Abstract
Fibroblast growth factor 5 (FGF5) is widely expressed in embryonic but scarcely in adult tissues. Here we report simultaneous overexpression of FGF5 and its predominant high-affinity receptor (FGFR1 IIIc) in astrocytic brain tumour specimens (N=49) and cell cultures (N=49). The levels of both ligand and receptor increased with enhanced malignancy in vivo and in vitro. Furthermore, secreted FGF5 protein was generally present in the supernatants of glioblastoma (GBM) cells. siRNA-mediated FGF5 down-modulation reduced moderately but significantly GBM cell proliferation while recombinant FGF5 (rFGF5) increased this parameter preferentially in cell lines with low endogenous expression levels. Apoptosis induction by prolonged serum starvation was significantly prevented by rFGF5. Moreover, tumour cell migration was distinctly stimulated by rFGF5 but attenuated by FGF5 siRNA. Blockade of FGFR1-mediated signals by pharmacological FGFR inhibitors or a dominant-negative FGFR1 IIIc protein inhibited GBM cell proliferation and/or induced apoptotic cell death. Moreover, rFGF5 and supernatants of highly FGF5-positive GBM cell lines specifically stimulated proliferation, migration and tube formation of human umbilical vein endothelial cells. In summary, we demonstrate for the first time that FGF5 contributes to the malignant progression of human astrocytic brain tumours by both autocrine and paracrine effects.
Keywords: FGF5, FGF receptor 1, astrocytic brain tumours, cell growth and migration, autocrine and paracrine activities
Introduction
Fibroblast growth factors (FGFs) represent a family of homologous heparin-binding polypeptides consisting a family of 22 members (McKeehan et al., 1998). FGFs regulate growth, differentiation, survival and migration of a wide variety of cell types and have been implicated in the development of several human neoplasms (Eswarakumar et al., 2005; Grose and Dickson, 2005) including human primary brain tumours (Jensen, 1998; Todo et al., 1998). FGF-mediated signals are predominantly transduced by interaction with four transmembrane receptor tyrosine kinases (FGFR1–4). Based on alternative mRNA splicing, FGFR1–3 are translated into a variety of different receptor molecules with altered ligand-binding specificities (Eswarakumar et al., 2005; Grose and Dickson, 2005).
With respect to oncogenic activities, most studies have been performed on acidic and basic FGF (FGF1 and FGF2 respectively) and the known proto-oncogenes FGF3 (INT-2) and FGF4 (HST-1) (Grose and Dickson, 2005). However, also other FGF family members as for example FGF7 (keratinocyte growth factor), FGF8, FGF9, FGF10 and FGF18 have been linked to the development and/or progression of several tumour types (Galzie et al., 1997; McKeehan et al., 1998; Grose and Dickson, 2005).
FGF5 was originally identified by Zhan et al. (1988) based on a screening approach for transforming oncogenes. Subsequently, FGF5 was characterized as a regulator of the hair growth cycle (Hebert et al., 1994; Suzuki et al., 1998, 2000; Taniguchi et al., 2003) leading to an angora phenotype in knockout mice (Sundberg et al., 1997). FGF5 serves as a muscle-derived trophic factor for motoneurons in the peripheral nervous system (Hughes et al., 1993) and as a regulator of neuron differentiation and survival in the brain (Lindholm et al., 1994). Moreover, FGF5 affects astroglial properties in vitro (Reuss et al., 2000), and shows neurotrophic activity in vivo (Ozawa et al., 1998). While murine FGF5 is widely expressed during embryogenesis, it is in large measure restricted to the central nervous system in adult mice (Haub et al., 1990; Haub and Goldfarb, 1991). Human and murine FGF5 genes contain consensus sequences for secretion and their translation is stringently controlled by several upstream AUGs (Bates et al., 1991).
Malignant gliomas, including the most devastating type glioblastoma (GBM), are characterized by local growth and aggressive infiltration of the normal brain. Consequently, GBM is lethal within 1 year in the majority of patients. Several growth factors and their respective receptors have been implicated in GBM development and progression including most prominently the epidermal growth factor receptor and the platelet-derived growth factor receptor (Puputti et al., 2006). With regard to the FGF/FGFR system, a correlation between expression patterns of FGF2 and FGF9 as well as FGFR1 and FGFR4 and enhanced malignancy of astrocytic brain tumours has been described (Morrison et al., 1994b; Jensen, 1998; Todo et al., 1998; Yamada et al., 2002). In this study we demonstrate that FGF5 is frequently overexpressed in human astrocytic brain tumours correlating with increasing malignancy and suggest that FGF5 exerts autocrine and paracrine pro-tumourigenic functions especially in human GBM.
Results
Overexpression of FGF5 in astrocytic brain tumour tissues and cell cultures
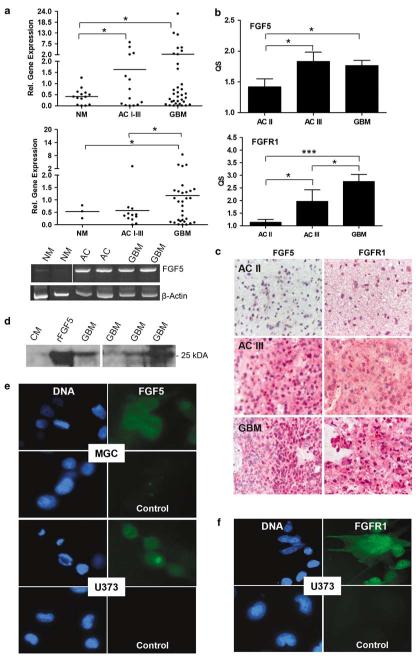
mRNA was obtained from tissue specimen and cell cultures of human astrocytic brain tumours and non-malignant brain samples and analysed for FGF5 mRNA expression by quantitative real-time RT–PCR. FGF5 mRNA levels were found to be significantly higher in astrocytomas (N=14) and GBM (N=35) tissue specimens as compared to non-malignant brain (epilepsy, N=10; schizophrenia, N=5; Figure 1a, upper panel). Correspondingly, FGF5 mRNA was generally expressed in cell cultures established from astrocytic brain tumours (representative reverse transcription (RT)–PCR examples are shown in Figure 1a, lower panel). Despite a high variability in the expression levels, GBM-derived cell cultures (N=37) contained significantly more FGF5 mRNA as compared to non-malignant brain (N=3) as well as astrocytoma-derived cell cultures (N=12; Figure 1a, middle panel).
Figure 1.
