaciens carrying both pCVA and pCVB developed mild leaf curling at 3 weeks post agroinoculation, while plants infiltrated with Agrobacterium carrying the empty binary vector remained symptomless (Fig. 1B). Viral DNA accumulated in the upper systemic leaves and was readily detectable by PCR using CaLCuV DNA A-specific primers (Fig. 1C). These results demonstrate that CaLCuV is infectious once delivered into plants by agroinoculation.
Figure 1.
Agroinfection of plants with the modified CaLCuV vector. A, Construction of CaLCuV T-DNA vector. CR, Common region; LB, left border; RB, right border. B, Agroinfection of N. benthamiana plants with CaLCuV pCVA and pCVB (right), but not the control vector pCAMBIA1301 (left), causes the leaf-curl symptom. Photographs were taken of N. benthamiana plants at 3 wpi. C, PCR showing the existence of CaLCuV infection in plants infiltrated with Agrobacterium containing pCVA and pCVB (right lane) but not pCAMBIA1301 (middle lane). The positions of primers OTY77/OTY78 for PCR detection are indicated. M, Marker.
MIR VIGS of Marker Plant Genes
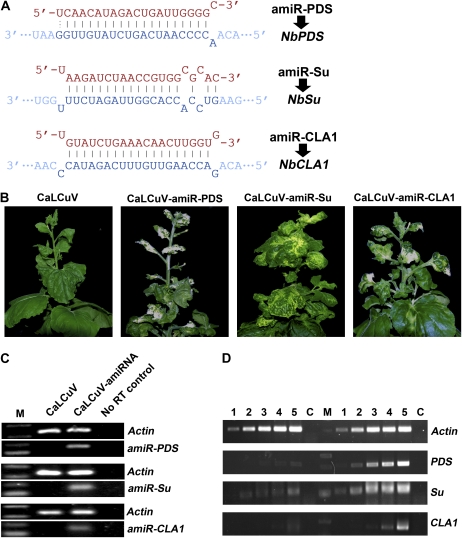
To test whether the modified CaLCuV vectors could express amiRNAs to induce gene silencing, we first identified three N. benthamiana genes: Phytoene desaturase (NbPDS; EU165355), Sulfur (NbSu; AJ571699), and Cloroplastos alterados1 (NbCLA1; N. benthamiana EST NbGI-3.0 Annotator TC16528). We then used the Web MicroRNA Designer, a Web-based tool (http://wmd2.weigelworld.org), to aid our amiRNA design and cloned the precursor sequences of amiRNAs targeting NbPDS, NbSu, and NbCLA1 (Fig. 2A) into pCVA in the same orientation of the AR1 gene to produce pCVA-amiR-PDS, pCVA-amiR-Su, and pCVA-amiR-CLA1, respectively. The CaLCuV AR1 gene promoter would drive the transcription of the amiRNA precursors in the nucleus, where these amiRNA precursors are expected to be processed to mature amiRNAs to induce silencing of target genes. It has been reported that SIR VIGS of PDS and Su resulted in photobleached leaves (Kumagai et al., 1995) and yellow-colored leaves (Hiriart et al., 2002, 2003). CLA1 encodes 1-deoxy-d-xylulose-5-phosphate synthase, and Arabidopsis plants with a CLA1 null mutation are albino (Mandel et al., 1996; Estevez et al., 2000, 2001). Similarly, in three separate experiments (four plants were used for each amiRNA in a single repeat experiment), all N. benthamiana plants coinfiltrated with Agrobacterium mixtures containing pCVA-amiR-PDS and pCVB (CaLCuV-amiR-PDS) developed a typical photobleached phenotype at approximately 3 weeks post infiltration (wpi). Plants coinfiltrated with Agrobacterium mixtures containing pCVA-amiR-Su and pCVB (CaLCuV-amiR-Su) resulted in yellowing leaves at about 3 wpi. Albino appeared in the freshly grown part of the plants coinfiltrated with Agrobacterium cultures containing pCVA-amiR-CLA1 and pCVB (CaLCuV-amiR-CLA1) in 2 to 3 wpi. However, none of CaLCuV-infected control plants showed any silencing phenotypes (Fig. 2B).
Figure 2.
MIR VIGS of marker genes. A, The designed amiRNA sequences and their related target mRNA sequences. B, Silencing phenotypes of marker genes in N. benthamiana plants coinfiltrated with CaLCuV, CaLCuV-amiR-PDS, CaLCuV-amiR-Su, or CaLCuV-amiR-CLA1. Photographs were taken at 35 dpi. C, End-point RT-PCR (30 cycles) analyses showing the existence of mature amiRNAs (amiR-PDS, amiR-Su, and amiR-CLA1) in inoculated plants. D, Semiquantitative RT-PCR analysis of the effects of MIR VIGS on the levels of the PDS,Su, and CLA1 mRNAs. Lanes 1 to 5 represent PCRs with 18, 21, 24, 27, and 30 cycles, respectively. PCR products to the left of the marker (M) are from the silenced plants, and those to the right are the nonsilenced controls. Actin mRNA was used as an internal control, and a no-RT control (C) was also included.
