Abstract
ABF2 is a basic leucine zipper protein that regulates abscisic acid (ABA)-dependent stress-responsive gene expression. We carried out yeast two-hybrid screens to isolate genes encoding ABF2-interacting proteins in Arabidopsis (Arabidopsis thaliana). Analysis of the resulting positive clones revealed that two of them encode an AP2 domain protein, which is the same as AtERF48/DREB2C. This protein, which will be referred to as DREB2C, could bind C-repeat/dehydration response element in vitro and possesses transcriptional activity. To determine its function, we generated DREB2C overexpression lines and investigated their phenotypes. The transgenic plants were ABA hypersensitive during germination and seedling establishment stages, whereas primary root elongation of seedlings was ABA insensitive, suggesting developmental stage dependence of DREB2C function. The DREB2C overexpression lines also displayed altered stress response; whereas the plants were dehydration sensitive, they were freezing and heat tolerant. We further show that other AP2 domain proteins, DREB1A and DREB2A, interact with ABF2 and that other ABF family members, ABF3 and ABF4, interact with DREB2C. Previously, others demonstrated that ABF and DREB family members cooperate to activate the transcription of an ABA-responsive gene. Our result implies that the cooperation of the two classes of transcription factors may involve physical interaction.
Plants are continually exposed to changing environments in nature and frequently encounter harsh environmental conditions, such as drought, high salinity, and extreme temperatures. Although they vary widely in their adjustability, plants are nonetheless able to respond adaptively to these and other “abiotic stresses” (Bohnert et al., 1995). This adaptive response is in major part controlled by the phytohormone abscisic acid (ABA; Finkelstein et al., 2002; Xiong et al., 2002).
Numerous genes, which are involved in various aspects of adaptive stress response, are up- or down-regulated under stress conditions (Kreps et al., 2002; Ramanjulu and Bartels, 2002; Takahashi et al., 2004). Promoter analyses of these ABA-regulated genes revealed a number of cis-elements known as “ABA response elements” (ABREs; Busk and Pages, 1998; Gomez-Porras et al., 2007). Among the ABREs, those sharing the ACGTGGC core sequence are found to be ubiquitous, and their role in ABA-responsive gene expression has been characterized in detail (Hattori et al., 2002). The ABREs are similar to the G-box, CACGTG, which is found in light-regulated gene promoters (Menkens et al., 1995). Several studies showed that the G-box-type ABRE, which will be referred to as G-ABRE, is necessary but not sufficient for ABA-induced gene expression. An additional element is usually required for high level ABA induction. These elements, generally known as “coupling elements,” constitute the ABA response complex together with the G-ABRE (Shen et al., 2004). For example, CE1 (CCACC) functions as a coupling element to the G-ABRE in barley (Hordeum vulgare) HVA22 promoter, and the two elements constitute the ABA response complex ABRC1 (Shen and Ho, 1995). Another coupling element, CE3, constitutes, with a G-ABRE, the ABA response complex (ABRC3) present in barley HVA1 promoter (Shen et al., 1996). It is also known that G-ABRE itself can function as a coupling element to another copy of G-ABRE (Hobo et al., 1999a).
A subfamily of basic leucine zipper (bZIP) class transcription factors that interact with the G-ABRE has been reported. Referred to as ABFs (ABF1–ABF4; Choi et al., 2000) or AREBs (AREB1–AREB3; Uno et al., 2000), they not only bind G-ABRE in vitro but also regulate ABA/stress response in planta (Fujita et al., 2005; Furihata et al., 2006; Kim, 2006). Overexpression of ABF3 or ABF4 in Arabidopsis (Arabidopsis thaliana), for instance, confers ABA hypersensitivity and drought tolerance (Kang et al., 2002; Kim et al., 2004a, 2004c), whereas their knockout mutants are partially insensitive to ABA and susceptible to drought (Kim et al., 2004c; Finkelstein et al., 2005). Several studies indicate that the in vivo functions of ABFs/AREBs are modulated by various kinases (Choi et al., 2005; Furihata et al., 2006; Chae et al., 2007). ABFs/AREBs are highly homologous to ABI5 (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000), sunflower (Helianthus annuus) and Arabidopsis DPBFs (Kim et al., 1997; Kim and Thomas, 1998; Kim et al., 2002), and rice (Oryza sativa) TRAB1 (Hobo et al., 1999b).
There is a group of stress-responsive genes whose expression is not regulated by ABA. Promoter analyses of these ABA-independent genes have revealed the presence of dehydration-responsive element (DRE; TACCGACAT) and C-repeat (CRT) element (G/ACCGAC) in drought-responsive and cold-regulated gene promoters, respectively (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994). The two elements share the consensus sequence CCGAC, and their cognate transcription factors are known as DREB (for DRE-binding protein; Liu et al., 1998) and CBF (for CRT-binding factor; Stockinger et al., 1997), respectively. DREB and CBF belong to the same subfamily of AP2/ERF domain transcription factors, and some members of the DREB family (i.e. DREB1A, DREB1B, and DREB1C) are identical to CBF family members (i.e. CBF3, CBF1, and CBF2, respectively).
Pathways leading to stress-responsive gene expression are generally divided into two broad categories, ABA-dependent and ABA-independent pathways, depending on the involvement of ABA (Yamaguchi-Shinozaki and Shinozaki, 2005). The pathway(s) leading to ABF/AREB-dependent gene expression via G-ABRE belongs to the former category. Meanwhile, the pathway(s) leading to DREB1/CBF-dependent cold/drought-responsive gene expression belongs to the ABA-independent pathways. Several studies, however, suggest that the two categories of signaling pathways may cross talk to each other; thus, they may be interdependent. For instance, the expression of Arabidopsis CBF family members is ABA inducible (Haake et al., 2002; Knight et al., 2004). ICE1, which is a regulator of CBF expression, is also up-regulated by ABA (Chinnusamy et al., 2003). In the case of maize (Zea mays), DRE-binding AP2/ERF domain factors, DBF1 and DBF2, are not only ABA inducible but also regulate ABA response in vivo (Kizis and Pages, 2002). These observations suggest that DRE/CRT-regulated expression of some genes may be ABA dependent. Furthermore, Narusaka et al. (2003) have demonstrated that a DRE may function as a coupling element to G-ABRE in the RD29A promoter. They also have shown that ABFs/AREBs and DREBs/CBFs synergistically activate the RD29A promoter in a transient assay, although the mechanism of synergistic activation has not been reported yet.