Expression of fibroblast growth factor 5 (FGF5) and FGFR1 in astrocytic brain tumour specimens and cell cultures. (a) FGF5 mRNA expression in tissue specimens (upper panel) and cell cultures (middle panel) was quantified by real-time RT-PCR. FGF5 mRNA expression was normalized to the housekeeping gene β-2 microglobulin. Data are given relative to an internal control (the GBM cell line MGC) included in all experiments and set arbitrarily as 1. Means are indicated by horizontal bars. (Lower panel) Representative examples of FGF5 and β-actin reverse transcription (RT)–PCR products amplified from the indicated tissue specimens are shown. NM, non-malignant brain; AC II, low-grade astrocytoma; AC III, anaplastic astrocytoma; GBM, glioblastoma. (b) Formalin-fixed, paraffin-embedded tissue specimens from AC II, AC III and GBM were immunostained using FGF5- and FGFR1-specific antibodies. Staining was evaluated as described under ‘Materials and methods’ and results given as quantitative scores (QS, means±s.d.) of all samples analysed. Significant differences are indicated (Student’s t-test, *P<0.05; ***P<0.0001). (c) Examples of FGF5 and FGFR1 immunohistochemical stainings in the indicated tissue specimens are shown. (d) FGF5 secretion by selected GBM cell lines was investigated by western blots. Serum-free culture medium (CM) and medium spiked with 150 ng ml−1 rFGF5 (rFGF5) were used as controls. (e, f) Immunofluorescence staining of FGF5 (e) and FGFR1 (f) was performed in the indicated GBM cell lines grown on chamber slides. DAPI (46-diamidino-2-phenyl indole) was used for staining of DNA. For control purposes, primary antibodies were either omitted (control panels) or replaced by isotype-specific unrelated control antibodies (data not shown).
FGF5 and FGFR1 protein expression was detected by immunohistochemistry in formalin-fixed tissue sections from astrocytoma and GBM. Representative examples of FGF5 and FGFR1 immunostainings in astrocytoma II, III and GBM are shown in Figure 1c and the mean quantitative score (QS)±s.d. for all investigated samples is given in Figure 1b. While in the non-malignant brain FGF5 express is restricted to the neurons (Haub et al., 1990), tumour cells emerged as the predominantly stained cell compartment with increasing malignancy. In all cases endothelial cells of brain capillaries stained positively for FGF5 and were used as internal references (set as 1). Enhanced levels of FGF5 were found in anaplastic astrocytomas (AC III) and GBM as compared to low-grade astrocytomas (AC II). While comparable amounts of FGF5 were found in AC III and GBM, FGFR1 expression continuously increased throughout malignant progression (Figure 1b) corresponding to data obtained by an earlier study (Morrison et al., 1994b).
With regard to GBM cell cultures, differing amounts of secreted FGF5 were detected in culture supernatants by western blot (Figure 1d). In contrast, FGF5 was hardly found in total cellular protein extracts (data not shown) arguing for a rapid secretion of the growth factor. FGF5 and FGFR1 proteins in tumour cell lines were also detected by immunofluorescence staining. Relatively weak but specific FGF5 immunoreactivity was found in the cytoplasm of both U373 and MGC cells (Figure 1e) while FGFR1 was readily detectable on the surface of U373 cells (Figure 1f).
Growth stimulation by exogenous FGF5
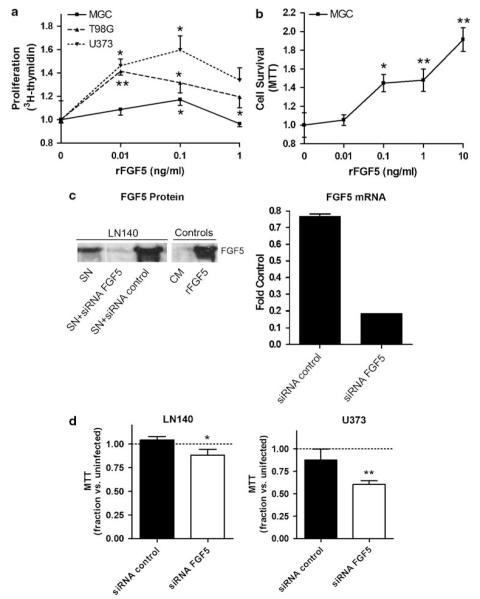
To investigate the effects of exogenous FGF5 (rFGF5) on the growth of GBM cells expressing relatively low (T98G, U373) or high (MGC) FGF5 mRNA levels, the recombinant growth factor was added to short-term serum-starved cell cultures. After 72 h, the level of DNA synthesis was measured by [3H]-thymidine incorporation assay. rFGF5 significantly stimulated GBM cell proliferation in all three cell lines with different potency (Figure 2a). While the effect was only weak in MGC cells producing high levels of endogenous FGF5, DNA synthesis was distinctly enhanced in T98G and U373 cells producing comparably low endogenous FGF5 expression.
Figure 2.
Effects of fibroblast growth factor 5 (FGF5) on growth and survival of human glioblastoma (GBM) cells. (a) Mitogenic effects of FGF5 were determined by treating the indicated GBM cell lines (after 48 h of serum starvation) for 72 h with increasing concentrations of rFGF5. DNA synthesis was measured by [3H]-thymidine incorporation assays and data are given relatively to serum-starved controls. (b) Cell death rescue effects of exogenous FGF5 were determined in MGC cells serum starved for 8 days (leading to around 50% cell death in the control group). The impact of increasing concentrations of rFGF5 on cell survival was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and data are given relatively to the control group without growth factor addition. (c) Downmodulation of endogenous FGF5 expression was performed using siRNA oligonucleotides as described in ‘Materials and methods’ and efficacy was determined after 72 h by western blot (left panel) and real-time PCR (right panel). Data for LN140 cells are shown representatively. (d) The indicated GBM cell lines were transfected with FGF5 or control siRNA, cultured for 72 h and analysed by MTT assay. Data are given relative to uninfected controls. Experiments were performed at least three times in triplicates. Significant differences are indicated as follows: Student’s t-test, *P<0.05; **P<0.01
Cell death rescue by exogenous FGF5
To determine the consequences of FGF5 on cell death induction by prolonged serum starvation, GBM cells were maintained in medium without fetal calf serum (FCS) for an extended period of time (6–8 days) leading to significant induction of apoptosis (around 50% of cells in the control group; data not shown). The impact of rFGF5 on this process was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The number of cells surviving long-term starvation was significantly increased by the presence of rFGF5 as representatively shown for MGC cells in Figure 2b.
Impact of endogenous FGF5 on tumour cell proliferation
To analyse the autocrine contribution of FGF5 to GBM cell growth, FGF5 siRNA oligonucleotides were used. Figure 2c demonstrates the potent siRNA-mediated downregulation of FGF5 expression at the level of mRNA (right panel) and protein (left panel) representatively for LN140 cells. Downmodulation of FGF5 resulted in moderately but significantly reduced numbers of viable cells following a 72 h exposure in both GBM cell lines tested (Figure 2d).