The end-point stem-loop reverse transcription (RT)-PCR is a highly sensitive and specific method to detect miRNA and has been widely used for the detection and quantification of miRNAs (Chen et al., 2005; Tang et al., 2006; Varkonyi-Gasic et al., 2007). To investigate whether CaLCuV-based expression of amiRNA genes can generate mature amiRNAs in plants, we did end-point stem-loop RT-PCR. Indeed, the corresponding mature amiRNAs were detected in plants infected with CaLCuV-amiR-PDS, CaLCuV-amiR-Su, or CaLCuV-amiR-CLA1, respectively, but not in CaLCuV-infected plants (Fig. 2C).
To confirm the silencing of NbPDS, NbSu, and NbCLA1 genes by CaLCuV-delivered amiRNAs at the molecular level, we performed semiquantitative RT-PCR. As shown in Figure 2D, the RNA levels of NbPDS, NbSu, or NbCLA1 in leaf tissues with silencing phenotypes decreased when compared with that in CaLCuV-based vector-infected leaf tissues. In all RNA samples, the endogenous actin mRNA levels, as an internal control, were similar (Fig. 2D). Taken together, our results suggest that CaLCuV-based vector can be used to express amiRNA to induce efficient gene silencing in N. benthamiana plants.
MIR VIGS of NbSGT1 Compromises N-Mediated Resistance to Tobacco mosaic virus
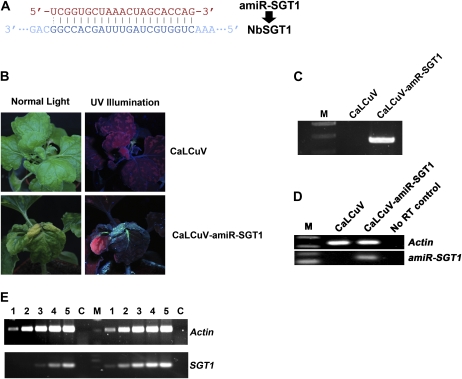
In order to test whether the modified CaLCuV-based miRNA expression vector can be used to study plant defense signaling, we decided to use this system to silence the SGT1 gene (for suppressor of G-two allele of Skp1) and investigated the effect of silencing of SGT1 on N gene-mediated resistance to Tobacco mosaic virus (TMV). SGT1 is a highly conserved eukaryotic protein and has been shown to interact directly with RAR1 and HSP90 to form a protein complex that plays an important role in R protein-mediated host resistance and nonhost resistance in plants (Shirasu and Schulze-Lefert, 2003; Schulze-Lefert, 2004). Silencing of SGT1 led to the loss of N-mediated resistance to TMV (Liu et al., 2002b; Peart et al., 2002; Xu et al., 2010). We designed an amiRNA targeting NbSGT1 (amiR-SGT1; Fig. 3A) and used the CaLCuV vector to express its precursor gene to induce miRNA-mediated silencing of NbSGT1 in N-containing transgenic N. benthamiana (NN plants; Liu et al., 2002a). As shown in Figure 3B, NN plants infected by CaLCuV vector alone remained resistant to TMV. GFP-tagged TMV (TMV-GFP; Liu et al., 2002a) was unable to move into the upper leaves. In contrast, plants infiltrated with Agrobacterium mixtures carrying pCVA-amiR-SGT1 and pCVB (CaLCuV-amiR-SGT1) became susceptible to TMV infection. TMV-GFP moved into the upper uninoculated leaves, and strong GFP fluorescence was observed (Fig. 3B). We then confirmed the presence of TMV RNA in the upper uninfected leaves by RT-PCR analysis using primers that anneal to the TMV movement protein gene (Fig. 3C). No TMV RNA was detected in NN control plants infected with CaLCuV alone. We repeated this experiment four times, with four plants for each construct in each experiment, and obtained similar results.
Figure 3.
TMV response to MIR VIGS of NbSGT1. A, The amiR-SGT1 sequence and its cognate target sequence. B, N. benthamiana plant coinfiltrated with TMV-GFP and CaLCuV (top row) or CaLCuV-amiR-SGT1 (bottom row). Photographs were taken at 20 dpi. C, RT-PCR detection of TMV in plants agroinfected by CaLCuV-amiR-SGT1. D, End-point stem-loop RT-PCR (30 cycles) analysis showing the existence of mature amiRNAs (amiR-SGT1) in inoculated plants. E, Semiquantitative RT-PCR analysis of the effects of MIR VIGS on NbSGT1 mRNA. Lanes 1 to 5 represent PCRs with 18, 21, 24, 27, and 30 cycles, respectively. PCR products to the left of the marker (M) are from the silenced plants, and those to the right are the nonsilenced controls. Actin mRNA was used as an internal control, and a no-RT control (C) was also included.