In this report, we show that ABF/AREB family transcription factors physically interact with DREB/CBF family members. To identify proteins that may modulate the ABF function, we carried out yeast two-hybrid screens employing ABF2 as bait. One of the resulting positive clones was AtERF48, which is identical to the DREB family member DREB2C (Sakuma et al., 2002; Nakano et al., 2006). The protein binds to DRE/CRT in vitro, and its overexpression affected both ABA sensitivity and stress tolerance. We further show that ABF2 interacts with other DREB family members, DREB1A and DREB2A, and that, conversely, DREB2C interacts with other ABF family members, ABF3 and ABF4. Our data suggest that the interaction between ABA-dependent and ABA-independent pathways may involve physical interaction between ABF and DREB family transcription factors.
RESULTS
Isolation of ABF2-Interacting Proteins
To identify ABF2-interacting proteins, we carried out yeast two-hybrid screens (Kim et al., 2004b). Using a partial fragment of ABF2 as bait, we screened approximately 6.5 million yeast transformants and were able to isolate five positive clones that specifically interact with the bait protein (Fig. 1A). Sequence analysis of the clones revealed that two of them encoded a protein with an AP2/ERF domain. The three other clones encoded an ARM repeat protein, which have been reported elsewhere (Kim et al., 2004b).
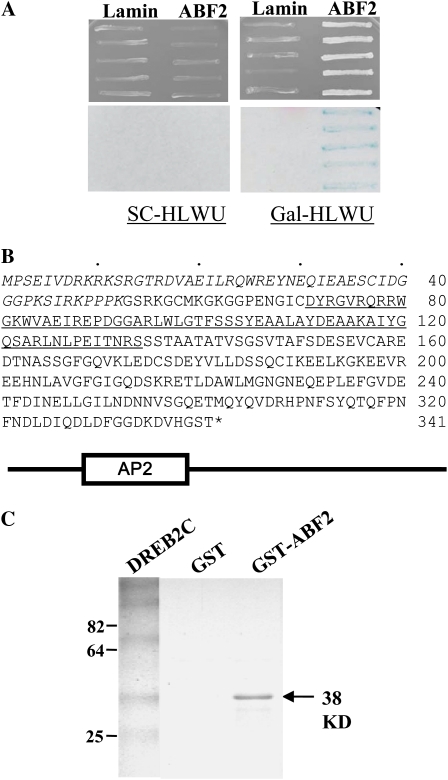
Figure 1.
Isolation of an ABF2-interacting protein gene by yeast two-hybrid screening. A, Specificity of interaction. Interaction between a positive clone (clone 27) isolated from the initial screen and ABF2 or nuclear lamin is shown. Reporter yeast containing the bait construct (ABF2 or lamin) was transformed with the positive clone, and the transformants were grown on SC-His-Leu-Trp-Ura (SC-HLWU; left) or Gal/raffinose-His-Leu-Trp-Ura (Gal-HLWU; right) medium. The bottom panels show the results of filter lift β-galactosidase assays of the transformants shown in the top panels. B, Deduced amino acid sequence of DREB2C. The first 52 amino acids missing in clone 27 are indicated by italicized characters. The schematic diagram at the bottom depicts the overall structure of DREB2C. C, In vitro interaction between ABF2 and DREB2C was investigated by GST pulldown assay. In vitro-translated DREB2C, labeled with [35S]Met, was incubated with GST alone or with GST-ABF2 fusion protein, the bound protein was eluted and electrophoresed on a polyacrylamide gel, and autoradiography was carried out to visualize the protein. The arrow indicates the position of DREB2C, and the numbers on the left indicate the positions of size markers. [See online article for color version of this figure.]
The AP2 domain protein (At2g40340) encoded by the two positive clones was partial, missing the N-terminal 52 amino acids. Subsequently, a full-length cDNA clone was isolated by PCR utilizing the sequence information available on the Arabidopsis database (http://www.arabidopsis.org). The encoded protein (Fig. 1B), which was originally named AAP (for ABF2-interacting AP2 domain protein), consisted of 341 amino acids with a calculated mass of 37.8 kD. The protein was found to be DREB2C and belongs to the group IV/A-2 subfamily of AP2/ERF proteins (Sakuma et al., 2002; Nakano et al., 2006), which includes DREB2 family proteins.
The interaction between ABF2 and the full-length DREB2C was confirmed by glutathione S-transferase (GST) pulldown assay. Recombinant ABF2 was prepared as a fusion to the GST. Subsequently, its interaction with DREB2C was determined using in vitro-translated DREB2C. Figure 1C shows that DREB2C was retained by the GST-ABF2 fusion protein but not by GST alone, indicating that DREB2C interacted with ABF2 in vitro.
Expression Patterns of DREB2C
The effects of exogenous ABA and various stress conditions on DREB2C expression were investigated by reverse transcription (RT)-PCR analysis. As shown in Figure 2A, ABA had little effect on DREB2C expression. However, a relatively high level of DREB2C RNA was detected after salt treatment. Lower level induction was observed after mannitol and cold treatments.
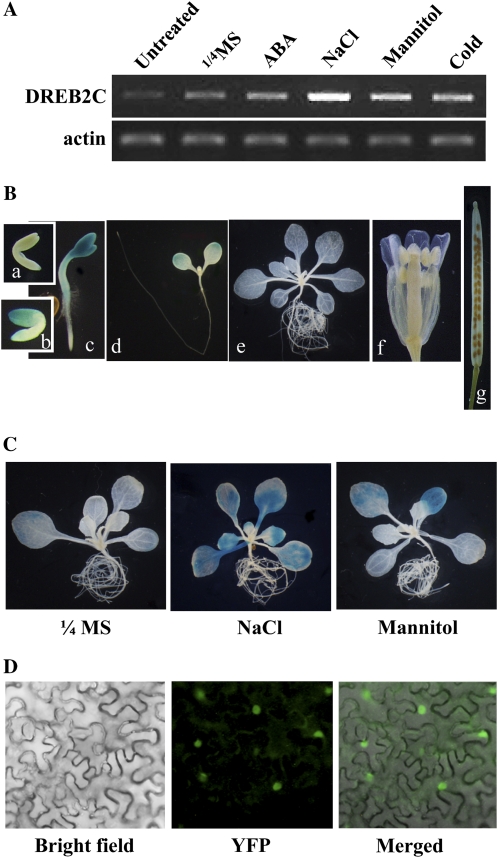
Figure 2.