Impact of FGF5 on tumour cell migration
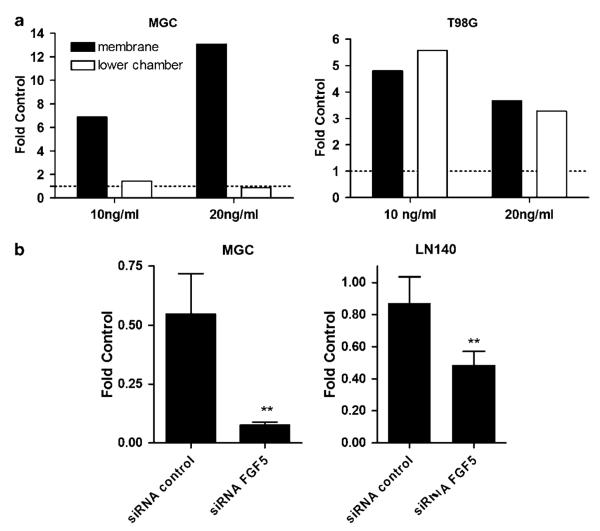
The impact of recombinant and endogenous FGF5 on GBM cell migration was tested by transwell chamber assays. Two GBM cell lines were treated under serum-reduced conditions (1% FCS) with rFGF5 for 12 h. Figure 3a depicts the number of those cells having migrated through the pores to the lower side of the membrane or the lower chamber. In the case of MGC cells, rFGF5 induced a dose-dependent increase of migration through the membrane (up to 14-fold; Figure 3a, left panel). Generally, only a few MGC cells managed to move through the culture medium to the lower chamber. On the contrary, T98G cells were able to cross the membrane, float through the medium and re-attach to the bottom of the lower chamber. Both processes were significantly stimulated by the presence of FGF5 (Figure 3a, right panel).
Figure 3.
Effects of fibroblast growth factor 5 (FGF5) on the migratory potential of human glioblastoma (GBM) cells. (a) GBM cell migration in culture medium containing 1% fetal calf serum (FCS) without and with rFGF5 at the indicated concentrations was studied by transwell chamber assays. After a migration period of 12 h, cells at the lower side of the membranes and in the lower chambers were fixed and counted microscopically. Data are given relative to the 1% FCS control without FGF5 set as 1. At least two experiments were performed delivering comparable results. (b) Migration ability of GBM cells treated with FGF5 or control siRNA was determined as described under (a). Data are shown relative to uninfected controls. Three experiments were performed and significant differences are indicated as follows: Student’s t-test, **P<0.01.
To investigate the impact of endogenous FGF5 the migratory potential of GBM cells, we knocked down FGF5 mRNA by siRNA. In both FGF5-positive cell lines tested, FGF5 inhibition significantly decreased the ability to migrate through the micropore membrane as compared to cells treated with control siRNA (Figure 3b).
Blockade of FGFR-mediated signals in GBM cells
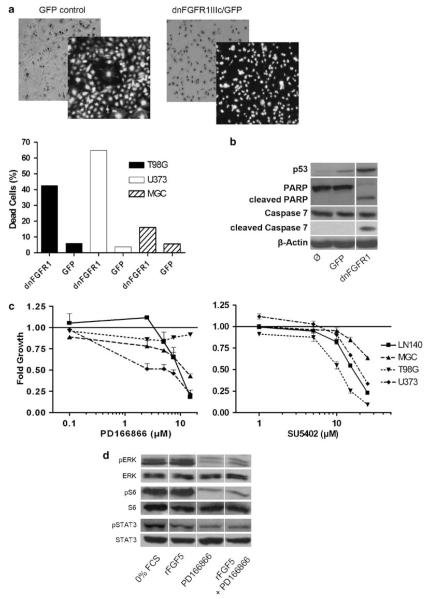
FGF5 signals are transduced predominantly through the FGFR1 receptor splice variant IIIc (Ornitz et al., 1996). Expression of a dominant-negative (dn)FGFR1 IIIc/GFP (green fluorescent protein) adenoviral construct caused specific blockade of FGFR1-mediated signals (data not shown). Infection with dnFGFR1 IIIc/GFP virus resulted in distinct apoptosis induction within 48–72 h in all GBM cell lines investigated (photomicrographs for U373 are representatively shown in Figure 4a, upper panel). U373 and T98G cells were significantly more sensitive than MGC cells (Figure 4a, lower panel). FGFR signal blockade and subsequent cell death induction were accompanied by upregulation of p53 and cleavage of caspase 7 as well as the caspase substrate poly(ADP-ribosyl)polymerase (PARP; Figure 4b).
Figure 4.
Blockade of FGFR1-mediated signal transduction in glioblastoma (GBM) cells. (a) Effects of a dnFGFR1 IIIc/GFP adenovirus on the survival of GBM cell lines were evaluated microscopically. Phase-contrast and fluorescence photomicrographs of U373 cells infected with the dnFGFR1 IIIc/GFP as compared to the green fluorescent protein (GFP) control adenovirus are shown representatively (upper panel). The number of dead cells induced by infection with the indicated adenoviruses was determined after 48 h by Trypan blue exclusion assay (lower panel). (b) The impact of infection with the indicated adenoviruses on p53 expression and caspase substrate cleavage (poly(ADP-ribosyl)polymerase (PARP), caspase 7) is representatively shown for U373 cells. Proteins were isolated 48 h after infection. (c) Effects of a 72 h exposure to the specific FGFR1 inhibitor PD166866 (left panel) and the general FGFR inhibitor SU5402 (right panel) on viability of GBM cell lines were determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay performed at least three times in triplicates. (d) The impact of PD166866 on phosphorylation of extracellular signal-regulated kinase (ERK), S6, and STAT3 in serum-starved MGC cells stimulated with recombinant fibroblast growth factor 5 (rFGF5, 30 min) was determined by western blot.
Additionally, the effects of two pharmacological FGFR inhibitors on GBM cell proliferation were tested by MTT assays. PD166866, an FGFR1 inhibitor (Panek et al., 1998), and the general FGFR inhibitor SU5402 reduced significantly and dose dependently growth of almost all GBM cell lines tested (Figure 4c).
Additionally, the impact of the FGFR1-specific inhibitor on the activity of the mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K) and JAK/STAT pathways was investigated under serum-starved conditions. Residual phosphorylations of extracellular signal-regulated kinase (ERK), S6 and STAT3 after 48 h serum starvation were distinctly reduced in the presence of the FGFR1 inhibitor. Concomitant treatment with rFGF5 was not able to recover downstream molecule phosphorylation in the presence of PD166866 (Figure 4d).