To verify the production of amiRNAs targeting NbSGT1 RNA, we performed end-point stem-loop RT-PCR. As shown in Figure 3D, amiR-SGT1 was detected in CaLCuV-amiR-SGT1-infected plants but not in CaLCuV vector-infected plants. In both RNA samples, the internal control RNA levels of endogenous actin were similar (Fig. 3D). These results showed that amiR-SGT1 was indeed produced in CaLCuV-amiR-SGT1-infected plants. Moreover, we performed semiquantitative RT-PCR to examine the effect of amiR-SGT1 on the endogenous mRNA level of NbSGT1. In CaLCuV-amiR-SGT1-infected plants, the mRNA level of NbSGT1 was reduced compared with that in CaLCuV vector alone-infected control plants. However, Actin mRNA levels were similar (Fig. 3E). These results clearly suggest that the CaLCuV-based MIR VIGS can be used to study plant defense.
Comparison between MIR VIGS and SIR VIGS
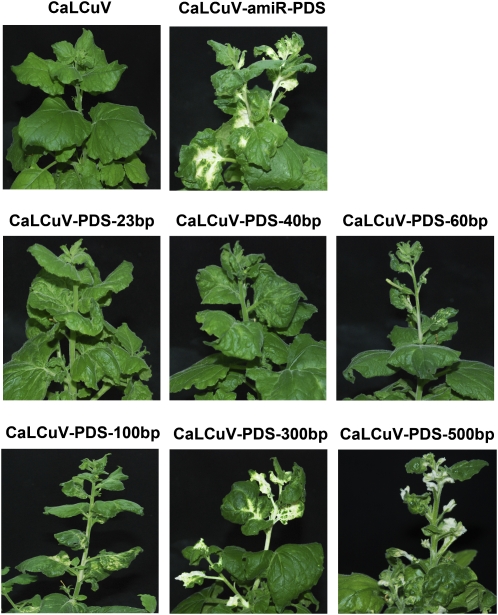
CaLCuV-based SIR VIGS using long insertion of the target gene has been proved to induce efficient gene silencing. Furthermore, it has been reported that potato virus X-based SIR VIGS using 23 nucleotides of a GFP fragment can induce weak GFP silencing in the GFP-overexpressing transgenic plants (Thomas et al., 2001). To test whether the MIR VIGS system has any advantage over SIR VIGS, we used a range of insert sizes and selected NbPDS as the target gene to compare their silencing efficiency. For this purpose, we cloned 23-bp (nucleotides 1,572–1,594), 40-bp (nucleotides 1,561–1,600), 60-bp (nucleotides 1,555–1,614), 100-bp (nucleotides 1,555–1,653), 300-bp (nucleotides 1,508–1,787), and 500-bp (nucleotides 1,190–1,689) fragments of NbPDS (EU165355) into pCVA to obtain pCVA-PDS-23, pCVA-PDS-40, pCVA-PDS-60, pCVA-PDS-100, pCVA-PDS-300, and pCVA-PDS-500, respectively. The insertion in each of these constructs included a common 21-bp target NbPDS sequence (nucleotides 1,572–1,592) of amiR-PDS. N. benthamiana plants were coinfiltrated with Agrobacterium carrying pCVB and pCVA or its derivatives. Plants coinfiltrated with pCVB and pCVA-amiR-PDS, pCVA-PDS-100, pCVA-PDS-300, or pCVA-PDS-500 began to show white color in their upper young leaves at 3 wpi. In contrast, no plants by SIR VIGS using shorter insertions (60, 40, and 23 bp) showed any leaf color change at 3 wpi (data not shown). At 4 wpi, SIR VIGS using the shortest PDS insertions (40 and 23 bp) did not result in any NbPDS silencing phenotype (Fig. 4). When the size of the SIR VIGS insertion was 60 bp, only some leaves of some infiltrated plants (four out of eight plants) showed weak white color. However, once the size of the SIR VIGS insertion increased to 100, 300, and 500 bp, all infiltrated plants developed a clear PDS gene silencing phenotype (Fig. 4). More importantly, although amiRNA-PDS targeted only one site on PDS mRNA, all plants infected with CaLCuV-miR-PDS showed the same level of photobleached phenotype as the plants infected with CaLCuV with insertion of an over 300-bp PDS fragment. These results suggest that 21-bp miRNA-based MIR VIGS is sufficient to trigger the same level of gene silencing induced by SIR VIGS with insertion of over 300 bp in length. We repeated this experiment twice, with four plants for each construct in each experiment, and obtained similar results.
Figure 4.
Comparison of MIR VIGS and SIR VIGS with different insertions. Phenotypes of N. benthamiana plants coinfiltrated with Agrobacterium carrying pCVB and pCVA or its derivatives are shown. Photographs were taken at 4 wpi.
