Abstract
Circadian clocks provide temporal coordination by synchronizing internal biological processes with daily environmental cycles. To date, study of the plant circadian clock has emphasized Arabidopsis (Arabidopsis thaliana) as a model, but it is important to determine the extent to which this model applies in other species. Accordingly, we have investigated circadian clock function in Brassica rapa. In Arabidopsis, analysis of gene expression in transgenic plants in which luciferase activity is expressed from clock-regulated promoters has proven a useful tool, although technical challenges associated with the regeneration of transgenic plants has hindered the implementation of this powerful tool in B. rapa. The circadian clock is cell autonomous, and rhythmicity has been shown to persist in tissue culture from a number of species. We have established a transgenic B. rapa tissue culture system to allow the facile measurement and manipulation of clock function. We demonstrate circadian rhythms in the expression of several promoter:LUC reporters in explant-induced tissue culture of B. rapa. These rhythms are temperature compensated and are reset by light and temperature pulses. We observe a strong positive correlation in period length between the tissue culture rhythm in gene expression and the seedling rhythm in cotyledon movement, indicating that the circadian clock in B. rapa tissue culture provides a good model for the clock in planta.
Circadian rhythms, the subset of endogenous rhythms with a period of approximately 24 h, are widely encountered in most organisms from cyanobacteria to humans. Although eukaryotes employ a common mechanistic logic of interlocked negative feedback loops to generate robust circadian oscillations, different components have been recruited to form the clock in different taxa (Bell-Pedersen et al., 2005; McClung, 2006; Wijnen and Young, 2006). Thus, fungal, animal, and plant clocks share a common architectural plan yet are composed of largely distinct components. This suggestion of polyphyletic origins of clocks implies strong selection for clock function. Indeed, in many organisms, including cyanobacteria, fruit fly, ground squirrel, and Arabidopsis (Arabidopsis thaliana), experimental evidence indicates that a robust circadian clock whose period resonates with the environmentally imposed diurnal cycle confers a fitness advantage (Yerushalmi and Green, 2009).
Among plants, most that is known about the molecular basis of circadian clock function comes from the study of Arabidopsis (McClung, 2008; Harmer, 2009). The current model of the Arabidopsis clock includes multiple interlocked negative feedback loops. In the central loop, two partially redundant single Myb domain transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), form the negative arm, binding to the TIMING OF CAB EXPRESSION1 (TOC1) promoter to inhibit expression (Alabadí et al., 2002; Mizoguchi et al., 2002). TOC1 is a positive regulator of both CCA1 and LHY (Alabadí et al., 2001). Although TOC1 lacks defined DNA-binding domains, it is recruited to the CCA1 promoter, possibly through interaction with other DNA-binding protein(s) such as CCA1 HIKING EXPEDITION (CHE), which binds to the CCA1 promoter to negatively regulate CCA1 expression (Pruneda-Paz et al., 2009). In a second interlocked loop, termed the “evening” loop, TOC1 is predicted to repress a component, Y, that in turn activates TOC1 expression (Locke et al., 2005, 2006). GIGANTEA (GI) is hypothesized to be a component of Y (Locke et al., 2006). In a third interlocked loop, termed the “morning” loop, CCA1 and LHY positively regulate two TOC1 paralogs, PSEUDO-RESPONSE REGULATOR7 (PRR7) and PRR9 (Harmer and Kay, 2005; Mizuno and Nakamichi, 2005). PRR5 is also implicated in this loop because, while the prr7prr9 double mutant exhibits an extremely long period (Farré et al., 2005; Nakamichi et al., 2005; Salomé and McClung, 2005a) and is conditionally arrhythmic, the prr5prr7prr9 triple mutant is completely arrhythmic, with CCA1 and LHY expressed at a high constitutive level, indicating that PRR5, PRR7, and PRR9 are repressors of CCA1 and LHY (Nakamichi et al., 2005). Consistent with this role, PRR5, PRR7, and PRR9 bind to the CCA1 and LHY promoters (Nakamichi et al., 2010).
Posttranscriptional regulation, in particular the temporally regulated proteasomal degradation of specific clock proteins, is critical to the clock mechanism (Gallego and Virshup, 2007). In Arabidopsis, the clock regulates the stability of many clock proteins, including GI (David et al., 2006), LHY (Song and Carré, 2005), ZEITLUPE (ZTL; Kim et al., 2003), and members of the TOC1/PRR family (Más et al., 2003b; Farré and Kay, 2007; Ito et al., 2007; Kiba et al., 2007; Para et al., 2007; Fujiwara et al., 2008; Harmon et al., 2008). An E3 ubiquitin ligase Skp, Cullin, F-box-containing complex including the F-box protein ZTL is crucial for clock-regulated proteasomal degradation of TOC1 (Más et al., 2003b; Han et al., 2004; Harmon et al., 2008). ZTL also targets PRR5 for proteasomal degradation (Kiba et al., 2007; Fujiwara et al., 2008), and proteasome activity is implicated the degradation of PRR7 and PRR9 (Farré and Kay, 2007; Ito et al., 2007).
It is important to determine the extent to which the Arabidopsis clock serves as a model for all plant clocks. To this end, a number of studies have identified orthologs of Arabidopsis clock genes in order to probe clock function in diverse species, including rice (Oryza sativa; Murakami et al., 2007), duckweed (Lemna gibba; Serikawa et al., 2008), the common ice plant (Mesembryanthemum crystallinum; Boxall et al., 2005), and chestnut (Castanea sativa; Ramos et al., 2005). Brassica species offer an excellent system in which to study clock function. Since the separation of their common ancestor from Arabidopsis some 16 to 21 million years ago (Koch et al., 2001), Brassica genomes have triplicated (O'Neill and Bancroft, 2000; Rana et al., 2004; Parkin et al., 2005; Town et al., 2006; Yang et al., 2006) and, in some cases, further undergone polyploidization through hybridization (U, 1935), offering opportunity for novel functional diversification, including in circadian timekeeping. In this context, it is worth recalling that altered expression of clock genes has been implicated in heterosis, the increased growth vigor often seen in hybrids and allopolyploids (Ni et al., 2009).
Brassica oleracea exhibits circadian rhythmicity in cotyledon movement that has allowed mapping of quantitative trait loci affecting period length (Salathia et al., 2007). We sought to extend this study through investigation of the genetic basis of circadian rhythmicity in Brassica rapa. Genomic studies have confirmed that B. rapa orthologs of many Arabidopsis clock genes are present in one or two, although to date in no cases three, copies (Kim et al., 2007).