Paracrine, neoangiogenic effects of FGF5
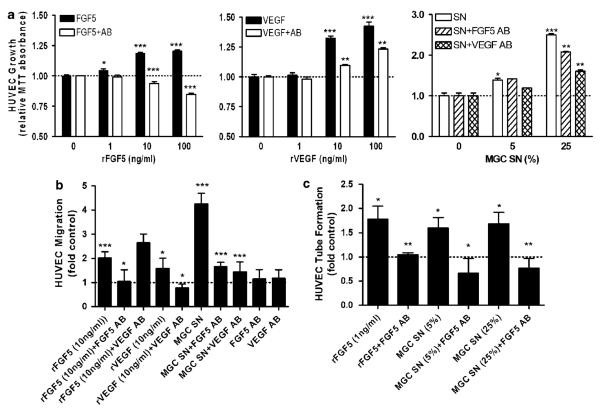
To analyse whether FGF5 exerts paracrine effects on endothelial cells, rFGF5 or conditioned media of FGF-expressing GBM cell lines were added to human umbilical vein endothelial cells (HUVEC). FGF5 exerted a dose-dependent, proliferative effect on endothelial cells (Figure 5a, left panel), which was slightly weaker than the one exerted by the known proangiogenic growth factor VEGF (vascular endothelial growth factor; Figure 5a, middle panel). Co-administration of neutralizing FGF5 or VEGF antibodies led to a significant attenuation of the respective growth-promoting effects. Interestingly, the FGF5 antibody reduced HUVEC proliferation even below control levels suggesting a contribution of endogenously produced FGF5. To directly demonstrate angiogenic activity of tumour-derived FGF5, culture supernatants obtained from the highly FGF5-expressing GBM cell line MGC were used. Addition of MGC-conditioned medium significantly stimulated growth of HUVEC (Figure 5a, right panel). Concomitant exposure to a neutralizing FGF5 antibody significantly decreased the pro-proliferative activity as did a VEGF-neutralizing antibody.
Figure 5.
Impact of recombinant and glioblastoma (GBM) cell-derived fibroblast growth factor 5 (FGF5) on human umbilical vein endothelial cells (HUVEC). (a) Effects of rFGF5 (left panel) as compared to rVEGF (middle panel) at the indicated concentrations and the respective neutralizing antibodies (Abs) on HUVEC growth were measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Additionally, HUVEC were exposed to culture supernatants (SN) of highly FGF5-positive MGC cells with and without antibody for FGF5 and vascular endothelial growth factor (VEGF, right panel). (b) The effects of FGF5, VEGF and MGC-conditioned medium (24 h exposure) with and without blocking antibodies on HUVEC migration were studied by micropore transwell chamber assays. To investigate a possible cross-reactivity of the neutralizing antibody, rFGF5 was combined with VEGF antibody. Cells at the lower side of the membranes were counted and data are given relative to the control (1% fetal calf serum, FCS). (c) HUVEC were suspended in matrigel and tube formation was induced with rFGF5 as well as with SN of MGC cells at the indicated dilutions with and without FGF5 blocking antibody. Tube formation was quantified as described in ‘Materials and methods’ and means±s.d. of at least three experiments are given. Significant differences to controls are indicated as follows: Student’s t-test, *P<0.05; **P<0.01; ***P<0.0001. In case of the antibody-containing groups ( + Ab) in panels a–c, the statistical comparison was performed to the respective groups without antibody.
To investigate the impact of rFGF5 on the migration potency, HUVEC were treated with rFGF5, rVEGF or culture supernatants of MGC cells under serum-reduced conditions (1% FCS). Both, rFGF5 and rVEGF led to a significant increase of the migratory ability of HUVEC cells which could be specifically suppressed by the respective neutralizing antibodies (Figure 5b). Also MGC cell-derived condition medium significantly stimulated HUVEC migration up to about fourfold. This effect was markedly attenuated to <2-fold by a neutralizing antibody against FGF5 comparable to the VEGF antibody (Figure 5b).
To examine the impact of FGF5 on endothelial cell differentiation, a tube formation assay with HUVEC was performed (Figure 5c). rFGF5 increased tube lengths about twofold, which was comparable to rVEGF (data not shown). Conditioned media obtained from MGC cells had similar effects. Stimulating activities of both rFGFs and the GBM cell supernatant were completely inhibited by the neutralizing FGF5 antibody (Figure 5c).
Discussion
In this study we present strong evidence that FGF5 exerts oncogenic activities in astrocytic brain tumours by growth-, survival- and migration-promoting effects on tumour cells and by supporting neoangiogenesis. Although originally identified as a transforming proto-oncogene, the data on oncogenic functions of endogenously expressed FGF5 have been relatively limited so far. Increased expression of FGF5 was associated with occurrence of human pancreatic cancer (Kornmann et al., 1997, 2001) and FGF5 was identified as an overexpressed antigen in multiple human adenocarcinomas (Hanada et al., 2001). Also the predominant FGF5-binding FGFR1 variant IIIc (Reuss and von Bohlen und Halbach, 2003) was found to be upregulated in various cancer types (Grose and Dickson, 2005), thus generating the conditions for autocrine loops. These data together with our observations indicate that the oncogenic potential of FGF5 might have been under-estimated so far.
Although FGF5 overexpression in astrocytic brain tumours is reported for the first time in this study, several previous reports suggested oncogenic contributions of other FGF/FGFR molecules to this tumour type. For instance elevated levels of FGF1, FGF2 and FGFR1 were shown in human gliomas (Morrison et al., 1994a, b). Furthermore, FGF2 stimulated proliferation of GBM cells (Engebraaten et al., 1993). Similar effects have also been observed in case of FGF9, a factor frequently overexpressed in GBM (Todo et al., 1998). Several publications of the Stachowiak group have indicated an oncogenic function of nuclear FGF2 and FGFR1 molecules (reviewed in Stachowiak et al., 1996).
