Abstract
Fibroblast growth factors (FGF) and their high-affinity receptors (FGFR) represent an extensive cellular growth and survival system. Aim of this study was to evaluate the contribution of FGF/FGFR-mediated signals to the malignant growth of non-small cell lung cancer (NSCLC) and to assess their potential as targets for therapeutic interventions. Multiple FGFR mRNA splice variants were coexpressed in NSCLC cells (n = 16) with predominance of FGFR1. Accordingly, both expression of a dominant-negative FGFR1 (dnFGFR1) IIIc-green fluorescent protein fusion protein and application of FGFR small-molecule inhibitors (SU5402 and PD166866) significantly reduced growth, survival, clonogenicity, and migratory potential of the majority of NSCLC cell lines. Moreover, dnFGFR1 expression completely blocked or at least significantly attenuated s.c. tumor formation of NSCLC cells in severe combined immunodeficient mice. Xenograft tumors expressing dnFGFR1 exhibited significantly reduced size and mitosis rate, enhanced cell death, and decreased tissue invasion. When FGFR inhibitors were combined with chemotherapy, antagonistic to synergistic in vitro anticancer activities were obtained depending on the application schedule. In contrast, simultaneous blockage of FGFR- and epidermal growth factor receptor-mediated signals exerted synergistic effects. In summary, FGFR-mediated signals in cooperation with those transmitted by epidermal growth factor receptor are involved in growth and survival of human NSCLC cells and should be considered as targets for combined therapeutic approaches.
Introduction
Despite recent improvements in diagnosis and treatment, lung cancer is the leading cause of cancer death worldwide. This highlights the urgent need for new therapeutic strategies against this tumor type. Over 75% of all lung cancers are non-small cell lung cancer (NSCLC), which consists of squamous cell carcinoma (SCC), adenocarcinoma, and large cell carcinoma. The still dismal prognosis is due to the prevalent diagnosis at advanced disease, the intrinsic resistance against chemotherapy, and the high rate of relapse following surgery (1).
During tumor development the dependency on exogenous growth factors is often lost/altered possibly due to endogenous overproduction of growth factors or abnormal expression as well as mutations of receptor molecules leading to uncontrolled, autocrine growth stimulation. Characterization of such key molecular alterations in lung cancer cells is believed to offer new chances for the development of tumor-specific systemic therapies (2). Indeed, several examples of new targeted drugs especially against growth factors and/or their receptor tyrosine kinases (RTK) have been developed for NSCLC successfully during the last years. Thus, the antiangiogenic vascular endothelial growth factor (VEGF) antibody bevacizumab and the epidermal growth factor receptor (EGFR) small-molecule inhibitor erlotinib have both been approved for treatment of advanced NSCLC (3). However, most of these compounds showed activity in patient subgroups only, suggesting that cancer cells can evade anticancer effects by activating alternative growth and survival pathways.
Evidence has accumulated that members of the fibroblast growth factor (FGF) family together with their four transmembrane tyrosine kinase receptors (FGFR1-4) might act as autocrine as well as paracrine (angiogenic) growth factors in many, if not all, solid tumors (4). In humans, the FGF family consists of 22 members, which vary in size but share a conserved sequence of 120 amino acids. FGF binds heparan sulfate proteoglycans and glycosaminoglycans with low affinity and FGFR with high affinity. FGFR mRNA molecules are extensively spliced leading to receptor isoforms differing in ligand binding specificities. In particular, alternate exon usage within the IgIII loop region, resulting in IIIb and IIIc variants of FGFR1-3, has a strong effect on ligand binding and signaling potency. In normal tissue, the expression of FGFR IIIb and IIIc isoforms is characteristic for epithelial or mesenchymal cells, respectively (5). Precisely controlled FGF-derived signals are key components in the regulation of vertebrate development during embryogenesis and also at later stages regarding growth and differentiation of various tissues and organs (6). FGF act as mitogens and some members induce cell migration, angiogenesis, neurite outgrowth, and cell survival (5). Strong indications for an important role of FGF/FGFR signals in malignant growth and probably malignant transformation have been published for several epithelial solid tumors including prostate, bladder, kidney, and breast cancer (4, 7). In a previous study, we showed coexpression of FGFR1 and FGF2 in malignant NSCLC cells in vitro and in tissue sections (8). Moreover, the expression levels of FGF2 indicated more aggressive growth behavior and correlated with insensitivity to growth stimulation by recombinant FGF2. Taken together, these data indicate the existence of autocrine FGF2-dependent growth signal loops in NSCLC cells.
In the present study, we show that coexpression of multiple FGFR IIIb and IIIc splice variants is characteristic for NSCLC cells. By usage of genetic and pharmacologic approaches, we show that FGFR-mediated signals contribute to growth, survival, and migration of NSCLC cells in vitro and in vivo and suggest FGFR inhibition as promising therapeutic strategy in NSCLC.