In this work, we first demonstrate a robust circadian rhythm in leaf movement in a number of B. rapa accessions. We next establish molecular assays of clock function, exploiting clock-regulated promoter:LUCIFERASE reporters. Agrobacterium tumefaciens-mediated transformation through explant-based callus cocultivation is an efficient method for many Brassica species and varieties, including Brassica napus, B. oleracea, and Brassica juncea (De Block et al., 1989; Akasaka-Kennedy et al., 2005; Arumugam et al., 2007; Bhalla and Singh, 2008). However, efforts to generate transgenic plants of B. rapa have been less successful, limited by low regeneration and rooting rates from callus and also by the inefficiency of vacuum infiltration (Zhang et al., 2000; Xu et al., 2008).
Recently, it has been established that mammalian tissue culture cells retain circadian rhythmicity (Balsalobre et al., 1998), but it has been known for nearly 60 years that plant tissue cultures can exhibit circadian rhythmicity. Rhythmic fluctuations in growth and turgidity of Daucus carota tissue culture persisted in prolonged darkness, although the assay required periodic microscopic examination in red light, which may have provided external time information to the culture (Enderle, 1951). However, a circadian rhythm in carbon dioxide emission was later established unambiguously in leaf-induced callus cultures of Bryophyllum daigremontianum (Wilkins and Holowinsky, 1965). Recently, Nakamichi and colleagues expanded the repertoire of clock assays for use in plant tissue culture to include promoter:LUCIFERASE (p:LUC) reporters and showed circadian rhythms in pCCA1:LUC and pTOC1:LUC expression in an Arabidopsis cell culture line (Nakamichi et al., 2003, 2004). Accordingly, we have established a transgenic B. rapa tissue culture system to allow the facile measurement and manipulation of clock function. We demonstrate circadian rhythms in the expression of several p:LUC reporters in explant-induced tissue culture of B. rapa. These rhythms are temperature compensated and are reset by light and temperature pulses. We observe a strong positive correlation in period length between the period of the tissue culture rhythm in gene expression and the period of seedling cotyledon movement, indicating that the circadian clock in B. rapa tissue culture provides a good model for the clock in planta. Finally, circadian rhythmicity in tissue culture is disrupted by overexpression of critical Arabidopsis clock genes, showing that the model of clock function that has been developed in Arabidopsis can be effectively applied to B. rapa.
RESULTS
B. rapa Expresses a Robust Circadian Rhythm in Cotyledon Movement in Intact Seedlings
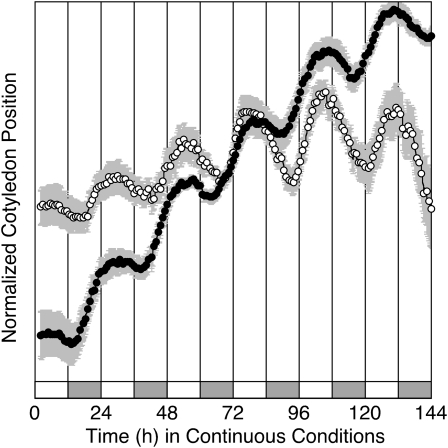
There is a robust circadian rhythm in cotyledon movement in B. rapa, as shown for two accessions: R500, a highly inbred annual yellow sarson seed oil genotype (subspecies trilocularis), and IMB211, a highly inbred rapid cycling Chinese cabbage (subspecies pekinensis; Fig. 1). There is a significant difference in period between the two accessions, with R500 having a shorter period than IMB211 (25.53 ± 0.17 h, n = 69 versus 26.47 ± 0.22 h, n = 81; P < 0.001 by Student's t test). The cotyledon movement rhythms are strong, as indicated by relative amplitude error (RAE; 0.13 ± 0.01 h, n = 69 for R500 versus 0.16 ± 0.0.01 h, n = 81 for IMB211). The RAE of a perfect sine wave is defined as 0, and a value of 1 defines the weakest rhythm considered to be statistically significant (Plautz et al., 1997), although frequently an upper bound of RAE = 0.5 to 0.7 is used to define the limit of rhythmicity.
Figure 1.
B. rapa seedlings exhibit a robust rhythm in cotyledon movement in continuous light. B. rapa seedlings of IMB211 (white symbols) or R500 (black symbols) were entrained to cycles of 12 h of light and 12 h of dark at 18°C for 7 d prior to release into continuous light and temperature (18°C) at time zero. Cotyledon position was monitored for 7 d. Data are presented as means ± se (in gray shading) for six seedlings of each genotype. For clarity, only every fifth data point is shown. The entraining light/dark cycles are indicated by the alternating white and gray bars below the data.
B. rapa Tissue Culture Expresses a Light-Entrainable and Temperature-Compensated Circadian Rhythm in Gene Expression
IMB211 and R500 were used to monitor the expression of circadian clock genes in explant-induced tissue culture lines. To increase the transformation efficiency, excised hypocotyls from B. rapa seedlings were preconditioned for 72 h before cocultivation with A. tumefaciens for 48 h. After 2 weeks of growth on callus induction medium, the transformed basta-resistant callus was green and healthy (Supplemental Fig. S1, A and B), while the basta-sensitive callus sectors turned white. After transfer to organogenesis medium, more than 90% of the green calli remained vigorous under continuous basta selection. However, very few (approximately 0.05%) of the green calli regenerated shoots, and none of the transgenic shoots were successfully rooted in our modified conditions (Supplemental Fig. S1C).
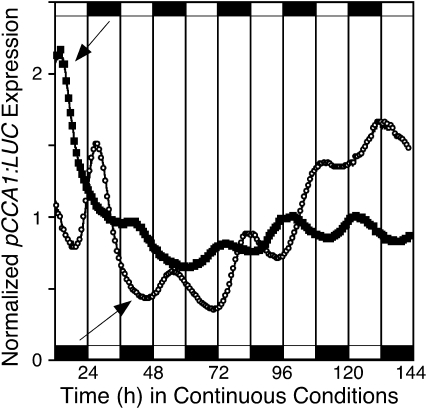
Callus from the shoot regeneration medium was used to characterize free-running rhythms by using the promoters of several Arabidopsis clock and clock-controlled genes, including CCA1 (Wang and Tobin, 1998), LHY (Schaffer et al., 1998), and COLD CIRCADIAN RHYTHM RNA-BINDING2 (CCR2; Carpenter et al., 1994) to drive LUC expression. We entrained two populations of callus to cycles of either 12 h of light and 12 h of dark (LD) or 12 h of dark and 12 h of light (DL) prior to release into continuous light (LL). The bioluminescence of CCA1:LUC maintained a robust circadian oscillation after transfer to LL conditions, peaking in the early morning as defined by the entraining LD or DL regimen, demonstrating that B. rapa callus expresses a robust light-entrainable circadian clock (Fig. 2).
Figure 2.