CaLCuV Vector Can Be Used to Evaluate Plant Endogenous miRNA Function
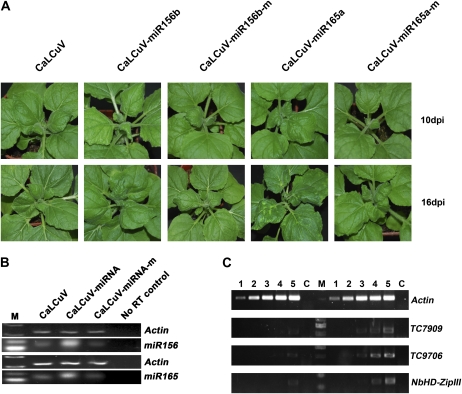
Overexpression of miRNAs is one of the most widely used approaches to study miRNA function in plants (Jones-Rhoades et al., 2006). In order to figure out whether a CaLCuV-based miRNA expression vector would be suitable to study the function of plant endogenous miRNAs, we used the CaLCuV vector to express precursors of Arabidopsis miR156b and miR165a (CaLCuV-miR156b and CaLCuV-miR165a, respectively) in N. benthamiana plants. As negative controls, we also expressed their inactive mutants (miR156b-m and miR165a-m) with mutations at positions 9 and 10. As shown in Figure 5A, plants infiltrated with Agrobacterium carrying CaLCuV-miR156b showed leaf-curl phenotype as early as 10 d post infiltration (dpi). In contrast, plants infected by CaLCuV-miR156b-m or CaLCuV appeared normal, and clear leaf-curl symptoms did not begin to develop until 16 to 20 dpi (Fig. 5A). Similarly, CaLCuV-based expression of the endogenous miR165a precursor gene also resulted in much earlier leaf-curl phenotype (Fig. 5A). Moreover, plants infected by CaLCuV-miR165a showed many chlorotic spots on leaves about 2 wpi (Fig. 5A). We repeated this experiment three times, with four plants for each construct in each experiment, and obtained similar results.
Figure 5.
CaLCuV-based expression of Arabidopsis miR156b and miR165a in N. benthamiana plants. A, Plant phenotypes associated with CaLCuV, CaLCuV-miR156, CaLCuV-miR156-m, CaLCuV-miR165, and CaLCuV-miR165-m at 10 dpi (top row) or 16 dpi (bottom row). B, End-point stem-loop RT-PCR (22 cycles) analysis showing the levels of mature miR156 and miR165 in inoculated plants. C, Semiquantitative RT-PCR analysis of the effects of CaLCuV-based expression of Arabidopsis miR156b and miR165a on their cognate target genes. Lanes 1 to 5 represent PCRs with 18, 21, 24, 27, and 30 cycles, respectively. PCR products to the left of the marker (M) are from the silenced plants, and those to the right are the nonsilenced controls. Actin mRNA was used as an internal control, and a no-RT control (C) was also included.
We used end-point stem-loop RT-PCR to evaluate the levels of miR156 and miR165. As shown in Figure 5B, the levels of miR156 and miR165 were elevated in CaLCuV-miR156b- and CaLCuV-miR165a-infiltrated plants, respectively, but were comparable in plants infiltrated with CaLCuV-miR156b-m or CaLCuV-miR165a-m and in control plants. The latter could be due to ineffective RT-PCR amplification of mutant miRNAs using active miRNA-specific primers containing two continuous mismatches near the 3′ end with the mutation form of miR156b or miR165a, and this result is consistent with the previous report that the stem-loop RT-PCR is able to discriminate miRNAs that differ by as little as a single nucleotide (Chen et al., 2005).
We used the miRNA target search tool included in Web MicroRNA Designer to look for the putative targets of miR156 and miR165 and found that miR156 could target at least five genes, including two putative Squamosa promoter-binding protein-like (SPL) genes (TC7909 and TC9706) in N. benthamiana, and miR165 could target a gene (TC12847) that encodes a class III HD-Zip transcription factor in Nicotiana tabacum. We monitored the expression of the above two putative SPL genes and the N. benthamiana homolog of class III HD-ZIP gene (NbHD-ZIPIII) by semiquantitative RT-PCR in order to evaluate the silencing effect of miR156 and miR165 that were processed from transiently expressed native miRNA precursors. In CaLCuV-miR156b-infected plants, there was an obvious reduction in the mRNA levels of TC7909 and TC9706 when compared with the CaLCuV-infected control (Fig. 5C). Similarly, the mRNA level of N. benthamiana homologs of TC12847 in CaLCuV-miR165a-infected plants decreased compared with the CaLCuV-infected control (Fig. 5C). These results show that TC7909 and TC9706 are the targets for miR156 in N. benthamiana plants and that miR165 has a target site in the N. benthamiana ortholog of N. tabacum TC12847.