Expression patterns of DREB2C. A, Induction patterns of DREB2C expression. ABA and stress induction patterns of DREB2C were determined by RT-PCR. RNA was isolated from seedlings treated with 100 μm ABA (4 h), 250 mm NaCl (4 h), or 600 mm mannitol (4 h) in quarter-strength Murashige and Skoog (MS) solution or with cold (24 h at 4°C). Untreated indicates control plants without any treatments, and ¼ MS indicates treatment with quarter-strength MS solution without any supplements. B, DREB2C promoter activity was determined by histochemical GUS staining. a, Immature embryo. b, Mature embryo. c, One-day-old seedling. d, Two-day-old seedling. e, Fifteen-day-old seedling. f, Flower. g, Silique. C, High-osmolarity effects on DREB2C promoter activity. Ten-day-old seedlings were treated with 250 mm NaCl or 600 mm mannitol for 4 h before GUS staining. GUS staining was for 12 h in B and C. D, Subcellular localization of DREB2C. Bright-field, fluorescence (YFP), and merged images of tobacco leaves infiltrated with Agrobacterium as described in “Materials and Methods” are shown.
To investigate the expression pattern of DREB2C in more detail, we generated transgenic plants harboring a promoter-GUS fusion construct and investigated its promoter activity by histochemical GUS staining. Figure 2B shows that the DREB2C promoter was active in mature embryo and the cotyledons of germinating seedlings. As the seedlings grew, the promoter activity gradually disappeared under normal growth condition. DREB2C promoter activity was induced, however, by treating seedlings with NaCl or mannitol, which is consistent with the results of our RT-PCR analysis (Fig. 2A).
We also determined the subcellular localization of DREB2C by Agrobacterium tumefaciens infiltration of tobacco (Nicotiana benthamiana) leaves. The coding region of DREB2C was fused to EYFP, and the construct was employed to transform Agrobacterium(C58C1). The Agrobacterium was then coinfiltrated with Agrobacterium containing p19 into tobacco leaves (Voinnet et al., 2003; Witte et al., 2004). Observation of the epidermal cells of the infiltrated leaves revealed that yellow fluorescent protein (YFP) signals were localized in the nuclei (Fig. 2D).
DNA-Binding and Transcriptional Activities of DREB2C
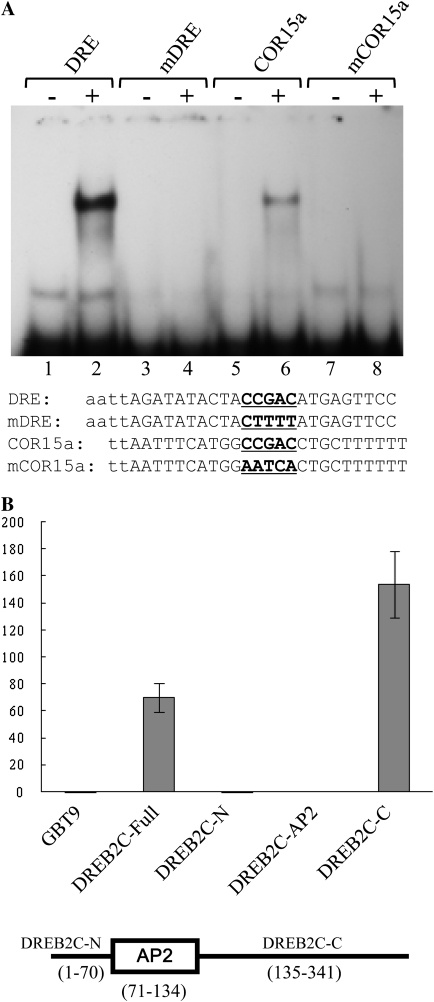
DREB2C belongs to the same subfamily of AP2/ERF proteins as DREB2A, which is one of the DRE-binding factors (Sakuma et al., 2002; Nakano et al., 2006). Therefore, we examined whether DREB2C can bind DRE. Recombinant DREB2C protein was prepared as a fusion to the GST, and its DNA-binding activity was determined by electrophoretic mobility shift assay. Figure 3A (lane 2) shows that a shifted band was observed with an oligonucleotide probe that contained a DRE core (i.e. CCGAC). In contrast, no shifted band was detected when an oligonucleotide with a mutated DRE core was employed as a probe (lane 4). In the same assay, a shifted band was observed with a Cor15a promoter fragment containing DRE sequence (lane 6) but not with the same promoter sequence containing a mutated DRE (lane 8). Thus, DREB2C could specifically bind DRE-containing sequences.
Figure 3.
DNA-binding and transcriptional activities of DREB2C. A, DNA-binding activity of DREB2C was determined by electrophoretic mobility shift assay. Oligonucleotides containing DRE, mutant DRE (mDRE), Cor15a promoter fragment, or mutant Cor15a fragment were employed as probes. The wild-type and mutant DRE/CRT core sequences in probe DNA are indicated by boldface. −, Probe without recombinant DREB2C; +, probe with recombinant DREB2C. B, Transcriptional activity of DREB2C. Transcriptional activity of DREB2C was determined employing a yeast assay system, as described in “Materials and Methods.” The various portions of DREB2C used in the assay are shown schematically at the bottom. GBT9, Empty vector without any insert; DREB2C-Full, full-length DREB2C; DREB2C-N, N-terminal portion of DREB2C; DREB2C-AP2, AP2 region of DREB2C; DREB2C-C, C-terminal portion of DREB2C. The numbers in parentheses indicate amino acid positions. The values represent β-galactosidase activity in Miller units, and the error bars denote se (n = 5 each).
The transcriptional activity of DREB2C was determined employing a yeast assay system. Full-length or partial DREB2C fragments were fused to the GAL4 DNA-binding domain, and the fusion constructs were introduced into a yeast strain (SFY526) harboring a lacZ reporter gene, which had GAL4-binding sites in its promoter. Transcriptional activity was then determined by measuring the β-galactosidase activity. With the full-length DREB2C, reporter activity much higher than the control activity observed with the GAL4-binding domain alone was detected (Fig. 3B). Even higher reporter activity was observed with the C-terminal fragment (DREB2C-C). On the other hand, no reporter activity was observed with the AP2 domain or with the N-terminal fragment (DREB2C-N). Thus, our results showed that DREB2C possesses transcriptional activity and that the activity resides within the C-terminal portion.
Overexpression of DREB2C Affects ABA Sensitivity
To investigate the in planta function of DREB2C, we generated DREB2C overexpression (OX) lines. Transgenic Arabidopsis plants expressing DREB2C under the control of a cauliflower mosaic virus 35S promoter were prepared, and after preliminary analysis of nine homozygous lines (T3 or T4), three representative lines with different DREB2C expression levels (Fig. 4A) were analyzed in more detail.