Materials and Methods
Cell Lines and Cell Culture
Ten cell lines were established in our laboratory from surgical specimens of primary or metastatic NSCLC (9). Histopathologically, cell lines were derived from 7 SCC (VL-3 and VL-5-VL-10), 2 large cell carcinoma (VL-2 and VL-4), and 1 adenocarcinoma (VL-1). Cells were grown in RPMI 1640 (Life Technologies; except VL-1 in DMEM) supplemented with 10% fetal bovine serum (Life Technologies). Additionally, 7 NSCLC cell lines (adenocarcinoma-derived) were obtained from other sources (A549, A427, Calu-3, Calu-6, SK-LU-1, and H2073 from the American Type Culture Collection and LXF from DKFZ) and grown in the respective culture medium with 10% fetal bovine serum. As nonmalignant cell counterparts, human, SV40 T-antigen-immortalized, bronchoepithelial cell lines (BEAS-2B, ATCC, LHC-9 medium), normal human bronchial epithelial cells (NHBE; Lonza), and two human embryonic lung fibroblast cell lines (F2000, Flow; RPMI and Wi38, ATCC, RPMI) were used. Cells were periodically checked for Mycoplasma contamination. Growth characteristics, ploidy, and patient as well as tumor data including the mutation status of K-Ras have been presented previously (9, 10).
Reagents and Antibodies
The FGFR1-specific inhibitor PD166866 (11) was generously supplied by Pfizer Global Research and Development, the pan-FGFR inhibitor SU5402 (12) obtained from Calbiochem. Lapatinib and erlotinib were generously supplied by GlaxoSmithKline and Roche, respectively. All other reagents were obtained from Sigma. Matched cDNA panels derived from normal and respective malignant human lung tissues were derived from BD Biosciences Clontech.
The expression vector pcHisCtrFGFR, encoding a kinasetruncated, dominant-negative FGFR1 (dnFGFR1) IIIc (13), was kindly provided by Dr. Francis Kern (Georgetown University Medical Center). The truncated FGFR1 was tagged with enhanced green fluorescent protein (GFP) at the COOH terminus by insertion into pEGFP-N3 (Clontech) to generate a dnFGFR1 IIIc-GFP protein chimera and subcloned into pShuttle-CMV (Stratagene). The adenoviral expression vector was created by a double-recombination event in bacteria between pAdEasy-1 (Stratagene) and shuttle vector pShuttle-CMV. An adenovirus expressing a dnEGFR-GFP was constructed in a similar way. Briefly, a cDNA of EGFR without the cytoplasmic tyrosine kinase domain (CD533; kind gift of A. Ullrich, Max-Planck Institute) was fused in-frame to GFP by insertion into pEGFP-N1 and the resulting dnEGFR-GFP chimera was subcloned into pShuttle-CMV. An enhanced GFP adenoviral expression vector was used as control (14). Virus titers were determined by Adeno-X Rapid Titer Kit (Clontech) according to the instructions of the manufacturer and by GFP fluorescence-activated cell sorting analysis (FACSCa-libur, BD Biosciences Clontech).
Anti-phosphorylated ERK1/2 (pERK1/2), anti-pS6, anti-S6, were from Cell Signaling Technology. Anti-ERK1/2 was from Upstate, and anti-cyclin A (clone H-432) was from Santa Cruz Biotechnology. Anti-human pan-cytokeratin (clones AE1/AE3) and anti-human Ki-67 (clone MiB-1) antibodies were purchased from DAKO and anti-GFP was from Roche Diagnostics.
Reverse Transcription-PCR and Real-time Reverse Transcription-PCR
Presence of the FGFR1-3 mRNA splice variants IIIb and IIIc were analyzed by reverse transcription-PCR. Total cellular RNA was prepared and reverse transcription-PCR was done as described previously (8). The primer sets were designed to be indicative for the composition of the third IgG loop (15). The amount of PCR products was semi-quantitatively determined by scanning densitometry and a cutoff value of 0.1 compared with the respective controls was counted as positive. Real-time reverse transcription-PCR for relative quantification of FGFR1 IIIc mRNA was done using SYBR Advantage qPCR Premix (Clontech) on a 7500 Fast Real-time instrument (Applied Biosystems) following the respective instructions. Quantification of gene expression was calculated by the comparative Ct method using β-actin or glyceraldehyde 3-phosphate dehydrogenase as reference genes. Information about the primers is shown in Supplementary Table S1.4
Transfection
For stable expression, cells were transfected with the dnFGFR1 construct by electroporation under G418 selection using standard protocols. Transgene expression was determined by fluorescence-activated cell sorting. For transient expression, 2 × 105 cells were seeded on six-well plates and infected with adenoviruses (containing dnFGFR1, dnEGFR, or GFP) after 24 h.
Cell Death Analysis
Assessment of cell death was done either by trypan blue exclusion assay or propidium iodide staining. For evaluation of apoptosis, cells were stained with 4′,6-diamidino-2-phenylindole and scored microscopically as published (16).
Migration Assays
Cell migration was tested by scratch or Transwell chamber assays as published recently (17, 18). Briefly, for scratch assays, cells were transduced with the respective adenoviruses for 4 h and seeded in six-well plates. A scratch was applied with a yellow tip in the confluent cell monolayers. Closure of scratch was monitored microscopically at least at three fixed positions/scratch, photomicrographs were taken, and the gap widths were measured using Metamorph 6.1 software (Universal Imaging). In case of Transwell chamber assays, cells were seeded on 8 μm pore size PET track-etched membranes (BD Falcon) in 24-well chambers. Following 12 h recovery, small-molecule inhibitors were added. After the respective incubation periods, cells on the lower surface of the membrane were fixed and stained with crystal violet and counted microscopically.