In the astrocytic brain tumour panel analysed in this study, FGF5 expression increased with malignant progression. Nevertheless, surgery samples and short-term cell cultures of non-malignant brain tissues were not completely devoid of FGF5 mRNA expression. This is in accordance with previous studies, demonstrating brain residence of FGF5 (Haub et al., 1990; Reuss and von Bohlen und Halbach, 2003). However in the normal rat brain, FGF5 mRNA (Gomez-Pinilla and Cotman, 1993) is primarily located in the neurons. Nevertheless, FGF5 functions not only as a regulator of neuron differentiation and survival (Lindholm et al., 1994), but also as a paracrine modulator of astroglial properties including gap junctions (Reuss et al., 2000) and expression of glial fibrillary acidic protein (Reuss et al., 2003). In contrast to normal astrocytes (Gomez-Pinilla and Cotman, 1993), we found strong FGF5 expression during malignant progression of astrocytes. The reason for activation of FGF5 expression in malignant astrocytes is currently speculative and the subject of ongoing investigations. An in silico analysis of the FGF5 promoter revealed presence of several binding sites for transcription factors activated in GBM including HIF1α and Smad3 (data not shown). FGF5 is frequently expressed in embryonic tissues and has been recently described as a stem cell marker (Pelton et al., 2002). Consequently, upregulation during malignant progression might reflect dedifferentiation and acquisition of stem cell-like properties.
rFGF5 simulated GBM cell growth only moderately as compared to stronger effects on migration and survival. Reciprocally, knockdown of endogenous FGF5 by siRNA resulted in significant but moderate growth inhibition in contrast to a severe impact on GBM cell migration. This suggests that autocrine FGF5 is predominantly a survival and migration factor for GBM cells. These observations correspond to a rapid activation of FGF5 expression in GBM cells following serum starvation (manuscript in preparation).
In addition to autocrine effects, we present evidence that FGF5 supports neoangiogenesis in astrocytic brain tumours by promoting growth, migration and differentiation of endothelial cells. GBM is a tumour entity characterized by extreme endothelial hyperproliferation (Kargiotis et al., 2006). Besides VEGF, PDGF and others, also FGF1 and FGF2 have been implicated in glioma neoangiogenesis (Kargiotis et al., 2006). Here we add FGF5 as an additional player contributing to the pro-angiogenic characteristics of GBM. Moreover, FGFR1 IIIc, representing the major FGF5 receptor, is the predominant FGFR splice variant in endothelial cells (Antoine et al., 2005). While FGF1 and FGF2 lack a signal peptide for secretion, FGF5 is readily secreted. This implicates that FGF5 might cooperate with other FGFs to support glioma tissue neoangiogenesis.
As our data and previous studies (Morrison et al., 1994a; Stachowiak et al., 1996) suggest a significant contribution of FGFR1-mediated signals to malignant growth of GBM, this receptor tyrosine kinase might represent a promising therapy target in GBM. Indeed, inhibition of FGFR by small molecule inhibitors attenuated growth and survival of GMB cell lines. As the specificity of small molecule inhibitors is generally questionable (Carter et al., 2005; Rix et al., 2007), we also performed experiments with a dominant-negative receptor molecule. Infection of GBM cells with dnFGFR1 IIIc/GFP adenovirus resulted in massive cell death accompanied by induction of p53 and cleavage of caspase substrates. The stronger effects of dnFGFR1 IIIc/GFP as compared to FGF5 siRNA can be well explained by the expression of additional FGFR ligands in GBM cells (Joy et al., 1997; Todo et al., 1998). Accordingly, the more potent apoptosis induction of the dnFGFR1 molecule as compared to small molecule inhibitors might be based on inhibition of ligand-independent functions of FGFR in cell adhesion (Cavallaro et al., 2001). Indeed, expression of dnFGFR1 IIIc/GFP induced rapid detachment of GBM cells from the culture plastic. Consequently, cells might die via anoikis, a specific form of apoptosis also induced by PI3K pathway blockade of GBM cells (Davies et al., 1998).
In summary, our data indicate that FGF5 is an oncogenic and neoangiogenic growth factor contributing to astrocytic brain tumour progression. Together with previous reports on oncogenic functions of other FGF/FGFR molecules (Morrison et al., 1994a; Stachowiak et al., 1996), our study strongly suggests this growth factor system as a promising target for therapeutic interventions in human GBM.
Materials and methods
Patient material
Tissue specimens were obtained from astrocytic brain tumour patients treated by surgery at the Wagner Jauregg Hospital, Linz. Additionally, non-malignant brain tissue samples from schizophrenia or epilepsy patients were derived from the Wagner Jauregg Hospital, Linz and the Department of Neurosurgery, Medical University, Vienna. All patients had given informed consent for this investigation.
Cell cultures
U373 and T98G GBM cells were obtained from the American Type Culture Collection (Manassas, VA, USA). MGC cells were kindly provided by Dr Kurata (Tokyo, Japan) and LN140 by Dr Tribolet (Lausanne, Switzerland). Additionally, cell cultures were established from 3 epileptic brain samples, 12 astrocytomas and 33 GBM surgery specimens as described (Spiegl-Kreinecker et al., 2007). HUVEC were isolated and cultured as described (Jaffe et al., 1973) (Cambrex, East Rutherford, NJ, USA). All cell cultures were periodically checked for Mycoplasma contamination (Mycoplasma Stain Kit; Sigma, St Louis, MO, USA).
Reagents
Recombinant FGF5 was obtained from Strathmann Biotec AG (Hamburg, Germany). The FGFR1-specific inhibitor PD166866 (Panek et al., 1998) was generously supplied by Pfizer Global Research and Development (Groton, CT, USA), the pan-FGFR inhibitor SU5402 obtained from Calbiochem (La Jolla, CA, USA). All other reagents were obtained from Sigma. The expression vector pcHisCtrFGFR, encoding a kinase-truncated dnFGFR1-IIIc (Zhang et al., 1998), was kindly provided by Dr Francis Kern (Georgetown University Medical Center, Washington, DC, USA). The truncated FGFR1 was tagged at the C terminus with enhanced green fluorescent protein (EGFP) by insertion into pEGFP-N3 (Clontech, Mountain View, CA, USA) to generate a dnFGFR1-IIIc/GFP protein chimera. The adenoviral expression vector was created using pAdEasy-1 (Stratagene, La Jolla, CA, USA) and shuttle vector pShuttle-cytomegalovirus (Stratagene). An EGFP adenoviral expression vector was used as control (Steiner et al., 2006). Virus titres were determined by Adeno-X Rapid Titer Kit (Clontech) and by GFP fluorescence-activated cell sorting (FACS) analyses (FACScalibur; BD Biosciences, Franklin Lakes, NJ, USA).
Real-time RT-PCR and RT–PCR
Total RNA was isolated using Trifast (PeqLab Biotechnologie, Erlangen, Germany) and cDNA synthesized as published (Berger et al., 1999). The expression of FGF5 and β-2 microglobulin mRNA was quantified by real-time RT-PCR using TaqMan assays (Applied Biosystems, Foster City, CA, USA). Quantification of gene expression was calculated by a standard curve method using β-2 microglobulin for normalization. Selected cDNA samples were additionally investigated by RT–PCR as described (Brachner et al., 2006). Oligonucleotide primer sequences were for FGF5 sense 5′-CCCGGATGGCAAAGTCAATGG-3′ and anti-sense 5′-TTCAGGGCAACATACCACTCCCG-3′. Amplifications were done for 40 and 35 cycles, respectively. Amplification of β-actin (sense 5′-CTCCTTAATGTCACGCACGATTTC-3′, anti-sense 5′-GTGGGGCGCCCCAGGCACCA-3′) was terminated after 25 cycles. Cycles consisted of 30 s denaturation at 94 °C, 50 s annealing at 60 °C and 40s extensionat 72 °C.