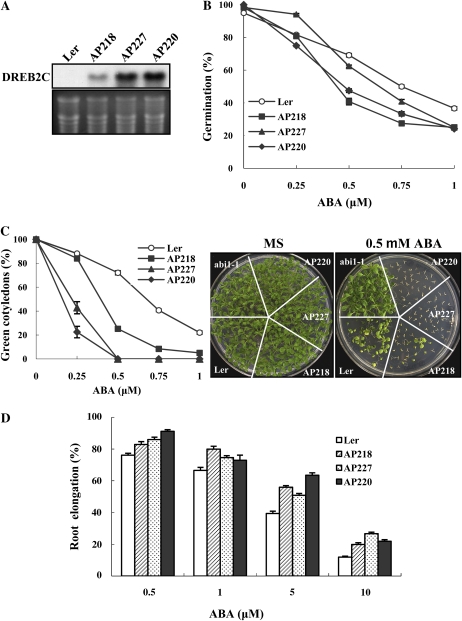
Figure 4.
ABA sensitivity of DREB2C overexpression lines. A, RNA gel-blot analysis of DREB2C expression in transgenic lines. The bottom panel shows an ethidium bromide-stained gel. B, ABA dose response during germination. Germination (full emergence of radical) was scored 5 d after plating. Experiments were done in quadruplicate (n = 30 each), and the error bars indicate se. C, ABA dose response during seedling development. Seeds were germinated and grown on ABA-containing medium for 2 weeks, and seedlings with green cotyledons and/or true leaves were counted. Experiments were carried out in quadruplicate (n = 30 each), and se values are indicated by error bars. Representative plants are shown in the right panels. D, ABA dose response of root elongation. Seeds were germinated on ABA-free medium for 4 d, the seedlings were transferred to medium containing various concentrations of ABA, and root elongation was measured 5 d after the transfer. The data represent relative root elongation rates compared with those on ABA-free medium. Experiments were done in quadruplicate (n = 12 each), and the error bars indicate se. Ler, Landsberg erecta.
The DREB2C OX lines germinated and grew normally, although they displayed minor growth retardation (data not shown). Because DREB2C interacts with ABF2, a regulator of ABA and stress responses, we asked if DREB2C overexpression would affect ABA sensitivity. When the seeds of the 35S-DREB2C plants with high DREB2C expression levels (AP220 and AP227) were germinated in the presence of ABA, their germination rates were lower than those of untransformed plants at ABA concentrations greater than 0.5 μm (Fig. 4B). The ABA effect was more pronounced during postgermination growth. When seeds were germinated and grown in the continuous presence of ABA, seedling development was almost completely inhibited after radicle emergence at 0.5 μm ABA (Fig. 4C). With the AP218 line, in which DREB2C expression level is relatively low, inhibition of shoot growth was observed in approximately 80% of the seedlings. At the same ABA concentration, only 20% of the wild-type seedlings were affected. Thus, 35S-DREB2C plants were hypersensitive to ABA during germination and seedling growth.
ABA sensitivity of 35S-ABF2 plants is developmental stage dependent (Kim et al., 2004c). We asked, therefore, whether 35S-DREB2C plants also exhibit development stage dependence in ABA response. 35S-DREB2C transgenic seeds were first germinated in ABA-free medium for 4 d. Subsequently, seedlings were transferred to medium containing ABA, and root elongation was measured 5 d after the transfer. Figure 4D shows that primary roots of all three DREB2C transgenic lines grew faster than those of wild-type plants in the presence of various concentrations of ABA. Thus, unlike young seedling shoot development, root elongation of 35S-DREB2C plants was partially insensitive to ABA, suggesting that ABA sensitivity of 35S-DREB2C plants is developmental stage dependent.
Overexpression of DREB2C Affects Stress Response
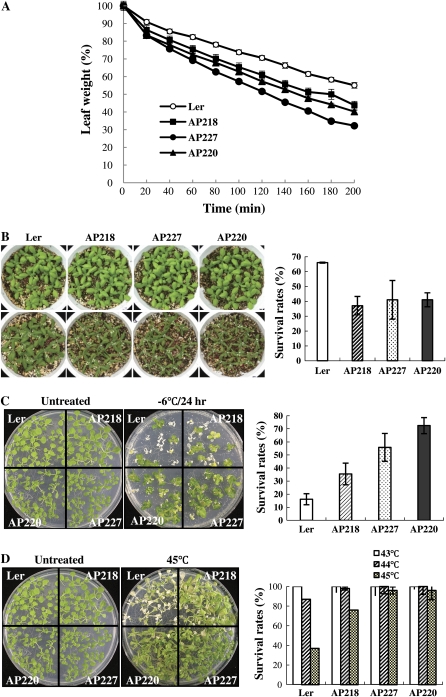
One of the major functions of ABA is the control of stomatal movement under water deficit conditions. To address if DREB2C is involved in the process, we determined the transpiration rates of 35S-DREB2C plants by measuring the rates of weight loss of detached leaves. As shown in Figure 5A, relative fresh weight of the transgenic leaves was consistently lower than that of the wild-type leaves, indicating that their transpiration rates are higher. Consistent with this result, survival rates (i.e. approximately 40%) of soil-grown plants under water deficit conditions were lower than the wild-type rate (i.e. 66%; Fig. 5B).
Figure 5.
Transpiration rates and stress tolerance of DREB2C overexpression lines. A, Transpiration rates of DREB2C overexpression lines. Leaves were detached from 3-week-old plants (fifth to eighth leaves) and weighed at 20-min intervals. Relative leaf weights (%) compared with initial weights are shown. Each data point represents the mean of quadruplicate measurements (n = 12 each), and the error bars indicate se. B, Drought tolerance of DREB2C overexpression lines. Nine-day-old seedlings were withheld from water for 10 d (top row) or 15 d (bottom row) before photographs were taken. Survival rates were determined 3 d after rewatering at the end of the test. The data represent means of three independent experiments (n = 100 each, P = 0.087), and the error bars indicate se. C, Freezing tolerance of DREB2C overexpression lines. Two-week-old plants were placed at −6°C for 24 h without any pretreatment, returned to normal growth temperature, and grown for 5 d. The data show mean survival rates of six independent measurements (n = 13 each). D, Heat tolerance of DREB2C overexpression lines. Two-week-old plants were placed at the indicated temperatures for 1 h and grown at normal conditions for 4 d before photographs were taken. The survival rates are means of five to 15 independent measurements (n = 13 each), and the error bars indicate se. In B to D, representative plants are shown with treatment conditions and line numbers. In A and D, error bars are generally smaller than the symbols. Ler, Landsberg erecta.
Both ABF and CBF/DREB family transcription factors control the abiotic stress response. Hence, we investigated whether DREB2C overexpression affected other abiotic stress tolerance. We first examined the freezing tolerance of DREB2C OX lines by determining their survival rates after exposing them to subfreezing temperature. When plants were exposed to −6°C for 24 h, 16% of the untransformed plants survived. At the same condition, the survival rates of AP218, AP227, and AP220 were 36%, 56%, and 73%, respectively (Fig. 5C). Next, we investigated the heat tolerance of the plants by determining their survival rates under high-temperature conditions. Figure 5D shows that both untransformed plants and DREB2C OX lines survived at 43°C and 44°C. At 45°C, however, DREB2C OX lines exhibited much higher survival rates; whereas 37% of the wild-type plants survived the temperature, the survival rates of AP218, AP227, and AP220 were 76%, 96%, and 96%, respectively. Thus, DREB2C OX lines were freezing and heat tolerant. We also investigated cold tolerance by growing plants at low, nonfreezing temperature (i.e. 4°C), but we did not observe differences in growth between wild-type and DREB2C OX lines.