Immunoblotting
In case of stimulation experiments, total proteins were isolated by washing cells with PBSand adding 150 μL of 2 × Laemmli sample buffer/well of a six-well plate. Lysates were sonicated in a water bath for 7 min followed by heating to 95°C for 7 min. Cell extracts (25 μL) were loaded. All other protein isolation and immunoblotting procedures were done as described previously (18). Following SDS-PAGE, proteins were transferred onto a polyvinylidene difluoride membrane and probed with the respective antibodies. Western blot signals were quantified using Multi-Analyst 1.1 software (Bio-Rad Laboratories).
Proliferation/Viability Assays
The effects of the FGFR inhibitors PD166866 and SU5402 as well as of the EGFR inhibitors erlotinib and lapatinib on proliferation of the lung cancer cell lines was measured by a MTT-based colorimetric assay as described (9). Cells (between 3,000 and 5,000 per well in 96-well plates) were seeded and drugs were added 24 h later. Exposure time was 96 h. IC50 values (concentrations causing a 50% reduction of viable cells) were estimated from whole dose-response curves done in triplicate. The effects of drug combinations were set up by exposing tumor cells in parallel to (a) five concentrations of the two investigated drugs (FGFR inhibitor and chemotherapeutic drug or EGFR inhibitor) as single agents and (b) the respective 25 concentration pairs. Synergism was determined by calculating the combination index (CI) according to Chou and Talalay (19) with CalcuSyn software (Biosoft). According to this method, CI < 1, CI = 1, or CI > 1 represent synergism, additive effects, and antagonism of the two investigated substances, respectively.
Tumor Formation in Severe Combined Immunodeficient Mice
Experiments were done as described previously (10). Briefly, cells were infected with dnFGFR1 or GFP adenovirus on cell culture plates. Subsequently, 1 × 106 cells in 50 μ L PBS were s.c. injected into the rear flanks of immunodeficient severe combined immunodeficient (SCID) BALB/c recipient mice (female, ages 4 weeks; Harlan Winkelmann). Tumor formation was measured periodically by palpation, and the tumor size was determined using a Vernier caliper. Tumor volume was calculated from tumor size using the formula: (smaller diameter2 × larger diameter) / 2. All experiments were done in triplicate and carried out according to the Austrian and FELASA guidelines for animal care and protection.
Immunohistochemistry
Immunohistochemistry was done as described previously (20). Fast Red (Ventana) was used as chromogen and hemalaun as counterstain. Observation was done with a Nikon Eclipse 80i light microscope. To evaluate programmed cell death, proliferating Ki-67-positive and non-proliferating Ki-67-negative regions were defined based on Ki-67 staining and apoptotic cells were counted in H&E- and Ki-67-stained tumor sections. At least six optical fields at a magnification of × 40 were counted.
Statistical Analysis
Data are presented as mean ± SD. Statistical significance between treatments and controls were analyzed either with Student’s t test or two-way ANOVA using GraphPad Prism 4.0. In all cases, P ≤ 0.05 was considered statistically significant.
Results
Expression Pattern of FGFR IgIII Splice Variants in NSCLC Cell Lines
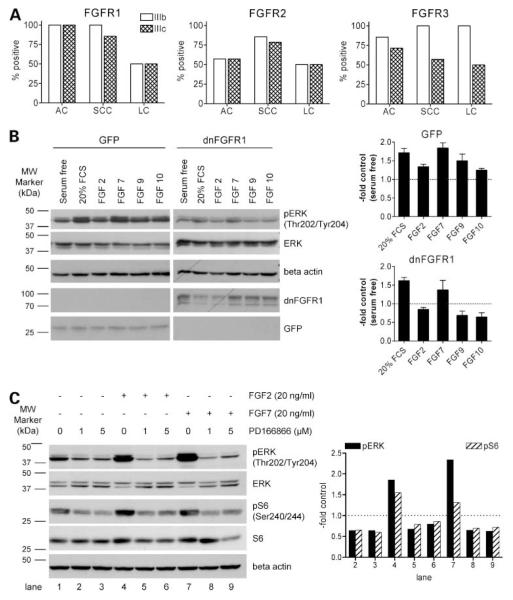
In a previous study, we have reported that mRNA for FGFR3 and FGFR4 are present in 100% and FGFR1 and FGFR2 in 93.7% and 75% of NSCLC cell lines, respectively (8). Here, we extended these data toward a reverse transcription-PCR analysis of the IgIII mRNA splice variants IIIb and IIIc of FGFR1-3 in NSCLC cell lines, normal cell counterparts, and matched normal and malignant lung tissue samples. FGFR4 was excluded as splice variants regarding the IgIII loop have not been reported. The vast majority (14 of 16) of normal and malignant cell cultures coexpressed significant amounts of FGFR1 IIIb and IIIc mRNA (Fig. 1A). Only one SCC and one large cell carcinoma cell line did not express the IIIc variant of FGFR1. Presence of both FGFR1 splice variants was confirmed in all normal and malignant tissue-derived cDNA (data not shown). Additionally, normal and immortalized human epithelial cell cultures and lung fibroblasts expressed both FGFR1 splice variants (Supplementary Table S2).4 Quantification of the FGFR1 IIIc levels by real-time PCR, however, showed that both epithelial cell cultures (NHBE and BEAS-2B) expressed the FGF1 IIIc variant at very low levels compared with malignant cells and lung fibroblasts (Supplementary Fig. S1).4 In case of FGFR2, the IIIb and IIIc variants were coexpressed in 9 of 16 NSCLC cell lines and another 4 cell lines expressed either the IIIb or the IIIc variant (Fig. 1A). Although both nonmalignant epithelial cell lines but none of the fibroblast models expressed FGFR2 IIIb, the IIIc isoforms was only present at low levels in F2000. For FGFR3, the splice variant IIIb was the dominant one, with >90% positivity in NSCLC cell lines, whereas only 62% of the cell lines expressed the IIIc variant (Fig. 1A). In the nonmalignant cultures, the FGFR3 IIIb variant was restricted to the epithelial cell types, whereas the IIIc variant was also expressed by both fibroblast lines. Based on the fact that presence of the tumor-related IIIc variant (21, 22) was predominantly observed in case of FGFR1, this receptor was used for further analyses.