Immunohistochemistry and immunofluorescence
Tissue sections were prepared and immunostained as described previously (Berger et al., 2005) using goat polyclonal FGF5 (AF-237-NA; R&D Systems, Minneapolis, MN, USA) and rabbit polyclonal FGFR1 antibodies (sc-121; Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:100, both). Staining was evaluated independently by two of the authors (SA and WB). Tumour cell staining intensity was evaluated in relation to the endothelial cells (which stained weakly positive in all cases analysed) and graded: 0.5=below endothelial cells; 1=resembling endothelial cells; 2=stronger than endothelial cells and 3=very strong staining. In case of different subgroups of tumour cells with different grading, the percentage of the cells in the respective group was counted. For each specimen the QS was calculated by the equation: Σ(grading × respective percentage of cells)/100.
For immunofluorescence staining cells were grown in chamber slides and processed as published (Steiner et al., 2006). Fixation was 10 min in either cold acetone/methanol (1:1) or 1 h in 3.6% formalin/phosphate-buffered saline (PBS) followed by 0.5% Triton X/PBS (5 min) for FGFR1 and FGF5 respectively. Primary antibodies were used at dilution 1:100 and secondary immunoglobulin G (Sigma) at 1:250. In all cases, controls without or with isotype-specific control antibodies instead of the first antibody (Sigma) were used.
Cell proliferation and cell death rescue assays
Cells (4×103 per well) were seeded into 96-well plates. After 24 h, cells were serum-starved for 48 h followed by stimulation with rFGF5 for 3 days. At this time point cell death induction was in all cases below 5% as determined by Trypan blue exclusion. DNA synthesis was determined by [3H]-thymidine incorporation assay as published (Heffeter et al., 2006). For cell death rescue analysis, cells were serum starved with and without rFGF5 for an extended time period (6–8 days) until substantial cell death (around 50% dead cells determined by Trypan blue exclusion) occurred in the control group without FGF5. Cell viability was assessed by MTT assay (EZ4U; Biomedica, Vienna, Austria).
Protein isolation and western blotting
Cellular proteins were extracted and processed for western blotting as described (Steiner et al., 2006). For preparation of secreted proteins, cells were seeded at a density of 1×106 cells per 25 cm2 flask. After 24 h, growth medium was changed to serum-free conditions and 48 h later supernatants were collected. Proteins were precipitated by acetone and directly applied to SDS–polyacrylamide gel electrophoresis. Western blots were performed with the following antibodies: phospho-S6 ribosomal protein (Ser240/244), S6, phospho-p44/42 MAPK (Thr202/Tyr204), p44/42 MAPK, phospho-STAT3 (Tyr705), STAT3, PARP, cleaved caspase 7, caspase 7 (all polyclonal rabbit contained in the respective sampler kits from Cell Signaling Technology, Beverly, MA, USA); p53 monoclonal mouse DO-1 (Neomarkers, CA, USA); goat polyclonal FGF5 (AF-237-NA; R&D Systems), β-actin monoclonal mouse AC-15 (Sigma).
Migration assays
Migration assays were performed in transwell chambers using 8 μm pore-size PET track-etched membranes (BD Biosciences) in 24-well plates. Cells were seeded at a density of 1× 105 cells cm−2. After a migration period of 12 h for GBM cells and 24 h for HUVEC, cells on the lower membrane surface and on the bottom of the lower chamber were stained with crystal violet and counted microscopically.
Knockdown of FGF5 expression
To knock down endogenous FGF5 expression, 2×105 cells were seeded into 6-well plates. After 24 h, cells were transfected with 8.25 μlml−1 ExGen 500 transfection reagent (Fermentas, Burlington, Canada) containing 100 pmol of the most effective FGF5 siRNA oligonucleotide (for MGC cells: no. 2824, for LN140 cells: no. 146518; for U373 cells: no. 2824 and no. 146518 mixed at a 1:1 ratio, Ambion, Austin, TX, USA) per well in culture medium without FCS. GAPDH siRNA was used as a control. After 48 h total RNA and proteins were isolated.
Dominant-negative FGFR1 IIIc expression in GBM cells
Cells were seeded at a density of 2×105 per well into 6-well plates containing 10% FCS, followed by dnFGFR1 IIIc/GFP adenoviral infection at multiplicity of infections between 34 and 200. A respective GFP adenovirus was used as control at concentrations yielding comparable proportions of green fluorescent cells as determined by FACS analyses. Viable and dead cells were counted by Trypan blue exclusion assay 2 days after transfection.
Cell proliferation and tube formation assay with HUVEC
HUVEC were seeded into 96-well plates (2×103 cells per well) coated with human fibronectin (Chemicon, Hampshire, UK). After 24 h growth in standard culture medium, the cells were serum starved for 24 h, then stimulated with rFGF5, rVEGF or conditioned media obtained from GBM cell cultures. For neutralization of rFGF5 or tumour cell-derived FGF5 or VEGF in the supernatants, neutralizing antibodies (AF-237-NA for FGF5: 2 μgml−1, MAb293 for VEGF: 1 μgml−1; R&D Systems) were used. Live cells were assessed by MTT assays after 48 h exposure. For tube formation assays, HUVEC were serum starved for 48 h. Cells (1.5×104 cells per well in 75 μl starvation medium) were seeded into 96-well plates coated with 50 μl growth factor-reduced matrigel (BD Biosciences). rFGF5 (1 ng ml−1), rVEGF (1 ng ml−1) or conditioned media (20 μl) were added and after 6 h tube lengths were quantified from digitized photo-micrographs using ImageJ software (freeware, NIH, Bethesda, ML, USA).
Acknowledgements
We thank Ninon Taylor, Paracelsus Medical University Salzburg, for the dnFGFR1-IIIc adenoviral construct and for reading the paper. Moreover, we are thankful to Vera Bachinger, Christian Balcarek, Maria Eisenbauer and Heidelinde Cantonati for skilful technical assistance as well as Stoffl Mayer and Hedwig Sutterlüty for discussion of the data.This study was supported by Austrian Science Fund FWF, project number P17630-B12, P19920-B12 (to WB) and the Jubiläums-fonds des Bürgermeisters der Bundeshauptstadt Wien, project number 2569 (to CM).