The circadian clock in B. rapa callus is entrained to light/dark cycles. Populations of callus derived from B. rapa R500 hypocotyl explants were grown in cycles of either 12 h of light and 12 h of dark (white circles) or 12 h of dark and 12 h of light (black squares) prior to release into continuous light at time zero. pCCA1:LUC expression was monitored for 6 d. For clarity, only every fourth data point is shown. The entraining light/dark cycles are indicated by the alternating white and black bars below (light/dark) or above (dark/light) the data.
One of the defining characteristics of a circadian rhythm is that the period is buffered against variations in temperature. Thus, the period of the clock is maintained across a range of physiologically relevant temperatures. Accordingly, to determine whether the rhythm in gene expression in B. rapa tissue culture exhibits temperature compensation, we measured the period length in pCCA1:LUC expression at 18°C and 22°C. The period of IMB211 was approximately 2 h longer than that of R500 at both temperatures; for both accessions, the period was consistent at 18°C and 22°C (increase in rate of process produced by raising temperature by 10°C [Q10] = 0.99 and 1.05, respectively; Table I), demonstrating robust temperature compensation in both genotypes.
Table I. The B. rapa circadian clock is temperature compensated.
| Genotype | Tissue Culture Luciferase Activity |
||
| Period ± se |
Q10 | ||
| 18°C | 22°C | ||
| h | |||
| IMB211 | 28.96 ± 0.35 (n = 8) | 28.87 ± 0.26 (n = 10) | 0.99 |
| R500 | 26.62 ± 0.26 (n = 6) | 27.11 ± 0.24 (n = 6) | 1.05 |
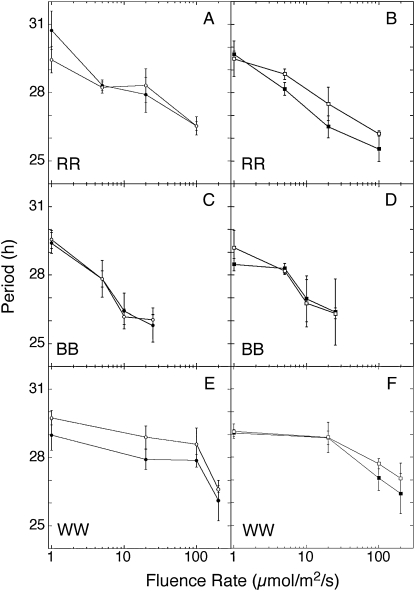
In plants and in day-active animals, the period of circadian rhythms is inversely proportional to light intensity, which is commonly known as Aschoff's rule (Aschoff, 1979). To determine whether the clock in B. rapa callus obeys Aschoff's rule, we examined the free-running period under various intensities of light. Callus expressing pCCA1:LUC or pLHY:LUC was entrained for 7 d in 12/12 LD cycles (using white light) before transfer to constant white, red, or blue light at different fluence rates. Circadian period shortens as fluence rate increases in both genotypes in each of the three light qualities (Fig. 3), indicating that the circadian clock in B. rapa is sensitive to light intensity.
Figure 3.
Circadian period in B. rapa callus is light sensitive and shortens with increasing light intensity. Expression of pCCA1:LUC (A, C, and E) and pLHY:LUC (B, D, and F) was measured in seedlings of B. rapa IMB211 (white symbols) and R500 (black symbols) growing in red (RR; A and B), blue (BB; C and D), or white (WW; E and F) light at the indicated fluence rates. Period was calculated with fast Fourier transform-nonlinear least squares analysis (Plautz et al., 1997).
Light Pulses Reset the Clock in B. rapa Callus
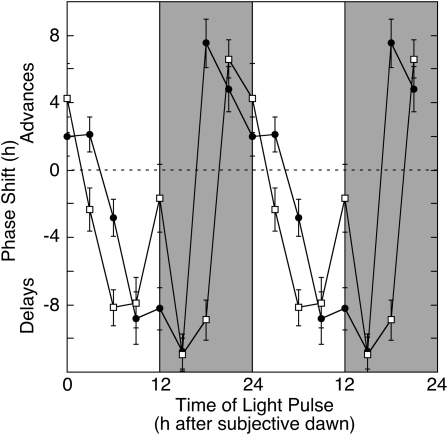
Environmental stimuli such as dawn or dusk and changes in temperature are important for the daily entrainment of the circadian clock (Bruce, 1960; Johnson, 1999; Salomé and McClung, 2005b). It is well established that the circadian clock modulates its own sensitivity to external stimuli such that the same stimulus administered at different times of day can evoke phase advances or delays or have no effect (Johnson, 1990, 1999). We constructed phase response curves (PRCs) to light pulses by entraining callus derived from B. rapa R500 to LD cycles, transferring into continuous dark (DD), exposing to red or blue light pulses (30 min) at 3-h intervals scanning one complete circadian cycle (24 h), and monitoring pCCR2:LUC expression over the ensuing 5 d. Changes in acrophase (peak phase) of pCCR2:LUC expression, expressed relative to callus not exposed to a light pulse, are plotted against the circadian time of the light pulse (Fig. 4). To emphasize the circadian oscillation in light sensitivity, phase shifts are double plotted. Strong (as great as 8 h) phase advances were observed in response to either red or blue light pulses in the early subjective morning and late subjective night (0–3 and 18–24 h after subjective dawn in callus transferred to DD), and strong (as great as 11 h) phase delays were observed in response to either red or blue light pulses in the subjective afternoon and evening (6–15 h after subjective dawn). Such PRCs are typical for plants exposed to light pulses (Johnson, 1990; Covington et al., 2001).
Figure 4.
PRC to light pulses. Callus derived from B. rapa R500 was entrained for 7 d to light/dark cycles followed by transfer into continuous dark. Callus was exposed to pulses (30 min) of red (100 μmol m−2 s−1; black circles) or blue (25 μmol m−2 s−1; white squares) light at 3-h intervals scanning one complete circadian cycle (24 h) and returned to continuous dark for measurement of pCCR2:LUC expression over the ensuing 4 d. Shading indicates subjective night. Changes in acrophase (peak phase), as determined by fast Fourier transform-nonlinear least squares analysis, are expressed relative to the acrophase in callus not exposed to a light pulse and are plotted against the time of onset of the light pulse. Data represent means ± se for three independent experiments. To emphasize the circadian oscillation in light sensitivity, phase shifts are double plotted.