Figure 1.
Expression of FGFR mRNA variants in NSCLC cells (A) and blockage of FGF-induced phosphorylation of downstream signaling molecules by dnFGFR1 (B) and small-molecule inhibitor PD166866 (C). A, expression of IgIII mRNA splice variants of FGFR1, FGFR2, and FGFR3 was analyzed by reverse transcription-PCR in 16 NSCLC cell lines. The percentage of positive cell lines separated into histologic sub-groups adenocarcinoma-derived, SCC-derived, and large cell carcinoma (LC)-derived cell lines is shown. FGFR primer pairs were designed to be variant specific with respect to the third IgG loop (variant FGFR-IIIb or FGFR-IIIc). B, GFP and dnFGFR1 (representing a GFP fusion protein) expression and effect on phosphorylation of ERK were analyzed after adenoviral transduction by Western blotting in VL-10 cells with respective antibodies. After infection, cells were serum starved for 48 h and stimulated with the indicated FGF for 15 min. Blots (left) were quantified densitometrically (mean and SD of three experiments; right). Values are expressed as pERK/ERK ratios normalized to the serum-free controls set arbitrarily as 1. C, effects of the indicated concentrations of PD166866 on phosphorylation of ERK and S6 were analyzed by Western blotting. VL-10 cells were serum starved for 48 h, incubated with PD166866 for 1 h, and stimulated with FGF2 and FGF7 (20 ng/mL) for 10 min. Blots (left) were quantified densitometrically. Values are expressed as pERK/ ERK and pS6/S6 ratios normalized to the serum-free controls set arbitrarily as 1 (right). One of three experiments delivering comparable results.
Blockage of FGFR1 Signaling by dnFGFR1 and Small-Molecule Inhibitors
To block FGF/FGFR-mediated signals in NSCLC cells, two approaches were used. The first one involved expression of a dnFGFR1 IIIc-GFP fusion protein by stable transfection with plasmid DNA or by transient transduction with an adenoviral construct. Specific inhibition of FGFR-mediated signals was confirmed by monitoring FGF-induced activation of the mitogen-activated protein kinase pathway (23). Transient expression of the adenoviral dnFGFR1 construct in the FGF-responsive NSCLC cell line VL-10 prevented FGF2-, FGF9-, and FGF10-induced phosphorylation of ERK, whereas the GFP control virus had no significant effect on mitogen-activated protein kinase activation. FCS- and FGF7-mediated ERK phosphorylation was not significantly influenced by dnFGFR1 expression (Fig. 1B), corresponding to predominant binding of FGF7 to the keratinocyte growth factor receptor FGFR2 IIIb (5).
The second approach to block endogenous FGF/FGFR signaling involved the small-molecule inhibitors SU5402 (ref. 12; data not shown) and PD166866 (ref. 11; Fig. 1C). FGF2- and FGF7-induced phosphorylation of ERK and S6 ribosomal protein was clearly suppressed by both FGFR inhibitors. Moreover, also the basal phosphorylation levels of these signaling molecules were significantly reduced in serum-starved NSCLC cells. This indicates a significant contribution of autocrine FGFR-mediated signals to maintain mitogen-activated protein kinase and phosphati-dylinostiol 3-kinase activities under serum-deprived conditions.