Overexpression of DREB2C Affects the Expression of Stress-Responsive Genes
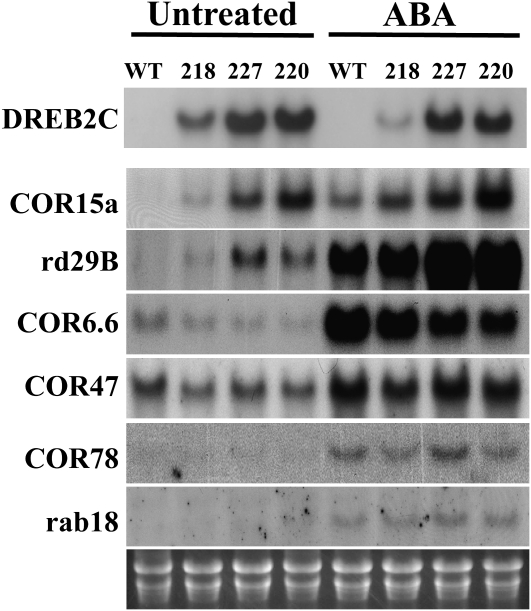
The altered ABA and abiotic stress responses suggested that DREB2C is likely to have a regulatory role in vivo. To test this possibility, we determined the expression levels of a number of stress-responsive, DRE/CRT-containing genes (Jaglo-Ottosen et al., 1998) in 35S-DREB2C lines. Figure 6 shows that the RNA level of COR15a gene, to whose promoter DREB2C binds (Fig. 3B), was elevated in the transgenic lines compared with the wild-type level. Similarly, RD29B(LTI65) gene expression was elevated in the transgenic plants. On the other hand, COR6.6 gene expression was reduced slightly in 35S-DREB2C plants, and the expression of COR47, COR78/RD29A, and RAB18 was not affected. Thus, DREB2C overexpression affected the expression of a subset of CBF/DREB-regulated genes.
Figure 6.
Expression of ABA-responsive genes in DREB2C overexpression lines. RNA was isolated from normal (untreated) or ABA-treated (100 μm for 4 h) plants, and the expression of ABA and/or cold-responsive genes was determined by RNA gel-blot analysis. The bottom panel shows an ethidium bromide-stained gel. WT, Wild type.
DREB Family Proteins Interact with ABF Family Proteins
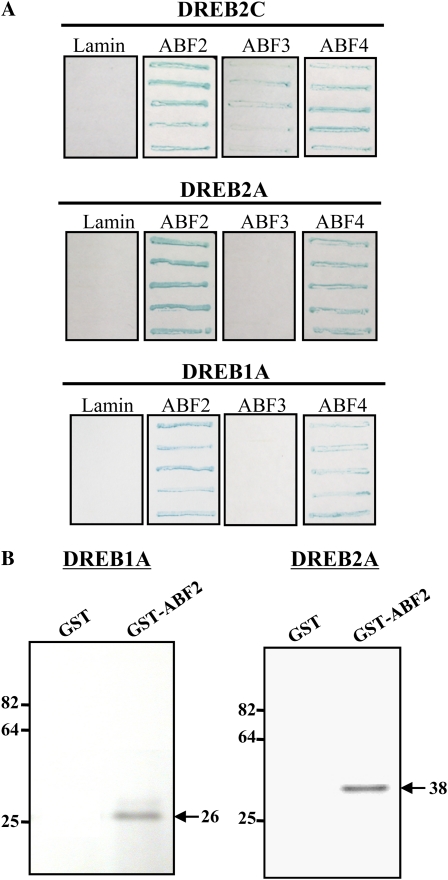
As mentioned earlier, DREB2C belongs to the same subfamily of AP2/ERF proteins as DREB2. Narusaka et al. (2003) previously demonstrated that DRE functions as a coupling element to ABRE to induce ABA-dependent gene expression. Therefore, we wanted to know whether ABF2 and other ABFs would interact with DREB family factors, especially with DREB1A and DREB2A, whose functions have been characterized in depth (Liu et al., 1998; Gilmour et al., 2000; Sakuma et al., 2006a, 2006b). We first carried out a yeast two-hybrid assay to address the question. Bait constructs containing partial fragments of ABF2, ABF3, or ABF4 were prepared, and their interactions with DREB2C, DREB2A, and DREB1A were determined. The results showed that DREB2C interacted with ABF3 and ABF4 as well as ABF2 (Fig. 7A, top). On the other hand, DERB2A interacted with ABF2 and ABF4 but not with ABF3 (Fig. 7A, middle). DREB1A displayed the same interaction specificity as DREB2A (i.e. it interacted with ABF2 and ABF4 but not with ABF3; Fig. 7A, bottom).
Figure 7.
Interactions between ABF and DREB family members. A, Two-hybrid assays were carried out to investigate the interactions between ABF and DREB family members. DREB2C, DREB2A, and DREB1A indicate prey constructs carried on pYesTrp-2. Lamin, ABF2, ABF3, and ABF4 denote bait constructs. Yeast containing each bait construct was transformed with individual prey constructs, transformants were grown on Gal/raffinose-His-Leu-Trp-Ura medium, and colony lift β-galactosidase assays were performed. B, In vitro interactions between ABF2 and DREBs. ABF2 interaction with DREB1A or DREB2A was investigated by GST pulldown assy. In vitro-translated DREB1A or DREB2A, each labeled with [35S]Met, was allowed to bind GST or GST-ABF2 fusion protein, and bound protein was visualized by autoradiography after SDS-PAGE. The numbers indicate protein size in kD. [See online article for color version of this figure.]
To confirm the result of the two-hybrid assay, ABF2 interaction with DREB1A and DREB2A was examined by GST pulldown assay. Recombinant GST-ABF2 fusion protein was prepared and allowed to bind the in vitro-translated DREB proteins labeled with [35S]Met. Bound proteins were then visualized by autoradiography after PAGE. Figure 7B shows that DREB1A bound the GST-ABF2 fusion protein but not GST alone. Likewise, DREB2A was retained by GST-ABF2 but it did not bind GST alone (Fig. 7B). Thus, our in vitro binding assay also showed that ABF2 interacts with DREB1A and DREB2A.