DISCUSSION
In this report, we have shown that the modified CaLCuV vector can be used to express amiRNAs and endogenous miRNA in plants. Using this viral system, we successfully silenced several endogenous genes using amiRNAs in N. benthamiana plants. MIR VIGS of NbSGT1 led to the loss of N-mediated resistance to TMV. Using PDS as a marker gene, we showed that MIR VIGS is more efficient than traditional SIR VIGS using short insertions and is as efficient as SIR VIGS using long insertions. In addition, we used the CaLCuV miRNA expression system to evaluate the functions of endogenous miR156 and miR165 in N. benthamiana. To the best of our knowledge, this is the first report to use viral vectors to induce miRNA-mediated silencing and to study the functions of endogenous miRNAs in plants.
Like the traditional SIR VIGS approach, the described CaLCuV-based MIR VIGS does not rely on plant transformation and can be carried out for gene silencing through simple agroinoculation. In addition, there are several advantages to the use of MIR VIGS compared with traditional SIR VIGS. First, MIR VIGS could be more accurate. During SIR VIGS, many unforeseen siRNAs are generated, which may cause nonspecific silencing (Jackson et al., 2003; Jackson and Linsley, 2004; Xu et al., 2006). Even the SIR VIGS insert is very short (e.g. 23 nucleotides), and many siRNAs can still be generated from the nontarget region of the target gene RNAs (Nishikura, 2001), which may cause nonspecific silencing. In contrast, during MIR VIGS, only one known mature miRNA is predicted to come out from an accurately designed miRNA precursor, which can highly specifically silence the predicted genes (Schwab et al., 2005; Qu et al., 2007). Second, MIR VIGS could be more efficient. Potato virus X-based SIR VIGS using 23 nucleotides of insertion has been demonstrated previously, but such SIR VIGS works only for GFP transgenes with high expression and not for endogenous PDS genes (Thomas et al., 2001). In our study, CaLCuV-based SIR VIGS using less than 100 nucleotides of PDS insertions only gave a weak photobleached silencing phenotype. In contrast, CaLCuV-based MIR VIGS producing only 21 nucleotides of mature miRNA induced a PDS silencing phenotype as triggered by CaLCuV-based VIGS using long PDS insertions (300 and 500 bp). This is consistent with the observation that miRNA could induce stronger silencing than siRNA (Qu et al., 2007). Third, MIR VIGS may have lower side effects. The traditional VIGS vector produces siRNAs and long double-stranded RNAs, and the latter may activate RNA-dependent protein kinase pathways and cause nonspecific cell death in plants, a phenomenon well documented in mammalian cells (Langland et al., 1995; Davis and Watson, 1996; Bridge et al., 2003; Sledz et al., 2003; Tang and Galili, 2004). By contrast, MIR VIGS uses DNA virus vector to express miRNA, and this may avoid triggering the protein kinase pathway. Fourth, MIR VIGS uses amiRNA to target genes for silencing; thus, it does not require cloning target gene fragments. Compared with the cDNA fragment used in SIR VIGS, amiRNAs could be easier to clone. The amiRNA precursor genes can be easily obtained by a simple and quick overlapping PCR (Ossowski et al., 2008) or a direct cloning of synthetic oligonucleotides into the stem region of pre-miR169d (Liu et al., 2010). Furthermore, it is simple to use the modified CaLCuV vector. Use of the previously described CaLCuV-based vector is time-consuming and difficult and needs particle bombardment, a process that requires care in the preparation of microprojectile particles and has the risk of carryover between experiments and the somewhat unpredictable introduction of the silencing vector (Turnage et al., 2002; Muangsan and Robertson, 2004; Burch-Smith et al., 2006). In contrast, we introduce the modified CaLCuV vectors into plants by agroinfiltration, a much simpler but effective technique with no need of complicated operations or expensive instruments.
Three major approaches to study the functions of miRNAs are overexpression of the target miRNAs, expression of the mutant target gene (Palatnik et al., 2003; Xie et al., 2005), and expression of miRNA target mimicries (Franco-Zorrilla et al., 2007) and usually need tedious and time-consuming work to generate the stable transgenic plants. We show here that a viral miRNA expression system could be used to study the functions of endogenous miRNA genes by agroinfiltration. The modified CaLCuV vector may be useful in high-throughput screening of miRNAs.