dnFGFR1 Induces NSCLC Growth Inhibition and Cell Death
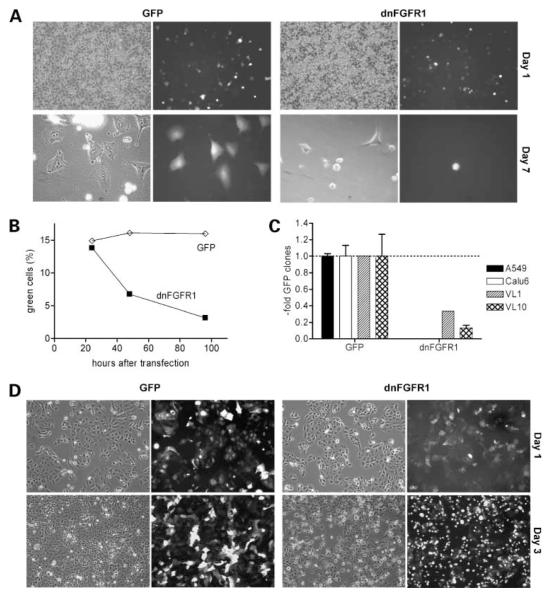
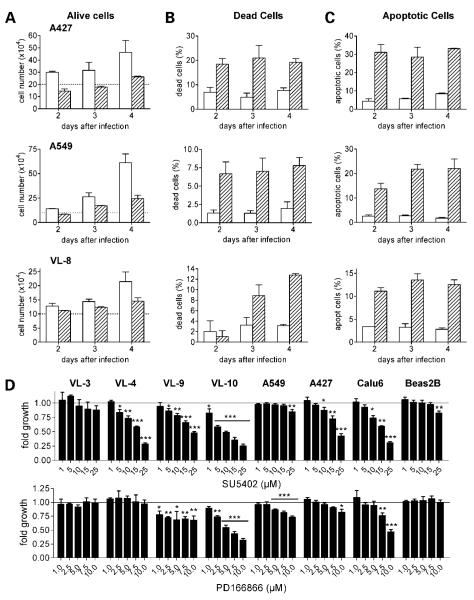
In stable transfection experiments, 8 of 10 investigated cell lines did not form clones exhibiting dnFGFR1 expression in contrast to the respective GFP control plasmid (Fig. 2A-C). dnFGFR1-expressing NSCLC cells either died within a few days after transfection (Fig. 2A and B) or, despite forming small green fluorescent colonies, did not survive the procedure of raising clones (Fig. 2C). Only in 2 of 10 investigated cell lines clones survived (VL-1 and VL-8) exhibiting very low but stable dnFGFR1 expression (data not shown). Consequently, we decided to use an adenoviral expression construct for further experiments, which enables transient transduction of a large fraction of the cells. In all NSCLC cell lines (n = 10) transduced with dnFGFR1 but not with the GFP control virus, a high proportion of cells detached from the plastic and rounded up within a few days (Fig. 2D, A427 cells are shown representatively). Percentages of viable and dead cells were determined by trypan blue exclusion assay. Although infection with the GFP virus had no significant effect, the dnFGFR1 virus resulted in decreased proliferation (Fig. 3A) and induced cell death (Fig. 3B). 4′,6-Diamidino-2-phenylindole staining showed that cells mainly died via apoptosis (Fig. 3C).
Figure 2.
Effects of dnFGFR1 expression on NSCLC cells. Cells were transfected with GFP or dnFGFR1 vectors by electroporation (A-C). A, photomicrographs were taken of A549 cells 24 h (note the similar proportion of green fluorescent cells showing comparable transfection rates) and 7 d after transfection as indicated. B, proportion of GFP- or dnFGFR1-expressing, viable A549 cells was analyzed by fluorescence-activated cell sorting at the indicated time points after transfection. One of three experiments yielding comparable results. C, after transfection of the indicated NSCLC cell lines and selection with G418 for about 3 wk, numbers of green clones were determined. Due to the high variation in the cloning potential of the analyzed cell lines, the data are given relatively to the number of controlGFP clones. D, A427 cells were transduced with GFP or dnFGFR1 adenoviruses and photomicrographs taken at the indicated time points.
Figure 3.
Expression of dnFGFR1 in NSCLC cells leads to apoptotic cell death. The indicated NSCLC cell lines were either transduced with GFP (open columns) or dnFGFR1 adenoviruses (hatched columns; A-C) or treated with small-molecule FGFR inhibitors (D). At the indicated time points after infection, the number of cells alive (A) and the percentage of dead cells (B) were assessed by trypan blue exclusion assay. Percentage of apoptotic cells (C) was analyzed by 4′,6-diamidino-2-phenylindole staining. D, NSCLC cell lines and nonmalignant bronchial epithelial cells BEAS-2B were treated with the indicated concentrations of the FGFR inhibitors SU5402 (top) and PD166866 (bottom) in growth medium containing 1% FCS. Following a 96 h exposure, cell number was analyzed by MTT assay. Representative results for each cell line are shown normalized to controls without drugs. Columns, mean; bars, SD. Significant differences to controls were determined by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
Blockage of FGFR1 Signaling Has an Antiproliferative Effect
FGFR signaling was additionally blocked by small-molecule inhibitors and effects on cell proliferation/viability were analyzed by MTT assays (Fig. 3D). Treatment with SU5402 under low serum conditions (1% FCS) inhibited proliferation in 5 of 7 analyzed NSCLC cells in a dose-dependent manner. The antiproliferative effect of FGFR1 inhibitor PD166866 on the investigated NSCLC cells was comparable. Proliferation/survival of nonmalignant lung epithelial cells (BEAS-2B) with either inhibitor was not significantly changed with exception of 25 μmol/L SU5402, which slightly but significantly reduced cell survival/proliferation by around 17% (Fig. 3D).