References
- Antoine M, Wirz W, Tag CG, Mavituna M, Emans N, Korff T, et al. Expression pattern of fibroblast growth factors (FGFs), their receptors and antagonists in primary endothelial cells and vascular smooth muscle cells. Growth Factors. 2005;23:87–95. doi: 10.1080/08977190500096004. [DOI] [PubMed] [Google Scholar]
- Bates B, Hardin J, Zhan X, Drickamer K, Goldfarb M. Biosynthesis of human fibroblast growth factor-5. Mol Cell Biol. 1991;11:1840–1845. doi: 10.1128/mcb.11.4.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W, Setinek U, Hollaus P, Zidek T, Steiner E, Elbling L, et al. Multidrug resistance markers P-glycoprotein, multidrug resistance protein 1, and lung resistance protein in non-small cell lung cancer: prognostic implications. J Cancer Res Clin Oncol. 2005;131:355–363. doi: 10.1007/s00432-004-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W, Setinek U, Mohr T, Kindas-Mugge I, Vetterlein M, Dekan G, et al. Evidence for a role of FGF-2 and FGF receptors in the proliferation of non-small cell lung cancer cells. Int J Cancer. 1999;83:415–423. doi: 10.1002/(sici)1097-0215(19991029)83:3<415::aid-ijc19>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Brachner A, Sasgary S, Pirker C, Rodgarkia C, Mikula M, Mikulits W, et al. Telomerase- and alternative telomere lengthening-independent telomere stabilization in a metastasis-derived human non-small cell lung cancer cell line: effect of ectopic hTERT. Cancer Res. 2006;66:3584–3592. doi: 10.1158/0008-5472.CAN-05-2839. [DOI] [PubMed] [Google Scholar]
- Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro U, Niedermeyer J, Fuxa M, Christofori G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat Cell Biol. 2001;3:650–657. doi: 10.1038/35083041. [DOI] [PubMed] [Google Scholar]
- Davies MA, Lu Y, Sano T, Fang X, Tang P, LaPushin R, et al. Adenoviral transgene expression of MMAC/PTEN in human glioma cells inhibits Akt activation and induces anoikis. Cancer Res. 1998;58:5285–5290. [PubMed] [Google Scholar]
- Engebraaten O, Bjerkvig R, Pedersen PH, Laerum OD. Effects of EGF, bFGF, NGF and PDGF(bb) on cell proliferative, migratory and invasive capacities of human brain-tumour biopsies in vitro. Int J Cancer. 1993;53:209–214. doi: 10.1002/ijc.2910530206. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Galzie Z, Kinsella AR, Smith JA. Fibroblast growth factors and their receptors. Biochem Cell Biol. 1997;75:669–685. [PubMed] [Google Scholar]
- Gomez-Pinilla F, Cotman CW. Distribution of fibroblast growth factor 5 mRNA in the rat brain: an in situ hybridization study. Brain Res. 1993;606:79–86. doi: 10.1016/0006-8993(93)91572-a. [DOI] [PubMed] [Google Scholar]
- Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005;16:179–186. doi: 10.1016/j.cytogfr.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Hanada K, Perry-Lalley DM, Ohnmacht GA, Bettinotti MP, Yang JC. Identification of fibroblast growth factor-5 as an overexpressed antigen in multiple human adenocarcinomas. Cancer Res. 2001;61:5511–5516. [PubMed] [Google Scholar]
- Haub O, Drucker B, Goldfarb M. Expression of the murine fibroblast growth factor 5 gene in the adult central nervous system. Proc Natl Acad Sci USA. 1990;87:8022–8026. doi: 10.1073/pnas.87.20.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haub O, Goldfarb M. Expression of the fibroblast growth factor-5 gene in the mouse embryo. Development. 1991;112:397–406. doi: 10.1242/dev.112.2.397. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Rosenquist T, Gotz J, Martin GR. FGF5 as a regulator of the hair growth cycle: evidence from targeted and spontaneous mutations. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- Heffeter P, Jakupec MA, Korner W, Wild S, von Keyserlingk NG, Elbling L, et al. Anticancer activity of the lanthanum compound [tris(1, 10-phenanthroline)lanthanum(III)]trithiocyanate (KP772; FFC24) Biochem Pharmacol. 2006;71:426–440. doi: 10.1016/j.bcp.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Hughes RA, Sendtner M, Goldfarb M, Lindholm D, Thoenen H. Evidence that fibroblast growth factor 5 is a major muscle-derived survival factor for cultured spinal motoneurons. Neuron. 1993;10:369–377. doi: 10.1016/0896-6273(93)90327-n. [DOI] [PubMed] [Google Scholar]
- Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest. 1973;52:2745–2756. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RL. Growth factor-mediated angiogenesis in the malignant progression of glial tumors: a review. Surg Neurol. 1998;49:189–195. doi: 10.1016/s0090-3019(97)00218-8. [DOI] [PubMed] [Google Scholar]
- Joy A, Moffett J, Neary K, Mordechai E, Stachowiak EK, Coons S, et al. Nuclear accumulation of FGF-2 is associated with proliferation of human astrocytes and glioma cells. Oncogene. 1997;14:171–183. doi: 10.1038/sj.onc.1200823. [DOI] [PubMed] [Google Scholar]
- Kargiotis O, Rao JS, Kyritsis AP. Mechanisms of angiogenesis in gliomas. J Neurooncol. 2006;78:281–293. doi: 10.1007/s11060-005-9097-6. [DOI] [PubMed] [Google Scholar]
- Kornmann M, Ishiwata T, Beger HG, Korc M. Fibroblast growth factor-5 stimulates mitogenic signaling and is overexpressed in human pancreatic cancer: evidence for autocrine and paracrine actions. Oncogene. 1997;15:1417–1424. doi: 10.1038/sj.onc.1201307. [DOI] [PubMed] [Google Scholar]
- Kornmann M, Lopez ME, Beger HG, Korc M. Expression of the IIIc variant of FGF receptor-1 confers mitogenic responsiveness to heparin and FGF-5 in TAKA-1 pancreatic ductal cells. Int J Pancreatol. 2001;29:85–92. doi: 10.1385/IJGC:29:2:085. [DOI] [PubMed] [Google Scholar]
- Lindholm D, Harikka J, da Penha Berzaghi M, Castren E, Tzimagiorgis G, Hughes RA, et al. Fibroblast growth factor-5 promotes differentiation of cultured rat septal cholinergic and raphe serotonergic neurons: comparison with the effects of neurotrophins. Eur J Neurosci. 1994;6:244–252. doi: 10.1111/j.1460-9568.1994.tb00267.x. [DOI] [PubMed] [Google Scholar]