Transient expression of Arabidopsis miR156b but not its mutant using CaLCuV vector resulted in earlier leaf-curl phenotype, suggesting that miR156 may play a role in leaf development in N. benthamiana plants. There are at least five putative targets of miR156, including two putative SPL genes and three uncharacterized genes in N. benthamiana. Overexpression of miR156 reduced the mRNA levels of two putative SPL genes (TC7909 and TC9706), which are the homologs of Arabidopsis SPL9 and SPL15, suggesting that these two SPL genes are the endogenous targets of miR156 in N. benthamiana. In Arabidopsis, miR156 has complementary sites on 11 of 17 SPL genes (Rhoades et al., 2002; Gandikota et al., 2007). Among these 11 putative Arabidopsis miR156 targets, AtSPL9 and AtSPL15 regulated by miR156 control shoot maturation (Schwab et al., 2005) and AtSPL3 acts as a positive transcriptional regulator modulating floral development in Arabidopsis (Cardon et al., 1997). In addition, miR156 has been reported to have functions in Arabidopsis and rice (Oryza sativa) development (Schwab et al., 2005; Xie et al., 2006). Therefore, it is possible that miR156 also regulates plant development through SPL genes in N. benthamiana plants.
The HD-ZIP III genes are thought to be targets of miR165 and to code for class III HD-Zip transcription factors, including Phabulosa, Phavoluta, and Revoluta, which regulate axillary meristem initiation and leaf development (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003; Juarez et al., 2004; Kidner and Martienssen, 2004). In Arabidopsis, overexpression of miR165 induces very similar phenotypes caused by loss-of-function mutations of HD-ZIP III genes (Zhou et al., 2007). Overexpression of miR165 but not its mutant in this research resulted in abnormal development of leaves and the appearance of many chlorotic spots in N. benthamiana plants. These phenotypes are very different from CaLCuV symptoms. Our results suggest that some HD-ZIP III genes may have a role in leaf development regulated by miR165.
In summary, the modified CaLCuV-based miRNA expression vector described in this report has great potential for the study of functions of not only protein-encoding genes but also miRNA genes.
MATERIALS AND METHODS
Plasmid Construction
Plasmids pCPCbLCVA.007 (GenBank accession no. AY279345) and pCPCbLCVB.002 (GenBank accession no. AY279344), containing partial direct repeats of the CaLCuV A and B components, were kindly provided by Prof. Dominique Robertson (Turnage et al., 2002). GFP-tagged TMV was described previously (Liu et al., 2002a).
To generate pCVA (Fig. 1A), a fragment containing partial repeats of the CaLCuV DNA-A was PCR amplified from pCPCbLCVA.007 using primers OYLT97 and OYLT98 (Supplemental Table S1). The resulting PCR product was digested with HindIII and cloned into HindIII-Ecl136II sites of pCAMBIA1301. pCVB (Fig. 1A) was generated by cloning a SalI-BamHI fragment containing the partial direct repeats of the CaLCuV B component from pCPCbLCVB.002 into SalI-BamHI sites of pCAMBIA1301. The cloned CaLCuV DNA fragments were confirmed by sequencing.
We use a Web-based tool called Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi; see also Supplemental Materials and Methods S1) to design 21-mer amiRNA sequences. The sequences of our chosen amiRNAs targeting NbPDS, NbSu, NbCLA1, and NbSGT1 are 5′-UCAACAUAGACUGAUUGGGGC-3′, 5′-UAAGAUCUAACCGUGGCGCAC-3′, 5′-UGUAUCUGAAACAACUUGGUG-3′, and 5′-UCGGUGCUAAACUAGCACCAG-3′, respectively. For each amiRNA sequence, four oligonucleotide sequences are suggested by Web MicroRNA Designer and then used to PCR amplify the designed amiRNA along with two common primers, oligo A and oligo B, using the Arabidopsis (Arabidopsis thaliana) endogenous miR319a precursor as backbone (At4g23713). The detailed protocol for designing and obtaining amiRNA gene can be found in Supplemental Materials and Methods S1. PCR products containing amiR-PDS, amiR-Su, amiR-CLA1, and amiR-SGT1 were digested by KpnI and XbaI and cloned into pCVA to produce pCVA-amiR-PDS, pCVA-amiR-Su, pCVA-amiR-CLA1, and pCVA-amiR-SGT1, respectively. All the cloned amiRNA sequences were confirmed by DNA sequencing.
Traditional CaLCuV SIR VIGS vectors pCVA-PDS-23, pCVA-PDS-40, pCVA-PDS-60, pCVA-PDS-100, pCVA-PDS-300, and pCVA-PDS-500 were generated by cloning 23-bp (nucleotides 1,572–1,594), 40-bp (nucleotides 1,561–1,600), 60-bp (nucleotides 1,555–1,614), 100-bp (nucleotides 1,555–1,653), 300-bp (nucleotides 1,508–1,787), and 500-bp (nucleotides 1,190–1,689) fragments of NbPDS (EU165355) into pCVA, respectively.
To generate pCVA-miR156b and pCVA-miR165a, Arabidopsis miR156b and miR165a (ath-miR156b and ath-miR165a) genes were PCR amplified from Arabidopsis genomic DNA and cloned into KpnI-XbaI sites of pCVA. Arabidopsis miR156b and miR165a mutant genes at positions 9 and 10 (ath-miR156b-m and ath-miR165a-m) were obtained by overlapping PCR and cloned into pCVA to generate pCVA-miR156b-m and pCVA-miR165a-m, respectively. The primers used in this study are listed in Supplemental Table S1.