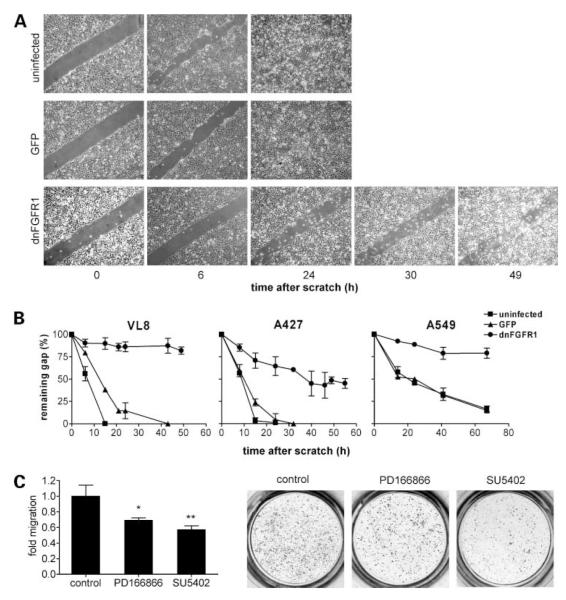
Blockage of FGFR1 Signaling Reduces Migration of NSCLC Cells
Expression of dnFGFR1 potently inhibited cell migration in all NSCLC cells tested. Representative scratch assay results are shown for VL-8, A549, and A427 (Fig. 4A and B). Although in the case of the parental and GFP-expressing cells the scratch was at least half-closed within 24 h, dnFGFR1-positive cells were unable to close the scratch even after 3 days. GFP expression had no or only a very minor effect on the migration potential of the tested NSCLC cells (Fig. 4A and B). Similar results were obtained when the effect of small-molecule FGFR inhibitors on cell migration was analyzed using Transwell chambers. Both SU5402 and PD166866 significantly reduced the migratory potential of all tested cell lines (VL-8 representatively in Fig. 4C).
Figure 4.
Blockade of FGFR leads to inhibition of NSCLC cell migration. After transduction with GFP or dnFGFR1 adenoviruses as indicated, cell migration was analyzed by scratch assays as described in Materials and Methods (A and B). Representative photomicrographs of scratch wounds in a VL-8 monolayer taken at the indicated time points are shown in A and quantitative evaluations for three NSCLC cell lines are shown in B. C, VL-8 cells were seeded on transmigration filters and treated with PD166866 (10 μmol/L) and SU5402 (15 μmol/L) for 72 h in medium with 10% FCS. Thereafter, cells on the lower surface of the filters were fixed, stained with crystalviolet, and counted. Left, columns, mean of three experiments; bars, SD. Significant difference to the controlwas determined by Student’s t test. *, P < 0.05; **, P < 0.01.
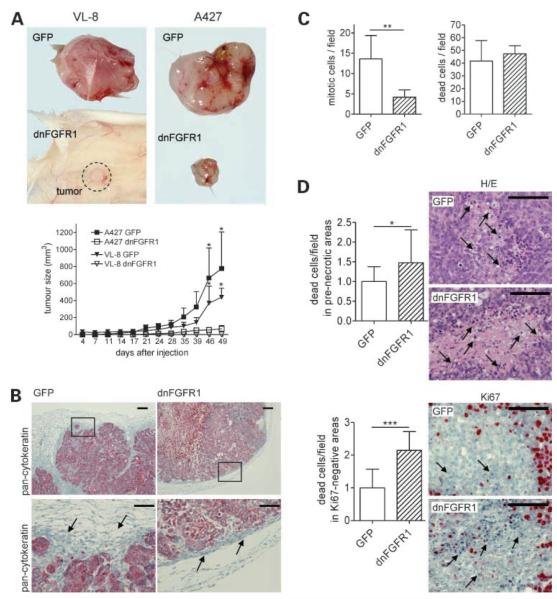
Expression of dnFGFR1 Inhibits NSCLC Growth In vivo
To assess whether expression of dnFGFR1 affects tumor growth in vivo, NSCLC cell lines (n = 5) infected with adenoviruses were injected s.c. into SCID mice. In Fig. 5, respective results for ras-wild type VL-8 (SCC) and ras-mutated A427 (adenocarcinoma) are shown representatively. No signs of systemic cytotoxicity were monitored following application of the cells transduced with the adenoviruses. All NSCLC cell lines infected with the control GFP virus rapidly formed tumors, whereas dnFGFR1 significantly reduced tumor growth in all cases. dnFGFR1-expressing cells almost completely lost in vivo tumorigenicity or formed significantly smaller tumors compared with the controls. As obvious from cytokeratin stainings (Fig. 5B), GFP control A427 tumors grew in an infiltrative manner, with a jagged and fissured margin and a high frequency of isolated tumor islets invading the surrounding stroma. In contrast, dnFGFR1-expressing tumors grew in a more encapsulated fashion and exhibited stronger compression of the surrounding stromal tissue. H&E- and Ki-67-stained tissue sections were employed to analyze the amount of proliferating and apoptotic cells within different parts of the tumor. In viable boundary regions, the mitosis rate was significantly reduced in dnFGFR1-expressing tumors, although no differences in the incidence of apoptotic cells were observed in these parts of the tumor (Fig. 5C). In contrast, a significantly enhanced cell death rate (based on evaluation of both H&E- and Ki-67-stained tissue sections) was detected in Ki-67-negative, prenecrotic areas of dnFGFR1-containing tumors (Fig. 5D).