The Circadian Clock in B. rapa Callus Is Entrained by Temperature Cycles
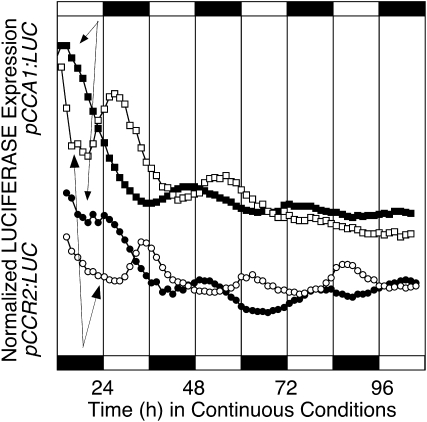
Temperature is an important environmental timing cue that can act independently of light to entrain the Arabidopsis circadian clock (Somers et al., 1998; Michael and McClung, 2002; Michael et al., 2003; Salomé and McClung, 2005b; Thines and Harmon, 2010). To test whether the circadian clock in B. rapa callus is temperature responsive, we exposed callus expressing either pCCA1:LUC or pCCR2:LUC and grown in LL to temperature cycles of either 12 h at 16°C followed by 12 h at 12°C or 12 h at 12°C followed by 12 h at 16°C for at least 3 weeks. Cultures were released into continuous light and temperature (16°C), and luciferase activity was measured. LUC activity continued to oscillate with phase determined by the entraining temperature cycle, with the warmer temperature interpreted as subjective day (Fig. 5). Similar results were obtained with callus entrained to 22°C/16°C cycles, although the subsequent rhythms were less robust (data not shown). These results demonstrate that the circadian clock in B. rapa callus is sensitive to and can be entrained by external temperature cycles.
Figure 5.
The circadian clock in B. rapa callus is entrained to temperature cycles. Populations of callus derived from B. rapa R500 hypocotyl explants growing in continuous light were exposed to cycles of either 12 h of hot (16°C) and 12 h of cold (12°C; white symbols) or 12 h of cold (12°C) and 12 h of hot (16°C; black symbols) prior to release into continuous light at 16°C at time zero. Expression of pCCA1:LUC (squares) and pCCR2:LUC (circles) was monitored for 5 d. The entraining light/dark cycles are indicated by the alternating white and black symbols below (hot/cold) or above (cold/hot) the data.
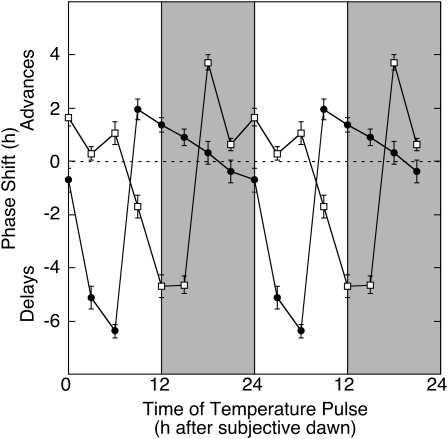
To further evaluate the temperature sensitivity of the B. rapa callus circadian clock, we examined phase resetting in response to pulses of either hot or cold temperature. Callus cultures were released to LL after entrainment to 12/12 LD cycles at 16°C. Pulses (3 h) of either hot (22°C) or cold (10°C) temperature were administered to the cultures at 3-h intervals scanning one circadian cycle, and pCCA1:LUC expression was monitored for the ensuing 4 d to generate PRCs to temperature pulses (Fig. 6). The PRC to hot temperature pulses is similar to that in response to light pulses, albeit with reduced amplitude of phase shifts, including advances of up to 4 h in the early subjective morning and late subjective night and delays of up to 5 h in the subjective afternoon and evening. The PRC to cold temperature pulses is roughly antiphase to that for hot pulses, with advances in the subjective afternoon and evening and delays in the subjective morning and late night. These PRCs are consistent with those seen in Arabidopsis and other plants (Johnson, 1990; Michael et al., 2003; Thines and Harmon, 2010) and demonstrate that the clock in B. rapa callus is responsive to temperature and can be entrained by temperature pulses and cycles.
Figure 6.
PRC to temperature pulses. Callus derived from B. rapa R500 was entrained for 7 d to light/dark cycles at 16°C followed by transfer into continuous light at 16°C. Callus was exposed to pulses (3 h) of hot (22°C; white squares) or cold (10°C; black circles) at 3-h intervals scanning one complete circadian cycle (24 h) and returned to continuous light at 16°C for measurement of pCCR2:LUC expression over the ensuing 4 d. Shading indicates subjective night. Changes in acrophase (peak phase), as determined by fast Fourier transform-nonlinear least squares analysis, are expressed relative to the acrophase in callus not exposed to a temperature pulse and are plotted against the time of onset of the temperature pulse. Data represent means ± se for three independent experiments. To emphasize the circadian oscillation in light sensitivity, phase shifts are double plotted.
The Circadian Clock in B. rapa Tissue Culture Provides a Good Model for the B. rapa Circadian Clock in Planta
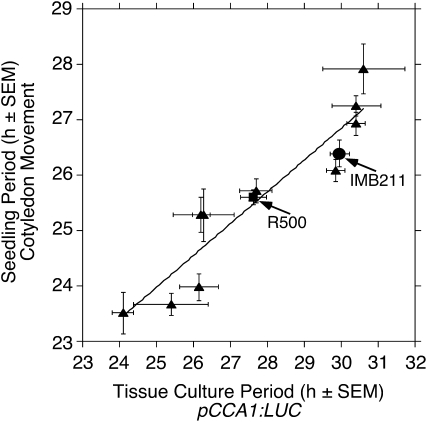
To test whether the circadian rhythm in gene expression in B. rapa callus provides an accurate indication of clock function in planta, we compared period length determined by gene expression (pCCA1:LUC expression) in callus with that determined by seedling cotyledon movement. We compared period determined by both methods in a set of 12 genotypes, including R500 and IMB211 as well as 10 recombinant inbred lines derived from a cross between R500 and IMB211 (Iniguez-Luy et al., 2009), that display a range in period lengths from approximately 23 to 30 h. We observed a strong positive correlation (Pearson product moment correlation coefficient of 0.914, P < 0.00005) in period length between the period of the tissue culture rhythm in gene expression and the period of seedling cotyledon movement (Fig. 7; Supplemental Table S1), indicating that clock function measured in tissue culture provides a good prediction of clock function in planta. However, it should be noted that in every genotype the period in tissue culture gene expression was longer than that in cotyledon movement.
Figure 7.
Period length of gene expression in B. rapa tissue culture predicts period length in planta. B. rapa seedlings as well as callus derived from B. rapa were entrained for 7 d to light/dark cycles at 18°C followed by transfer into continuous light at 18°C for measurement of cotyledon position for the ensuing 7 d or for measurement of pCCA1:LUC expression over the ensuing 4 d. Circadian period is plotted for IMB211 (circles), R500 (squares), and recombinant inbred lines derived from a cross between R500 and IMB211 (triangles). Error bars indicate se. The correlation between periods in the two assays was highly significant (P < 0.00005; Pearson product moment correlation coefficient = 0.914).