Plant Growth, Agroinfiltration, and GFP Imaging
Nicotiana benthamiana plants were grown in pots at 25°C under a 16-h-light/8-h-dark cycle. For the CaLCuV-based MIR VIGS and SIR VIGS assays, pCVB and pCVA or their derivatives were introduced into Agrobacterium tumefaciens strain GV3101. The Agrobacterium cultures were inoculated in 5 mL of Luria-Bertani medium containing antibiotics (30 mg L−1 rifampicin, 50 mg L−1 gentamycin, and 50 mg L−1 kanamycin) and grown overnight in a 28°C shaker. Agrobacterium cultures were harvested and resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES, and 200 μm acetosyringone) and then adjusted to an optical density at 600 nm of 2.0. After keeping at room temperature for 3 to 4 h, Agrobacterium cultures containing pCVB and pCVA or their derivatives were mixed at a 1:1 ratio and infiltrated into the stem nodes and petioles of six-leaf stage plants using a 1-mL syringe. In order to investigate the effect of expression of amiR-SGT1 on N-mediated TMV resistance, the N-containing N. benthamiana plants (Liu et al., 2002a) were infiltrated with mixture cultures of Agrobacterium containing pCVB, pCVA-amiR-SGT1, and TMV-GFP at a 1:1:2 ratio. GFP imaging was illuminated under long-wavelength UV light, and photographs were taken using a Nikon D80 digital camera.
DNA and RNA Isolation, PCR Detection, RT-PCR Analysis, and Mature miRNA Detection
Total DNA was extracted from CaLCuV-infected N. benthamiana plants following a standard cetyl-trimethyl-ammonium bromide extraction protocol. Primers used for CaLCuV DNA detection are OTY77 and OTY78 (Supplemental Table S1). Total RNA was extracted from leaves of silenced and nonsilenced N. benthamiana plants using TRNzol solution (Tiangen) and treated with RNase-free DNase I (MBI Fermentas). First-strand cDNA was synthesized using 10 μg of total RNA, oligo(dT) primer, and RevertAid Moloney murine leukemia virus reverse transcriptase (MBI Fermentas). Semiquantitative RT-PCR was performed as described previously (Liu et al., 2004). Mature miRNAs were detected using an end-point stem-loop RT-PCR method (Chen et al., 2005; Tang et al., 2006; Varkonyi-Gasic et al., 2007). A stem-loop-containing RT primer with its 5′ end complementary to target the miRNA's last six nucleotides at the 3′ end was designed. RT was performed at 16°C for 30 min, followed by 60 cycles of pulsed RT at 30°C for 30 s, 42°C for 30 s, and 50°C for 1 s using 10 μg of total RNA, the stem-loop RT primer, and RevertAid Moloney murine leukemia virus reverse transcriptase (MBI Fermentas). Semiquantitative RT-PCR was performed using a forward primer containing the 5′ part sequence of miRNA and a universal primer complementing the stem-loop part of the RT primer at 94°C for 2 min, followed by 30 cycles (for amiRNAs) or 22 cycles (for miR156 and miR165) of 94°C for 15 s and 60°C for 1 min. The reaction products (about 60 bp) were analyzed by electrophoresis on a 4% agarose gel in 1× Tris-acetate EDTA buffer. The primers used in this study are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. List of primers used in this work.
Supplemental Materials and Methods S1. The protocol for MIR VIGS.
Supplementary Material
Acknowledgments
We are very grateful to Dr. Dominique Robertson for CaLCuV vectors pCPCbLCVA.007 and pCPCbLCVB.002, to Dr. Detlef Weigel for pRS300, and to CAMBIA for T-DNA binary vector pCAMBIA1301.