DISCUSSION
The ABF subfamily of bZIP factors consists of four members, ABF1 to ABF4 (Choi et al., 2000). Analyses of their overexpression and knockout lines showed that ABF2, ABF3, and ABF4 play overlapping but distinct roles in ABA and abiotic stress responses (Kim, 2006). In particular, ABF2 plays an essential role in seedling growth regulation and Glc response, and its overexpression results in altered ABA/stress responses. ABF2 is unique in that, unlike ABF3 and ABF4, its overexpression phenotypes are developmental stage dependent (Kim et al., 2004c).
Our results indicate that ABF2 interacts with the AP2/ERF domain factor DREB2C. DREB2C is a member of the DREB2 subfamily of AP2/ERF domain transcription factors, which consists of eight members (i.e. DREB2A–DREB2H) and ABI4 (Finkelstein et al., 1998). Among the subfamily members, DREB2C exhibits highest sequence identity to DREB2H. The sequence identity (75%), however, is limited to the AP2 domain region, because DREB2H lacks the C-terminal half (i.e. it consists of only the AP2 domain). DREB2C does not show significant homology to other DREB2 members outside the AP2 domain.
Several AP2/ERF family proteins, which do not belong to the DREB2 subfamily, have been reported that are involved in ABA response. ABI4 (Finkelstein et al., 1998) is a positive regulator of ABA and sugar responses. On the other hand, ABR1 (Pandey et al., 2005) and AtERF7 (Song et al., 2005) are negative regulators. The maize AP2/ERF domain protein DBF1 (Kizis and Pages, 2002) is a DRE-binding factor that positively regulates ABA-responsive expression of RAB17. DREB2C is not homologous to any of these AP2/ERF family proteins.
Little is known about the functions of the DREB2 family transcription factors except those of DREB2A (Sakuma et al., 2006a, 2006b), which is involved in drought and heat responses. Others (Lim et al., 2007; Lee et al., 2009) previously showed that DREB2C OX lines are heat tolerant. They also observed DNA-binding and transcriptional activities of DREB2C and reported a number of DREB2C-regulated genes. Our results suggest that DREB2C is involved not only in heat response but also in other stress responses. Furthermore, we showed that DREB2C OX lines exhibit a number of phenotypes that suggest its regulatory role in ABA response. Germination of the DREB2C transgenic seeds and young seedling growth are ABA hypersensitive (Fig. 4, B and C). Postgermination root elongation, on the contrary, is ABA insensitive (Fig. 4D). Transpiration rates of the transgenic plants are higher than the wild-type rate, and the plants are more susceptible to water-deficit conditions (Fig. 5, A and B). DREB2C overexpression also affected the expression of several ABA/stress-responsive genes and promoted not only thermotolerance but also freezing tolerance (Figs. 5, C and D, and 6). Together, these observations strongly suggest that DREB2C may play a regulatory role in ABA and stress responses, probably by regulating a subset of ABA/stress-responsive genes via DRE/CRT.
Narusaka et al. (2003) showed that DRE/CRT in the RD29A promoter can function as a coupling element to ABRE; whereas a single copy of ABRE cannot direct ABA-responsive reporter gene activation, DRE/CRT and ABRE together can. Furthermore, they showed that coexpression of DRE-binding and ABRE-binding factors synergistically activates the RD29A promoter. A similar observation was also made by Xue and Loveridge (2004), who reported that the barley AP2 domain protein HvDRF1 cooperates with HvABI5 in the activation of the ABA-inducible HVA1 promoter. These results imply that ABFs/AREBs and DREBs may synergistically interact with each other in ABA-responsive gene expression. The nature of the interaction, however, was not yet known.
Our results suggest that the interaction between ABFs/AREBs and DREBs may involve physical interaction. ABF2 interacts not only with DREB2C (Fig. 7) but also with DREB1A and DREB2A in vitro and in yeast. Moreover, ABF4 also interacts with DREB1A and DREB2A. The DREB2 subfamily (i.e. group IV/A-2 subfamily) of AP2/ERF proteins consists of nine members including ABI4, and DREB1A belongs to another subfamily consisting of six members (Sakuma et al., 2002; Nakano et al., 2006). ABFs/AREBs, on the other hand, are members of a bZIP subfamily consisting of nine members (Kim et al., 2002). At present, we do not know whether other members of these bZIP and AP2/ERF subfamilies also interact with each other. Our limited analysis of interactions presented in this paper, however, suggests that, if they do, specificity would exist in their physical interactions. It remains to be determined what determines the putative interaction specificity between the two families of transcription factors. In addition, in vivo, their interactions would be further limited by their spatial and temporal expression patterns.
Although our data indicate that ABF2 interacts with DREB2C and other DREB family members, we do not know the biological significance of their interactions. In the case of ABF2 and DREB2C, their expression patterns are similar in several respects. ABF2 is nucleus localized and highly inducible by high salt and ABA (Kim et al., 2004c). Similarly, DREB2C is localized in the nucleus and inducible by high salinity and high osmolarity (Fig. 2), implying that they may interact in vivo under these abiotic stress conditions. Considering the diverse expression patterns of ABF and DREB family members, it would be possible that different members of the two protein families may interact with each other, depending on tissue type and environmental conditions.
Also, the interaction mechanism and the outcome of their interactions need to be determined in the future. Both ABF2 and DREB2C possess DNA-binding and transcriptional activities, and other members of the two protein families also possess DNA-binding and/or transcriptional activities. It may be speculated that ABFs/AREBs and DREBs might cooperate in their binding to respective cognate cis-elements, or their physical association might somehow modulate their transcriptional activities. In the transient assay by Narusaka et al. (2003), coexpression of ABFs/AREBs and DREBs enhanced transcription of the RD29A promoter. In our case, overexpression of DREB2C resulted in phenotypes that are different from those of ABF2: shoot development of DREB2C OX lines was hypersensitive to ABA, whereas root elongation was insensitive to ABA. ABF2/AREB1 activity is regulated by ABA-dependent phosphorylation (Furihata et al., 2006). Thus, part of the reason for the difference may be the requirement of posttranslational modification for ABF2 activity. Also, it may be speculated that equimolar amounts of ABF2 and DREB2C need to be present for cooperative interaction, and excess presence of one of the proteins may have an inhibitory effect.
MATERIALS AND METHODS
Manipulation of DNA, RNA Isolation, and Expression Analysis
Manipulation of DNA and RNA was according to the standard methods (Sambrook and Russell, 2001). RNA isolation, RNA gel-blot analysis, and RT-PCR were carried out as described previously (Kang et al., 2002; Kim et al., 2004b). RNA samples used for RT-PCR analysis were treated with DNase I to remove possible contaminating DNA. The first-strand cDNA synthesis was carried out employing SuperScript III (Invitrogen) according to the supplier's instruction, and amplifications were carried out within linear range. The primer set 5′-AGGATTGTAGCGATGAATATGTTCTC-3′ and 5′-CCAGTCAAACGAAATTTGAAATCTTATG-3′ was used for the RT-PCR analysis of DREB2C. Probes for northern-blot analysis were prepared by PCR, using primer sets that span gene-specific regions whenever possible, and gel purified. Primer sequences are available upon request.