Overexpression of Arabidopsis Clock Genes Perturbs the Circadian Clock in B. rapa Callus
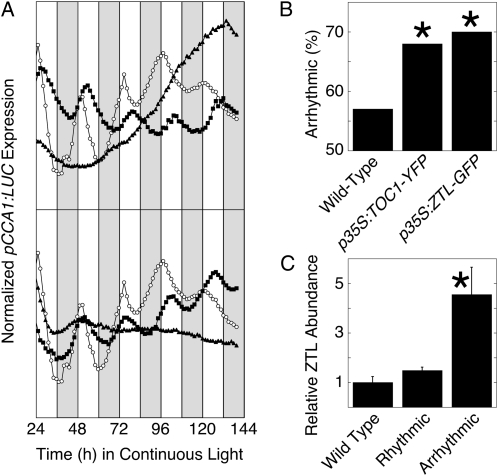
To further test whether the Arabidopsis circadian clock provides a good model for the clock in B. rapa callus, we perturbed the expression of key clock genes through overexpression. TOC1 and ZTL are both critical clock components, and strong overexpression of either results in arrhythmicity in Arabidopsis (Más et al., 2003a; Somers et al., 2004). In Arabidopsis, the response of period to ZTL overexpression shows dosage sensitivity and weak overexpression of ZTL shortens the circadian period (Somers et al., 2004), which is consistent with the period lengthening seen in loss-of-function ztl mutants (Somers et al., 2000). Similarly, weak overexpression of TOC1 lengthens the circadian period, consistent with the period shortening seen in loss-of-function toc1 mutants (Más et al., 2003a). We cotransformed Arabidopsis TOC1 (fused to yellow fluorescent protein [YFP]; Más et al., 2003a) or ZTL (fused to GFP; Somers et al., 2004), both driven by the strong constitutive cauliflower mosaic virus 35S promoter, together with pCCA1:LUC into B. rapa R500 callus. In each case, overexpression of the Arabidopsis clock gene perturbed B. rapa clock function (Table II; Fig. 8A). Overexpression of either TOC1-YFP or ZTL-GFP increased the proportion of callus arrhythmic for pCCA1:LUC activity (Table II; Fig. 8B). Where rhythmicity was detected, the period was lengthened and the rhythm was weaker, as indicated by an increase in RAE, relative to callus transformed only with the pCCA1:LUC reporter (Table II). B. rapa calli transformed with p35S:TOC1 that retain rhythmicity exhibit lengthened period, consistent with weak overexpression in Arabidopsis seedlings (Más et al., 2003a). To determine whether the ZTL overexpression construct yielded the expected overexpression of ZTL, we quantified ZTL expression by western-blot analysis using an antibody directed against Arabidopsis ZTL (Kim et al., 2003). The ZTL antibody detected both endogenous B. rapa ZTL protein and the transgenic Arabidopsis ZTL-GFP fusion protein. ZTL protein was detected at only slightly increased abundance in the rhythmic callus carrying the p35S:ZTL-GFP transgene relative to callus transformed only with the pCCA1:LUC reporter but was detected at greatly increased abundance in the arrhythmic callus (Fig. 8C), consistent with dosage-dependent perturbation of rhythmicity.
Table II. Effects of overexpression of Arabidopsis clock genes on the B. rapa callus clock.
| Transgene | Period ± se | RAE ± se | Arrhythmic | n |
| h | % | |||
| pCCA1:LUC | 23.93 ± 0.41 | 0.38 ± 0.03 | 56 | 62 |
| pCCA1:LUC | 25.51 ± 0.27a | 0.45 ± 0.01b | 68c | 290 |
| p35S:ZTL-GFP | ||||
| pCCA1:LUC | 24.59 ± 0.36d | 0.47 ± 0.03e | 70c | 169 |
| p35S:TOC1-YFP |
Significantly different (P < 0.005) than wild-type R500 transformed with pCCA1:LUC only as determined by Student's t test.
Significantly different (P = 0.005) than wild-type R500 transformed with pCCA1:LUC only as determined by Student's two-tailed t test.
Significantly different (P < 0.001) than wild-type R500 transformed with pCCA1:LUC only as determined by χ2 test.
Not significantly different (P > 0.25) than wild-type R500 transformed with pCCA1:LUC only as determined by Student's two-tailed t test.
Not significantly different (P = 0.0552) than wild-type R500 transformed with pCCA1:LUC only as determined by Student's two-tailed t test.
Figure 8.
Overexpression of Arabidopsis clock genes disrupts B. rapa clock function. A, Callus derived from hypocotyl explants of B. rapa R500 was transformed with pCCA1:LUC (white symbols) or cotransformed with pCCA1:LUC and p35S:TOC1-YFP (top; black symbols) or with pCCA1:LUC and p35S:ZTL-GFP (bottom; black symbols). Callus was entrained for 7 d to light/dark cycles at 16°C followed by transfer into continuous light at 16°C. Cotransformation with p35S:TOC1-YFP or p35S:ZTL-GFP results in either lengthened period (black squares) or arrhythmicity (black triangles). Data are presented as average traces for multiple calli (wild type, n = 5; p35S:TOC1-YFL, n = 21; p35S:ZTL-GFP, n = 31). B, Cotransformation with p35S:TOC1-YFP or with p35S:ZTL-GFP increases the proportion of arrhythmic calli as determined by pCCA1:LUC expression. C, Levels of endogenous ZTL and transgenic ZTL-GFP expression in B. rapa callus. ZTL expression was determined by western-blot analysis using a polyclonal antibody directed against Arabidopsis ZTL and was normalized to γ-tubulin expression and is expressed relative to expression in callus transformed only with pCCA1:LUC. *, Significantly different from the wild type (P < 0.05).
DISCUSSION
The circadian clock allows an organism to coordinate its biology with its temporal environmental conditions and thereby enhances fitness (Yerushalmi and Green, 2009). Thus, better understanding of the circadian clock in crop plants may permit its manipulation toward the goal of enhancing crop productivity. Most that is known about the mechanism of the plant circadian clock is based on Arabidopsis, and it is important to determine the extent to which the circadian clock model developed in Arabidopsis can be extended to other plants, especially to crop species. King and colleagues adapted an automated imaging system to measure cotyledon movement in B. oleracea and used this assay to demonstrate transgressive variation in period length and to map quantitative trait loci that contribute to this variation (Salathia et al., 2007). We herein extend this cotyledon movement assay to B. rapa. Not only is B. rapa an important cultivated crop, but it is closely related to and serves as a simpler genetic model for B. napus, a hybrid between B. rapa and B. oleracea (U, 1935), which is broadly cultivated and a leading source of vegetable oil and protein meal. Thus, the identification and characterization of genes that contribute to variation in clock function in B. rapa will enhance efforts to measure the effects of variation in clock function on field performance and yield in crops of global importance. Such efforts would be greatly facilitated by the development of genetic techniques to test putative clock gene function through transgenic manipulation of candidate gene expression. To that end, we have developed a high-throughput method to measure clock function in transgenic B. rapa tissue culture via the expression of LUC reporters driven by clock-regulated promoters of Arabidopsis clock and clock output genes.