Figure 5.
Inhibition of in vivo NSCLC growth by expression of dnFGFR1. Tumor formation of SCC-derived VL-8 and adenocarcinoma-derived A427 cells transduced with GFP or dnFGFR1 adenoviruses were analyzed by s.c. implantation in SCID BALB/c mice. Forty-nine days after s.c. injection, the mice were killed and the tumors were isolated and further analyzed. Photographs of representative tumors are shown (A, top). Tumor volumes (A, bottom) were calculated as described in Materials and Methods. Each experimental group contained three to six animals. Mean ± SD. Significant differences to controls were determined by two-way ANOVA with Bonferroni post-test. Formalin-fixed A427-derived tumor specimens were routinely processed for paraffin embedding, and sections were stained with pan-cytokeratin (B) or H&E and Ki-67 (D) as indicated. B, controltumors grew in an infiltrative manner, with a jagged and fissured margin and a high frequency of isolated tumor islets invading the surrounding stroma. In contrast, dnFGFR1-expressing tumors grew in a more encapsulated fashion and exhibited stronger compression of the surrounding stromal tissue (arrows). Boxed regions (top) are shown at higher magnifications (bottom). C, mitotic (left) and apoptotic (right) cells were counted in viable boundary regions of H&E-stained tumor sections. D, apoptotic cells were counted in at least 10 small prenecrotic (top, H/E stain) and Ki-67-negative (bottom) regions as representatively shown (right). Bar, 50 μm (B, bottom) and 100 μm (B, top, and D). Columns, mean of at least 15 optical fields (C) or 10 regions (D) analyzed; bars, SD. Significant differences to controls were determined by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
Combination of FGFR Inhibitors with Chemotherapeutics
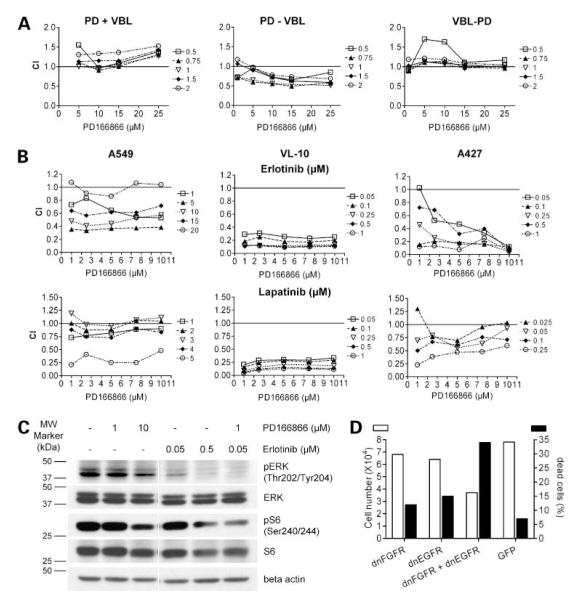
Small-molecule FGFR inhibitors as single agents exerted a moderate antiproliferative and cytostatic activity in the majority of investigated NSCLC cell lines (compare Fig. 3D). Thus, we combined these drugs with clinically used chemotherapeutics (doxorubicin, cisplatin, vinblastine, and Taxol). Concomitant administration revealed in most cases antagonistic effects as shown representatively for the combination of PD166866 with vinblastine (CI blot in Fig. 6A, left). However, when the administration scheme was changed to sequential application, the outcome depended largely on the schedule. In several cases, including drugs like cisplatin and vinblastine, distinctly synergistic effects were observed when the FGFR inhibitor was added before the chemotherapeutics. In contrast, weak synergistic/additive to antagonistic effects were observed by the opposite sequence (representatively PD166866 followed by vinblastine and the opposite sequence are shown in Fig. 6A, middle and right, respectively).
Figure 6.
Combination of small-molecule FGFR inhibitors with chemotherapeutic drugs and EGFR inhibitors. Concentrations of the drugs [vinblastine (VBL), erlotinib, and lapatinib] used for combination experiments (A and B, right) were selected from single substance dose-response curves (data not shown) and covered the range from ineffective to around 50% growth inhibition. A, A549 cells were simultaneously treated for 96 h with the indicated VBL + PD166866 combinations (left), 72 h with PD166866 followed by 72 h VBL (middle), or 72 h with VBL followed by 72 h with PD166866 (right) and cell viability was analyzed by MTT assays. B, NSCLC cell lines were simultaneously treated with the indicated combinations of the FGFR inhibitor PD166866 with the EGFR inhibitor erlotinib (top) or the EGFR/HER-2 dualtyrosine kinase inhibitor lapatinib (bottom) for 96 h in growth medium containing 1% FCS and cell survival was analyzed by MTT assay. Data in A and B present CI of the respective drug combinations calculated according to Chou and Talalay (19) with CalcuSyn software. Following this evaluation, CI < 1 indicates synergism, CI = 1 stands for additive effects, and CI >1 depicts antagonistic effects. C, effect of a 2 h exposure of the indicated RTK inhibitors on the phosphorylation of the downstream signaling molecules ERK and S6 was analyzed by Western blotting. Representatively, data for SCC-derived VL-10 cells are shown. D, VL-8 cells were transduced with dnFGFR1 or dnEGFR1 adenoviruses alone or in combination whereby the viral dose was kept constant by complementing with respective amounts of control virus. After 48 h exposure, numbers of viable (open columns) and dead cells (black columns) were determined by trypan blue exclusion assay.