References
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. (2003) Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet 34: 263–264 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. (2006) Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142: 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P. (1997) Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J 12: 367–377 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Watson JC. (1996) In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human alpha-tropomyosin. Proc Natl Acad Sci USA 93: 508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Estevez JM, Cantero A, Reindl A, Reichler S, Leon P. (2001) 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276: 22901–22909 [DOI] [PubMed] [Google Scholar]
- Estevez JM, Cantero A, Romero C, Kawaide H, Jimenez LF, Kuzuyama T, Seto H, Kamiya Y, Leon P. (2000) Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol 124: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Grimsley N, Hohn B, Hohn T, Walden R. (1986) “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proc Natl Acad Sci USA 83: 3282–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Strandberg JO, Hiebert E, Lazarowitz SG. (1998) Asymmetric infectivity of pseudorecombinants of cabbage leaf curl virus and squash leaf curl virus: implications for bipartite geminivirus evolution and movement. Virology 250: 283–292 [DOI] [PubMed] [Google Scholar]
- Hiriart JB, Aro EM, Lehto K. (2003) Dynamics of the VIGS-mediated chimeric silencing of the Nicotiana benthamiana ChlH gene and of the tobacco mosaic virus vector. Mol Plant Microbe Interact 16: 99–106 [DOI] [PubMed] [Google Scholar]
- Hiriart JB, Lehto K, Tyystjarvi E, Junttila T, Aro EM. (2002) Suppression of a key gene involved in chlorophyll biosynthesis by means of virus-inducing gene silencing. Plant Mol Biol 50: 213–224 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. (2004) Noise amidst the silence: off-target effects of siRNAs? Trends Genet 20: 521–524 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. (2004) MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Khraiwesh B, Ossowski S, Weigel D, Reski R, Frank W. (2008) Specific gene silencing by artificial microRNAs in Physcomitrella patens: an alternative to targeted gene knockouts. Plant Physiol 148: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. (2004) Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84 [DOI] [PubMed] [Google Scholar]
- Kumagai MH, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill LK. (1995) Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci USA 92: 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M. (2004) RNA interference in crop plants. Curr Opin Biotechnol 15: 139–143 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. (2003) New microRNAs from mouse and human. RNA 9: 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Jin S, Jacobs BL, Roth DA. (1995) Identification of a plant-encoded analog of PKR, the mammalian double-stranded RNA-dependent protein kinase. Plant Physiol 108: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang L, Sun J, Luo Y, Wang MB, Fan YL, Wang L. (2010) A simple artificial microRNA vector based on ath-miR169d precursor from Arabidopsis. Mol Biol Rep 37: 903–909 [DOI] [PubMed] [Google Scholar]
- Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP. (2004) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chem 279: 2101–2108 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. (2002a) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP. (2002b) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. (2003) Virus-induced gene silencing in plants. Methods 30: 296–303 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, Leon P. (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Mansoor S, Amin I, Hussain M, Zafar Y, Briddon RW. (2006) Engineering novel traits in plants through RNA interference. Trends Plant Sci 11: 559–565 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Muangsan N, Robertson D. (2004) Geminivirus vectors for transient gene silencing in plants. Methods Mol Biol 265: 101–115 [DOI] [PubMed] [Google Scholar]
- Nishikura K. (2001) A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107: 415–418 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Paximadis M, Idris AM, Torres-Jerez I, Villarreal A, Rey ME, Brown JK. (1999) Characterization of tobacco geminiviruses in the Old and New World. Arch Virol 144: 703–717 [DOI] [PubMed] [Google Scholar]
- Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jaggard DA, Xiao S, Coleman MJ, et al. (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Ye J, Fang R. (2007) Artificial microRNA-mediated virus resistance in plants. J Virol 81: 6690–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Robertson D. (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55: 495–519 [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P. (2004) Plant immunity: the origami of receptor activation. Curr Biol 14: R22–R24 [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Schulze-Lefert P. (2003) Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci 8: 252–258 [DOI] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. (2003) Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 5: 834–839 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G. (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Hajkova P, Barton SC, Lao K, Surani MA. (2006) MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res 34: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Galili G. (2004) Using RNAi to improve plant nutritional value: from mechanism to application. Trends Biotechnol 22: 463–469 [DOI] [PubMed] [Google Scholar]
- Tenllado F, Llave C, Diaz-Ruiz JR. (2004) RNA interference as a new biotechnological tool for the control of virus diseases in plants. Virus Res 102: 85–96 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Jones L, Baulcombe DC, Maule AJ. (2001) Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J 25: 417–425 [DOI] [PubMed] [Google Scholar]
- Turnage MA, Muangsan N, Peele CG, Robertson D. (2002) Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J 30: 107–114 [DOI] [PubMed] [Google Scholar]
- Vaghchhipawala ZE, Mysore KS. (2008) Agroinoculation: a simple procedure for systemic infection of plants with viruses. Methods Mol Biol 451: 555–562 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P. (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS One 3: e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Fusaro AF, Wang M, Waterhouse PM. (2005) RNA silencing platforms in plants. FEBS Lett 579: 5982–5987 [DOI] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GY, Sui N, Tang Y, Xie K, Lai YZ, Liu YL. (2010) One-step, zero-background ligation independent cloning intron-containing hairpin RNA constructs for RNAi in plants. New Phytol (in press) [DOI] [PubMed] [Google Scholar]
- Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS. (2006) Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol 142: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. (2006) Conservation and divergence of plant microRNA genes. Plant J 46: 243–259 [DOI] [PubMed] [Google Scholar]
- Zhao T, Wang W, Bai X, Qi Y. (2008) Gene silencing by artificial microRNAs in Chlamydomonas. Plant J 58: 157–164 [DOI] [PubMed] [Google Scholar]
- Zhou GK, Kubo M, Zhong R, Demura T, Ye ZH. (2007) Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol 48: 391–404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.