B. rapa callus derived from hypocotyl or cotyledon explants is readily transformed via cocultivation with A. tumefaciens. We introduced pCCA1:LUC, pLHY:LUC, and pCCR2:LUC reporters and showed that each expresses a strong circadian rhythm in light production with characteristic circadian phase in peak light production: morning for CCA1 and LHY (Schaffer et al., 1998; Wang and Tobin, 1998) and evening for CCR2 (Carpenter et al., 1994; Strayer et al., 2000). Thus, B. rapa expresses a circadian clock capable of regulating the expression of Arabidopsis clock genes, providing a high-throughput molecular assay with which to probe clock function. There are three defining characteristics of circadian clock function: an endogenous period of approximately 24 h, entrainment by environmental time cues such as light and temperature, and compensation of period length against changes in ambient temperature (Johnson et al., 2004). We have demonstrated that each of these characteristics is an attribute of the clock in B. rapa tissue culture. The circadian clock in B. rapa callus can be entrained to both light/dark cycles and to temperature cycles. Consistent with this, we have developed PRCs to light or temperature pulses. We show that the period in the rhythm shortens with increasing light intensity, which is generally true for plants and light-active animals. Finally, we show that the rhythm in tissue culture is temperature compensated, exhibiting a Q10 of approximately 1. Thus, we conclude that the clock in B. rapa callus is a bona fide circadian clock and provides a model in which to study circadian clock function in a crop.
Is the molecular constitution of the B. rapa callus clock consistent with that predicted from the Arabidopsis clock? Kim et al. (2007) have shown that the B. rapa genome includes orthologs of all Arabidopsis clock genes queried. We demonstrate that perturbation of the expression of two key Arabidopsis clock genes, TOC1 and ZTL, disrupts circadian clock function in B. rapa tissue culture. In Arabidopsis, strong overexpression of either TOC1 or ZTL results in arrhythmicity and weak overexpression alters period length (Más et al., 2003a; Somers et al., 2004). Consistent with this, we observed that an increased proportion of B. rapa calli transformed with either TOC1 or ZTL driven from the strong constitutive 35S promoter is arrhythmic. We confirmed that ZTL accumulation in the arrhythmic calli transformed with p35S:ZTL was approximately 4-fold greater than that seen in calli not transformed with the overexpression construct. Calli transformed with p35S:TOC1 that retain rhythmicity exhibit lengthened period, consistent with the results of weak overexpression in Arabidopsis (Más et al., 2003a). However, calli transformed with p35S:ZTL that retain rhythmicity also exhibit lengthened period, which is inconsistent with the shortened period associated with weak ZTL overexpression in Arabidopsis plants (Somers et al., 2004). Possibly the target(s) of ZTL differ in B. rapa from those in Arabidopsis, or possibly ZTL function is altered such that its targets are stabilized rather than destabilized, although we have no mechanistic data to support this explanation. We hypothesize that perhaps ZTL expression was reduced in the calli that retained rhythmicity, possibly through cosuppression, which would be consistent with the lengthened period of ztl loss-of-function mutants (Somers et al., 2000). However, ZTL abundance in those calli that retained rhythmicity was not reduced relative to the wild type. Although we cannot conclusively resolve this apparent contradiction, it is possible that only a small proportion of any individual callus is actually transformed with and expressing both the LUC reporter and the ZTL overexpression construct and that the expression of ZTL in those cells is reduced, but we are unable to detect this reduction due to a high background expression of ZTL in the surrounding cells.
The loss of fitness associated with impaired clock function (Yerushalmi and Green, 2009) suggests that the understanding of the molecular mechanism of the circadian clock may offer strategies to manipulate clock function toward the goal of enhancing crop performance. The implementation of this strategy will require much greater insight into the clock mechanism in crop plants. It is critical, therefore, to establish the degree to which the model of the clock in the model species Arabidopsis can be applied to crop species. The demonstration of circadian rhythmicity in a transgenic B. rapa tissue culture system offers a useful tool with which to probe clock function in this crop and represents an important step in testing the applicability of the Arabidopsis clock model to crop species.
MATERIALS AND METHODS
Cotyledon Movement
Brassica rapa seeds were planted in potting soil (Pro-mix), stratified at 4°C in the dark for 3 d, and entrained to cycles of 12 h of light (120 μmol m−2 s−1 white light provided by fluorescent and incandescent bulbs) and 12 h of dark at 18°C for 7 d prior to release into continuous light and temperature (18°C). Cotyledon position was monitored for 7 d using a Lariion Digital Video Surveillance System (www.lariion.com) that multiplexes 32 Sony color video cameras with 3.0- to 8-mm lenses, each of which captures data from 10 seedlings. Cotyledon position is recovered from the time series images using nktrace (Onai et al., 2004) and Metamorph software (Molecular Devices). The quantified leaf position series were then analyzed using the Biological Rhythms Analysis Software System (BRASS 2.1.4; www.amillar.org), which includes fast Fourier transform-nonlinear least squares analysis (Plautz et al., 1997).
Transformation of B. rapa Callus Culture
For tissue culture, seeds of B. rapa cv R500 and cv IMB211 were immersed in distilled water for 30 min, surface sterilized with 70% ethanol for 5 min, and rinsed four times with sterile distilled water. Seeds were then immersed in 20% commercial bleach (containing 5.25% sodium hypochlorite) for 10 min and rinsed four times with sterile distilled water. Then, the seeds were allowed to germinate in Magenta culture boxes on a seedling growth medium containing half-strength Murashige and Skoog (MS) salts, B5 vitamins, 3% (w/v) Suc, and 2 g L−1 Gelrite, pH 5.8. The cultures were incubated at 22°C under 12-h/12-h light/dark cycles using cool-white fluorescent lights at 120 μmol m−2 s−1.