Two-Hybrid Screen and Two-Hybrid Assay
Bait construction, two-hybrid screening, and the analysis of positive clones have been described in detail elsewhere (Kim et al., 2004b). A full-length DREB2C cDNA, which includes parts of 5′ and 3′ untranslated regions, was isolated from a cDNA plasmid library by PCR, using the primers 5′-GCTATCGTCTTTGCTACTACTACTAC-3′ and 5′-CTTGTTCATATGTCATATGTATTCACAAG-3′. The PCR product was cloned into the TOPO-TA cloning vector (Invitrogen), and the nucleotide sequence was confirmed by sequencing the entire insert fragment.
Bait constructs for the two-hybrid assays shown in Figure 7 were prepared by cloning various portions of ABF coding regions into pGilda (Clontech). The ABF2 fragment containing amino acids 65 to 337 was prepared by PCR and cloned into the EcoRI-XhoI sites of pGilda, whereas the PCR product of the ABF3 fragment (amino acids 274–373) was cloned into the SmaI-XhoI sites of the same vector. The ABF4 fragment (amino acids 89–352) was cloned into the EcoRI-XhoI sites of pGilda. Prey constructs were prepared using pYESTrp-2 (Invitrogen). For DREB2C, the original positive clone (no. 27) was used in the assay. For DREB1A and DREB2A, entire coding regions were amplified and cloned into the BamHI-NotI and the EcoRI-NotI sites of pYESTrp-2, respectively. Primer sequences used in the bait and the prey constructs are available upon request.
GST Pulldown Assay
To prepare the GST-ABF2 fusion construct, a partial ABF2 coding region containing the N-terminal 337 amino acids but lacking the C-terminal 79 amino acids (i.e. the bZIP region) was prepared by PCR using primers 5′-ATGGATGGTAGTATGAATTTGG-3′ and 5′-gagagctcgagCTCTACAACTTTCTCCACAGTG-3′ and, after XhoI digestion, cloned into the SmaI-XhoI sites of pGEX6P-2 (GE Healthcare). GST-ABF2 fusion protein was prepared using a GST purification kit (GE Healthcare) according to the supplier's instructions. Briefly, fresh culture (optical density at 600 nm = 0.6) of BL21(DE3) cells transformed with the GST-ABF2 fusion construct was induced with 1 mm isopropyl-β-d-thiogalactoside for 2 h at 37°C to express the recombinant protein, the cells were lysed by sonication, and the cell debris were removed by centrifugation. The cleared lysate was loaded onto the glutathione-Sepharose 4B column (GE Healthcare). The column was washed with 1× phosphate-buffered saline (PBS) buffer (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4, pH 7.4), and bound proteins were eluted with a glutathione elution buffer (50 mm Tris-HCl, pH 8.0, and 10 mm glutathione).
For in vitro translation, the open reading frame of DREB2C, DREB1A, or DREB2A was cloned into pGEM-T Easy vector (Promega). In vitro translation was performed using the TNT Quick coupled transcription/translation system (Promega) according to the manufacturer's instructions in the presence of [35S]Met.
For the GST pulldown assay, in vitro-translated DREB2C, DREB1A, or DREB2A product was incubated with 10 μg of GST and glutathione-Sepharose 4B resin for 1 h at room temperature in 1× PBS buffer. After brief centrifugation, the supernatant was incubated with GST or GST-ABF2 fusion protein (5 μg) in the presence of glutathione-Sepharose 4B resin for 1 h at room temperature with constant rotation. At the end of binding, resins were washed three times with 1 mL of 1× PBS buffer to remove unbound proteins, and bound proteins were eluted with a glutathione elution buffer. Proteins were separated on 12% SDS-polyacrylamide gels and visualized by autoradiography.
Electrophoretic Mobility Shift Assay
To prepare recombinant DREB2C, the entire DREB2C coding region was amplified using the primers 5′-ATGCCGTCGGAGATTGTTGACAG-3′ and 5′-ttttccttttgcggccgcAATCTTATGTAGATCCATGAACATC-3′ and cloned into pGEX 6P-2 (GE Healthcare) after NotI digestion. The GST-DREB2C fusion construct was used to transform BL21 cells, and GST-DREB2C fusion protein was prepared using a GST purification kit (GE Healthcare) from the transformed cells according to the supplier's instructions.
Electrophoretic mobility shift assay was carried out as described (Choi et al., 2000). Briefly, 2 μg of GST-AP2 fusion protein was incubated with each DNA probe for 30 min on ice in a binding buffer (25 mm HEPES, pH 7.6, 10% glycerol, 1 mm MgCl2, 1 mm dithiothreitol, 1 mm EDTA, and 50 mm KCl) containing 1 μg of poly(dI-dC). Sequences of the forward strand of probe DNA (Stockinger et al., 1997; Liu et al., 1998), which were prepared by Klenow fill-in reaction in the presence of [32P]dATP, are as follows: DRE, aattAGATATACTACCGACATGAGTTCC; mDRE, aattAGATATACTACTTTTATGAGTTCC; COR15a, ttAATTTCATGGCCGACCTGCTTTTTT; and mCOR15a, ttAATTTCATGGAATCACTGCTTTTTT.
Transcriptional Assay
Full-length or partial fragments of DREB2C were prepared by PCR using the following primer sets: 5′-CATGCCGTCGGAGATTGTTGACAG-3′ and 5′-AATCTTATGTAGATCCATGAA CATC-3′ for the full-length construct; 5′-ATGCCGTCGGAGATTGTTGACAG-3′ and 5′-CAAATCCCGTTTTCAGGTCCACC-3′ for DREB2C-N; 5′-TGACTATAGAGGAGTTAGACAGAG-3′ and 5′-GAGCGATTTGTGATCTCGGGAAG-3′ for DREB2C-AP2; and 5′-TTCTTCGACTGCTGCCACTGCC-3′ and 5′-AATCTTATGTAGATCCATGAACATC-3′ for DREB2C-C. The PCR products were cloned into GBT9 (Clontech) digested with SmaI.