- McKeehan WL, Wang F, Kan M. The heparan sulfate-fibroblast growth factor family: diversity of structure and function. Prog Nucleic Acid Res Mol Biol. 1998;59:135–176. doi: 10.1016/s0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- Morrison RS, Yamaguchi F, Bruner JM, Tang M, McKeehan W, Berger MS. Fibroblast growth factor receptor gene expression and immunoreactivity are elevated in human glioblastoma multiforme. Cancer Res. 1994a;54:2794–2799. [PubMed] [Google Scholar]
- Morrison RS, Yamaguchi F, Saya H, Bruner JM, Yahanda AM, Donehower LA, et al. Basic fibroblast growth factor and fibroblast growth factor receptor I are implicated in the growth of human astrocytomas. J Neurooncol. 1994b;18:207–216. doi: 10.1007/BF01328955. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Suzuki S, Asada M, Tomooka Y, Li AJ, Yoneda A, et al. An alternatively spliced fibroblast growth factor (FGF)-5 mRNA is abundant in brain and translates into a partial agonist/antagonist for FGF-5 neurotrophic activity. J Biol Chem. 1998;273:29262–29271. doi: 10.1074/jbc.273.44.29262. [DOI] [PubMed] [Google Scholar]
- Panek RL, Lu GH, Dahring TK, Batley BL, Connolly C, Hamby JM, et al. In vitro biological characterization and antiangiogenic effects of PD 166866, a selective inhibitor of the FGF-1 receptor tyrosine kinase. J Pharmacol Exp Ther. 1998;286:569–577. [PubMed] [Google Scholar]
- Pelton TA, Sharma S, Schulz TC, Rathjen J, Rathjen PD. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J Cell Sci. 2002;115:329–339. doi: 10.1242/jcs.115.2.329. [DOI] [PubMed] [Google Scholar]
- Puputti M, Tynninen O, Sihto H, Blom T, Maenpaa H, Isola J, et al. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in gliomas. Mol Cancer Res. 2006;4:927–934. doi: 10.1158/1541-7786.MCR-06-0085. [DOI] [PubMed] [Google Scholar]
- Reuss B, Dono R, Unsicker K. Functions of fibroblast growth factor (FGF)-2 and FGF-5 in astroglial differentiation and blood–brain barrier permeability: evidence from mouse mutants. J Neurosci. 2003;23:6404–6412. doi: 10.1523/JNEUROSCI.23-16-06404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss B, Hertel M, Werner S, Unsicker K. Fibroblast growth factors-5 and -9 distinctly regulate expression and function of the gap junction protein connexin 43 in cultured astroglial cells from different brain regions. Glia. 2000;30:231–241. [PubMed] [Google Scholar]
- Reuss B, von Bohlen und Halbach O. Fibroblast growth factors and their receptors in the central nervous system. Cell Tissue Res. 2003;313:139–157. doi: 10.1007/s00441-003-0756-7. [DOI] [PubMed] [Google Scholar]
- Rix U, Hantschel O, Durnberger G, Rix LL Remsing, Planyavsky M, Fernbach NV, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib and dasatinib reveal novel kinase and non-kinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- Spiegl-Kreinecker S, Pirker C, Marosi C, Buchroithner J, Pichler J, Silye R, et al. Dynamics of chemosensitivity and chromosomal instability in recurrent glioblastoma. Br J Cancer. 2007;96:960–969. doi: 10.1038/sj.bjc.6603652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Joy A, Mordechai E, Stachowiak EK. Nuclear localization of functional FGF receptor 1 in human astrocytes suggests a novel mechanism for growth factor action. Brain Res Mol Brain Res. 1996;38:161–165. doi: 10.1016/0169-328x(96)00010-1. [DOI] [PubMed] [Google Scholar]
- Steiner E, Holzmann K, Pirker C, Elbling L, Micksche M, Sutterluty H, et al. The major vault protein is responsive to and interferes with interferon-gamma-mediated STAT1 signals. J Cell Sci. 2006;119:459–469. doi: 10.1242/jcs.02773. [DOI] [PubMed] [Google Scholar]
- Sundberg JP, Rourk MH, Boggess D, Hogan ME, Sundberg BA, Bertolino AP. Angora mouse mutation: altered hair cycle, follicular dystrophy, phenotypic maintenance of skin grafts, and changes in keratin expression. Vet Pathol. 1997;34:171–179. doi: 10.1177/030098589703400301. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Kato T, Takimoto H, Masui S, Oshima H, Ozawa K, et al. Localization of rat FGF-5 protein in skin macrophage-like cells and FGF-5S protein in hair follicle: possible involvement of two Fgf-5 gene products in hair growth cycle regulation. J Invest Dermatol. 1998;111:963–972. doi: 10.1046/j.1523-1747.1998.00427.x. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Ota Y, Ozawa K, Imamura T. Dual-mode regulation of hair growth cycle by two Fgf-5 gene products. J Invest Dermatol. 2000;114:456–463. doi: 10.1046/j.1523-1747.2000.00912.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi F, Harada T, Sakamoto Y, Yamauchi N, Yoshida S, Iwabe T, et al. Activation of mitogen-activated protein kinase pathway by keratinocyte growth factor or fibroblast growth factor-10 promotes cell proliferation in human endometrial carcinoma cells. J Clin Endocrinol Metab. 2003;88:773–780. doi: 10.1210/jc.2002-021062. [DOI] [PubMed] [Google Scholar]
- Todo T, Kondo T, Kirino T, Asai A, Adams EF, Nakamura S, et al. Expression and growth stimulatory effect of fibroblast growth factor 9 in human brain tumors. Neurosurgery. 1998;43:337–346. doi: 10.1097/00006123-199808000-00098. [DOI] [PubMed] [Google Scholar]
- Yamada SM, Yamada S, Hayashi Y, Takahashi H, Teramoto A, Matsumoto K. Fibroblast growth factor receptor (FGFR) 4 correlated with the malignancy of human astrocytomas. Neurol Res. 2002;24:244–248. doi: 10.1179/016164102101199864. [DOI] [PubMed] [Google Scholar]
- Zhan X, Bates B, Hu XG, Goldfarb M. The human FGF-5 oncogene encodes a novel protein related to fibroblast growth factors. Mol Cell Biol. 1988;8:3487–3495. doi: 10.1128/mcb.8.8.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kharbanda S, Hanfelt J, Kern FG. Both autocrine and paracrine effects of transfected acidic fibroblast growth factor are involved in the estrogen-independent and antiestrogen-resistant growth of MCF-7 breast cancer cells. Cancer Res. 1998;58:352–361. [PubMed] [Google Scholar]