Hypocotyl segments (0.4–0.6 cm) cut from 8-d-old seedlings were preconditioned for 3 d on callus induction medium containing MS salts, B5 vitamins, 1 mg L−1 2,4-dichlorophenoxy acetic acid (Sigma), 3% (w/v) Suc, and 2 g L−1 Gelrite, pH 5.8. Agrobacterium tumefaciens strain GV3101 harboring the various constructs was grown overnight in Luria-Bertani medium containing the appropriate antibiotics (50 mg L−1 rifampicin, 100 mg L−1 gentamycin, and 100 mg L−1 spectinomycin) for selection of transformants. Cells were harvested from 50 mL of overnight culture by centrifugation at 4,000g for 10 min, resuspended in 20 to 25 mL of liquid callus induction medium containing 80 μm acetosyringone (Sigma), and incubated with shaking for 2 to 3 h to activate the A. tumefaciens virulence mechanism. The preconditioned hypocotyls were inoculated with A. tumefaciens for 30 min, transferred onto solid callus induction medium and cocultured for 48 h without antibiotic selection, and then transferred to callus induction medium with antibiotic selection (75 mg L−1 gentamycin or 15 mg −1L glufosinate-ammonium [basta]). Resistant callus typically appeared in 2 weeks, after which the segments with green callus were transferred to organogenesis medium containing MS salts, B5 vitamins, 4 mg L−1 6-benzyl-aminopurine, 2 mg L−1 zeatin, 5 mg L−1 silver nitrate, 3% (w/v) Suc, and 2 g L−1 Gelrite, pH 5.8, maintaining the antibiotic selection. After 2 weeks, the organogenic calli were transferred onto fresh medium containing reduced (2 mg L−1) 6-benzyl-aminopurine and lacking silver nitrate. Transgenic callus at this stage was used for circadian rhythm analysis. Transgenic calli were also maintained for several generations by subculturing every 2 weeks in the same culture medium with continuous selection. Under these conditions, few shoots are produced and none of the shoots develop roots.
Transformation Constructs
Firefly luciferase was driven from three Arabidopsis (Arabidopsis thaliana) clock gene promoters: pCCA1:LUC+ and pLHY:LUC+ in the pZPBAR vector (Salomé and McClung, 2005a) and pCCR2:LUC in pPZPC2LUC (Covington et al., 2001). TOC1 and ZTL were overexpressed using 35S:TOC1-YFP (Más et al., 2003a) and 35S:ZTL-GFP (Han et al., 2004).
Fluence Response Curves
Transgenic calli were entrained at 17°C in 12-h/12-h white light (provided with fluorescent lights at 70 μmol m−2 s−1)/dark cycles for 7 d before transfer to continuous red (670 nm) or blue (470 nm) light at indicated fluence rates in a Percival E30 LED color light chamber (Percival Scientific) or to indicated intensities of continuous white fluorescent light. On the 1st d in continuous conditions, calli were transferred onto Opti Plate-96 white microtiter plates (Perkin-Elmer) containing 200 μL of shoot regeneration medium plus 30 μL of 2.5 mm luciferin, and light production was measured with a Packard TopCount luminescence counter (Kim et al., 2010).
PRCs
For light pulse PRC experiments, pCCR2:LUC transgenic calli were entrained for 5 d at 17°C in 12-h/12-h light/dark cycles and then transferred to DD at 17°C. After 1 d in DD, calli were exposed to 30 min of red light (100 μmol m−2 s−1) or blue light (25 μmol m−2 s−1) at 3-h intervals and returned to continuous dark for luciferase measurement. For temperature pulse PRC experiments, pCCA1:LUC transgenic calli were entrained for 7 d at 16°C in 12-h/12-h light/dark cycles before transfer to LL at 16°C. After 1 d in LL, calli were subjected to a 3-h pulse of hot (22°C) or cold (10°C) at 3-h intervals. After the pulse, the calli were returned to LL at 16°C for luciferase measurement. Changes in acrophase (peak phase) of luciferase expression were calculated relative to callus not exposed to a light or temperature pulse. Phase advances and delays are shown as positive and negative values, respectively. The pooled se was calculated as described (Covington et al., 2001).
Data were assayed using the Biological Rhythms Analysis Software System (BRASS 2.1.4; www.amillar.org), which integrates the fast Fourier transform-nonlinear least squares analysis for circadian rhythms (Plautz et al., 1997). We considered luciferase expression in transgenic B. rapa calli to be rhythmic if period was between 20 and 30 h, the peak signal strength exceeded 100 photons callus−1 s−1, and the RAE, a measure of the strength of the rhythmicity, was less than 0.8 (RAE = 0 is an ideal cosine wave, RAE = 1 is the limit of statistical significance for any given rhythmic amplitude). At least 60 individual and independent transgenic calli were used for each analysis.
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: CCA1 (At2g46830), CCR2 (At2g21660), CHE (At5g08330), GI (At1g22770), LHY (At1g01060), PRR5 (At5g24470), PRR7 (At5g02810), PRR9 (At2g46790), TOC1 (At5g61380), and ZTL (At5g57360).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. B. rapa tissue culture.
Supplemental Table S1. Circadian period in B. rapa seedlings and tissue culture.
Supplementary Material
Acknowledgments
We thank L. Østergaard, C. Goldsack, and J. Irwin for helpful discussions on the transformation of B. rapa. We thank A.M. Citro and R.A. McConnell for assistance with B. rapa tissue culture. We thank S.A. Kay for pCCR2:LUC and D.E. Somers for the TOC1 and ZTL overexpression constructs and for the ZTL-specific antibody. We thank J. Hayes for keeping the TopCount running.