The constructs were individually introduced into SFY526 yeast strain by transformation, and the resulting transformants were kept on SC-Trp-Ura medium. For transcriptional assay, colonies from each transformant group were grown in a SC-Trp-Ura medium to A600 of approximately 1. The cultures were diluted five times with fresh medium and grown further for 4 h. A600 was measured at the end of the growth period, and 1.5-mL aliquots were processed according to the instructions for β-galactosidase assay provided by Clontech. For each construct, five colonies were assayed. β-Galactosidase activity was expressed in Miller units.
Plant Material, Preparation of Transgenic Plants, and Phenotype Analysis
To prepare DREB2C OX lines, the coding region of DREB2C was amplified using the primers 5′-ATGCCGTCGGAGATTGTTGACAG-3′ and 5′-AATCTTATGTAGATCCATGAACATC-3′ from the full-length DREB2C cDNA clone. The amplified fragment was then cloned into pBI121 (Jefferson et al., 1987), which was prepared by XbaI digestion followed by Klenow fill-in reaction after removal of the GUS coding region.
To prepare the DREB2C promoter-GUS construct, the 1.1-kb promoter fragment was amplified using the primer set 5′-acgcgtcgacGTTGTAGATTGAAAATGAGATAGCTC-3′ and 5′-GTAAGAAAGCACTCGAACAAAGTAGC-3′. The amplified fragment was then cloned into SmaI-SalI sites of pBI101.2 after digestion with SalI. Arabidopsis (Arabidopsis thaliana) plants were transformed according to the vacuum infiltration method (Bechtold and Pelletier, 1998) using Agrobacterium tumefaciens strain GV3101. Nine homozygous overexpression lines were recovered, and, after preliminary analysis, three representative lines were analyzed in detail. Phenotype analysis, ABA response, and various stress tests were carried out as described (Kang et al., 2002; Kim et al., 2004a, 2004c) using T4 homozygous lines. For promoter analysis, four transgenic lines were generated, which displayed the same GUS staining pattern. GUS staining was conducted as described (Kang et al., 2002) with T2 or T3 generation transgenic plants.
Subcellular Localization
The coding region of DREB2C was prepared by PCR using the primer set 5′-aaggagctcATGCCGTCGGAGATTGTTGACA-3′ and 5′-TGTAGATCCATGAACATCTTTGTCTC-3′, and after SacI digestion, the amplified fragment was cloned into the SacI-SmaI sites of p35S-FAST/EYFP in frame with the EYFP coding region. Tobacco (Nicotiana benthamiana) leaves were coinfiltrated with the Agrobacteriumstrains (C58C1) containing the DREB2C-EYFP construct and p19 according to Witte et al. (2004). The images of the tobacco epidermal cells were obtained with a fluorescence microscope (Olympus BX51) 30 to 40 h after infiltration.
Acknowledgments
We are grateful to the Kumho Life Science Laboratory of Chonnam National University for providing equipments and plant growth facilities.
References
- Baker SS, Wilhelm KS, Thomashow MF. (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701–713 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. (1995) Adaptations to environmental stresses. Plant Cell 7: 1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pages M. (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37: 425–435 [DOI] [PubMed] [Google Scholar]
- Chae MJ, Lee JS, Nam MH, Cho K, Hong JY, Yi SA, Suh SC, Yoon IS. (2007) A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol Biol 63: 151–169 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong JH, Kang J, Kim SY. (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 21: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Choi H, Park HJ, Park JH, Kim S, Im MY, Seo HH, Kim YW, Hwang I, Kim SY. (2005) Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of ABA-responsive gene expression, and modulates its activity. Plant Physiol 139: 1750–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SS, Lynch TJ, Thomas TL, Rock CD. (2005) Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE (ABI) 5 and ABRE-BINDING FACTOR (ABF) 3. Plant Mol Biol 59: 253–267 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Porras JL, Riano-Pachon DM, Dreyer I, Mayer JE, Mueller-Roeber B. (2007) Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics 8: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A. (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T. (1999a) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19: 679–689 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. (1999b) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. (1998) Arabidopsis CBF1 overexpression induces cor genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 20: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Choi H, Im M, Kim SY. (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Kang JY, Kim SY. (2004a) Over-expression of a transcription factor regulating ABA-responsive gene expression confers multiple stress tolerance. Plant Biotechnol J 2: 459–466 [DOI] [PubMed] [Google Scholar]
- Kim S, Choi HI, Ryu HJ, Park JH, Kim MD, Kim SY. (2004b) ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol 136: 3639–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY. (2004c) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40: 75–87 [DOI] [PubMed] [Google Scholar]
- Kim SY. (2006) The role of ABF family bZIP class transcription factors in stress response. Physiol Plant 126: 519–527 [Google Scholar]
- Kim SY, Chung HJ, Thomas TL. (1997) Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J 11: 1237–1251 [DOI] [PubMed] [Google Scholar]
- Kim SY, Thomas TL. (1998) A family of novel basic leucine zipper proteins bind to seed-specification elements in the carrot Dc3 gene promoter. J Plant Physiol 152: 607–613 [Google Scholar]
- Kim SY, Ma J, Perret P, Li Z, Thomas TL. (2002) Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol 130: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis D, Pages M. (2002) Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J 30: 679–689 [DOI] [PubMed] [Google Scholar]
- Knight H, Zarka DG, Okamoto H, Thomashow MF, Knight MR. (2004) Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol 135: 1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Han KS, Kwon YS, Lee JH, Kim SH, Chung WS, Kim Y, Chun SS, Kim HK, Bae DW. (2009) Identification of potential DREB2C targets in Arabidopsis thaliana plants overexpressing DREB2C using proteomic analysis. Mol Cells 28: 383–388 [DOI] [PubMed] [Google Scholar]
- Lim CJ, Hwang JE, Chen H, Hong JK, Yang KA, Choi MS, Lee KO, Chung WS, Lee SY, Lim CO. (2007) Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem Biophys Res Commun 362: 431–436 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH. (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. (1995) The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S. (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanjulu S, Bartels D. (2002) Drought- and desiccation-induced modulation of gene expression in plants. Plant Cell Environ 25: 141–151 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2006a) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. (2006b) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Shen Q, Ho TH. (1995) Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho TH. (1996) Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8: 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QJ, Casaretto JA, Zhang P, Ho TH. (2004) Functional definition of ABA-response complexes: the promoter units necessary and sufficient for ABA induction of gene expression in barley (Hordeum vulgare L.). Plant Mol Biol 54: 111–124 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Seki M, Ishida J, Satou M, Sakurai T, Narusaka M, Kamiya A, Nakajima M, Enju A, Akiyama K, et al. (2004) Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol Biol 56: 29–55 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Witte CP, Noël LD, Gielbert J, Parker JE, Romeis T. (2004) Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol Biol 55: 135–147 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP, Loveridge CW. (2004) HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J 37: 326–339 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salinity stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10: 88–94 [DOI] [PubMed] [Google Scholar]