References
- Akasaka-Kennedy Y, Yoshida H, Takahata Y. (2005) Efficient plant regeneration from leaves of rapeseed (Brassica napus L.): the influence of AgNO3 and genotype. Plant Cell Rep 24: 649–654 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA. (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12: 757–761 [DOI] [PubMed] [Google Scholar]
- Arumugam N, Gupta V, Jagannath A, Mukhopadhyay A, Pradhan AK, Burma PK, Pental D. (2007) A passage through in vitro culture leads to efficient production of marker-free transgenic plants in Brassica juncea using the Cre-loxP system. Transgenic Res 16: 703–712 [DOI] [PubMed] [Google Scholar]
- Aschoff J. (1979) Circadian rhythms: influences of internal and external factors on the period measured in constant conditions. Z Tierpsychol 49: 225–249 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla PL, Singh MB. (2008) Agrobacterium-mediated transformation of Brassica napus and Brassica oleracea. Nat Protoc 3: 181–189 [DOI] [PubMed] [Google Scholar]
- Boxall SF, Foster JM, Bohnert HJ, Cushman JC, Nimmo HG, Hartwell J. (2005) Conservation and divergence of circadian clock operation in a stress-inducible Crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiol 137: 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce VG. (1960) Environmental entrainment of circadian rhythms. Cold Spring Harb Symp Quant Biol 25: 29–48 [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE. (1994) Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol 104: 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David KM, Armbruster U, Tama N, Putterill J. (2006) Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett 580: 1193–1197 [DOI] [PubMed] [Google Scholar]
- De Block M, De Brouwer D, Tenning P. (1989) Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression of the bar and neo genes in the transgenic plants. Plant Physiol 91: 694–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle W. (1951) Tagesperiodische Wachstumsschwankungen und Turgorschwankungen an Gewebekulturen. Planta 39: 570–588 [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Farré EM, Kay SA. (2007) PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J 52: 548–560 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE. (2008) Post-translational regulation of the circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. (2007) Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–148 [DOI] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE. (2004) Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J 40: 291–301 [DOI] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA. (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F, Imaizumi T, Gray WM. (2008) CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J 55: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez-Luy FL, Lukens L, Farnham MW, Amasino RM, Osborn TC. (2009) Development of public immortal mapping populations, molecular markers and linkage maps for rapid cycling Brassica rapa and B. oleracea. Theor Appl Genet 119: 31–43 [DOI] [PubMed] [Google Scholar]
- Ito S, Nakamichi N, Kiba T, Yamashino T, Mizuno T. (2007) Rhythmic and light-inducible appearance of clock-associated pseudo-response regulator protein PRR9 through programmed degradation in the dark in Arabidopsis thaliana. Plant Cell Physiol 48: 1644–1651 [DOI] [PubMed] [Google Scholar]
- Johnson CH. (1990) An Atlas of Phase Response Curves for Circadian and Circatidal Rhythms. Vanderbilt University, Nashville, TN [Google Scholar]
- Johnson CH. (1999) Forty years of PRCs: what have we learned? Chronobiol Int 16: 711–743 [DOI] [PubMed] [Google Scholar]
- Johnson CH, Elliott J, Foster R, Honma KI, Kronauer R. (2004) Fundamental properties of circadian rhythms. Dunlap JC, Loros JJ, DeCoursey P, , Chronobiology: Biological Timekeeping. Sinauer, Sunderland, MA, pp 67–105 [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH. (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by a SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Yang TJ, Kim JS, Park JY, Kwon SJ, Lim MH, Jin M, Lee SC, Lee SI, Choi B-S, et al. (2007) Isolation of circadian-associated genes in Brassica rapa by comparative genomics with Arabidopsis thaliana. Mol Cells 23: 145–153 [PubMed] [Google Scholar]
- Kim WY, Geng R, Somers DE. (2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci USA 100: 4933–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Salomé PA, Fujiwara S, Somers DE, McClung CR. (2010) Characterization of pseudo-response regulators in plants. Methods Enzymol 471: 359–380 [DOI] [PubMed] [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T. (2001) Molecular systematics of the Brassicaceae: evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot 88: 534–544 [PubMed] [Google Scholar]
- Locke JCW, Kozma-Bognár L, Gould PD, Fehér B, Kevei É, Nagy F, Turner MS, Hall A, Millar AJ. (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ. (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Sys Biol 1: 0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA. (2003a) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA. (2003b) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- McClung CR. (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. (2008) Comes a time. Curr Opin Plant Biol 11: 514–520 [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR. (2002) Phase-specific circadian clock regulatory elements in Arabidopsis thaliana. Plant Physiol 130: 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, McClung CR. (2003) Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA 100: 6878–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G. (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nakamichi N. (2005) Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol 46: 677–685 [DOI] [PubMed] [Google Scholar]
- Murakami M, Tago Y, Yamashino T, Mizuno T. (2007) Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol 48: 110–121 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Ito S, Oyama T, Yamashino T, Kondo T, Mizuno T. (2004) Characterization of plant circadian rhythms by employing Arabidopsis cultured cells with bioluminescence reporters. Plant Cell Physiol 45: 57–67 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. (2010) PSEUDO-RESPONSE REGULATORS 9, 7 and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T. (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46: 686–698 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Matsushika A, Yamashino T, Mizuno T. (2003) Cell autonomous circadian waves of the APRR1/TOC1 quintet in an established cell line of Arabidopsis thaliana. Plant Cell Physiol 44: 360–365 [DOI] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai K, Okamoto K, Nishimoto H, Morioka C, Hirano M, Kami-ike N, Ishiura M. (2004) Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput real-time bioluminescence monitoring system. Plant J 40: 1–11 [DOI] [PubMed] [Google Scholar]
- O'Neill C, Bancroft I. (2000) Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J 23: 233–243 [DOI] [PubMed] [Google Scholar]
- Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA. (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. (2009) A functional genomics approach reveals CHE as a novel component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Perez-Solis E, Ibanez C, Casado R, Collada C, Gomez L, Aragoncillo C, Allona I. (2005) Winter disruption of the circadian clock in chestnut. Proc Natl Acad Sci USA 102: 7037–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana D, Van Den Boogaart T, O'Neill CM, Hynes L, Bent E, MacPherson L, Park JY, Lim YP, Bancroft I. (2004) Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J 40: 725–733 [DOI] [PubMed] [Google Scholar]
- Salathia N, Lynn JR, Millar AJ, King GJ. (2007) Detection and resolution of genetic loci affecting circadian period in Brassica oleracea. Theor Appl Genet 114: 683–692 [DOI] [PubMed] [Google Scholar]
- Salomé PA, McClung CR. (2005a) PRR7 and PRR9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, McClung CR. (2005b) What makes Arabidopsis tick: light and temperature entrainment of the circadian clock. Plant Cell Environ 28: 21–38 [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Serikawa M, Miwa K, Kondo T, Oyama T. (2008) Functional conservation of clock-related genes in flowering plants: overexpression and RNAi analyses of the circadian rhythm in the monocotyledon Lemna gibba. Plant Physiol 146: 1952–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R. (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA. (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494 [DOI] [PubMed] [Google Scholar]
- Song HR, Carré IA. (2005) DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol Biol 57: 761–771 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Thines B, Harmon FG. (2010) Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, et al. (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7: 389–452 [Google Scholar]
- Wang ZY, Tobin EM. (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wijnen H, Young MW. (2006) Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet 40: 409–448 [DOI] [PubMed] [Google Scholar]
- Wilkins MB, Holowinsky AW. (1965) The occurrence of an endogenous circadian rhythm in a plant tissue culture. Plant Physiol 40: 907–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang X, Zhao H, Liu F. (2008) An intensive understanding of vacuum infiltration transformation of pakchoi (Brassica rapa ssp. chinensis). Plant Cell Rep 27: 1369–1376 [DOI] [PubMed] [Google Scholar]
- Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi BS, Kim JA, Jin M, Park JY, Lim MH, Kim HI, et al. (2006) Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi S, Green RM. (2009) Evidence for the adaptive significance of circadian rhythms. Ecol Lett 12: 970–981 [DOI] [PubMed] [Google Scholar]
- Zhang FL, Takahata Y, Watanabe M, Xu JB. (2000) Agrobacterium-mediated transformation of cotyledonary explants of Chinese cabbage (Brassica campestris L. ssp pekinensis). Plant Cell Rep 19: 569–575 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.