Combined Blockage of FGFR and EGFR
EGFR-specific inhibitors are already used in targeted therapy of lung cancer, but only patient subpopulations benefit from these treatments (24). Thus, we set out to investigate whether the combined blockage of the EGFR and FGFR might exert synergistic antiproliferative effects. NSCLC cells were treated simultaneously with the FGFR-specific inhibitors SU5402 or PD166866 and the EGFR-targeting drugs erlotinib (EGFR = HER1-specific) and lapatinib (EGFR/HER-2 dual tyrosine kinase inhibitor; ref. 25). In all investigated NSCLC cell lines, mostly additive to distinctly synergistic antiproliferative effects were observed (examples in Fig. 6B). For example, all drug combinations synergistically inhibited VL-10 cell proliferation, whereas in case of A549 and A427 cells the extent of synergism varied predominantly with the concentration of the EGFR inhibitor. Accordingly, the combination of PD166866 and erlotinib synergistically decreased the activation of RTK downstream signals detected as phosphorylation of ERK and S6 (Fig. 6C). As a second approach for targeting the two RTK molecules, dominant-negative receptor constructs were employed (Fig. 6D). As mentioned above, the introduction of dnFGFR1 in NSCLC cells led to induction of cell death (compare Figs. 2 and 3). A comparable number of dead cells were also induced by infection of VL-8 cells with dnEGFR adenovirus. However, when the same viral dose was added as a mixture of both constructs, the proportion of dead cells was synergistically increased (Fig. 6D).
Discussion
Despite the effort of constantly improving treatment modalities such as chemotherapy, radiotherapy, and surgery, the prognosis of lung cancer remains poor. Therefore, new therapeutic options targeting tumor-specific growth and survival signals are urgently needed. Thus far, RTK-mediated protumorigenic signals are among the most successful targets in modern anticancer drug development. In case of NSCLC, RTK targeting focused mainly on EGFR and several agents inhibiting EGFR have been clinically approved (24). Unfortunately, only a subgroup of patients including mainly those harboring certain EGFR mutations benefit from these treatments (26). A possible explanation is that, in addition to EGFR, other RTK are actively contributing to NSCLC cell survival.
In this study, we present strong evidence that FGF/FGFR-mediated loops are active and cooperate with EGFR-mediated signals to promote growth and survival of NSCLC cells. Correspondingly, FGFR1 was recently shown to belong to the most frequently (over)expressed RTK molecules in early-stage NSCLC (27). Moreover, the expression of FGFR1 was connected to an about 3.5-fold, of EGFR to a 3.1-fold, and of FGFR4 to a 2-fold increased hazard ratio for developing metastatic disease (27). Zhao et al. (28) reported occasional high-level amplifications of the FGFR1 gene in NSCLC. In a previous study, we have already shown constitutive overproduction of FGF2 (basic FGF) and several FGFR molecules in NSCLC cells in vitro and in vivo (8). However, activation of oncogenic FGF/FGFR-mediated signals can be based not only on overexpression of ligand and/or receptor molecules but also on alternative splicing of FGFR mRNA (5, 29). Consequently, we have extended our analysis to detection of epithelial IIIb and mesenchymal IIIc variants of FGFR1-3 in NSCLC cells and nonmalignant lung cell counterparts. In case of FGFR4, almost constitutively expressed in NSCLC (8), no such splice variants are generated (5). Frequent coexpression of several IIIb and IIIc FGFR mRNA variants in the majority of NSCLC cells was observed. Whereas in the case of FGFR1 and FGFR2 both variants were expressed in a comparable percentage, in case of FGFR3 the IIIb variant was prevalent. With regard to expression of the mesenchymal, carcinoma-associated IIIc splice variants (21, 22), FGFR1 was dominating in our NSCLC cell panel and in respective tissue specimen.5 Nonmalignant human bronchial epithelial cells expressed very low levels of FGFR1 IIIc mRNA compared with both malignant cell and lung fibroblasts (Supplementary Fig. S5).4 No extensive analysis of FGFR splice variants in NSCLC has been reported before this study. However, resembling our results, FGFR1 was expressed as IIIc variant, whereas FGFR3 was predominantly present as IIIb variant in prostate cancer cells (22). Accordingly, pancreatic cancer cells expressed FGFR1 IIIc but not the IIIb mRNA (21). Taken together, all these observations suggest that FGFR-mediated signals might be (hyper)activated in NSCLC cells by several molecular mechanisms. Accordingly, we have recently shown that reduced expression of Sprouty 2, an endogenous FGFR downstream signal inhibitor, supports malignant growth of NSCLC (10).
In accordance with these data, we present strong evidence that FGFR represents a promising therapeutic target in NSCLC. Expression of a dominant-negative FGFR1 (dnFGFR1) molecule was almost incompatible with proliferation and/or survival of the vast majority of NSCLC cell lines. Although not investigated in lung cancer before, comparable effects were also observed in other cancer cell lines derived from prostate cancer (30), glioma (31), and melanoma (32). dnFGFR1 expression also prevented or significantly inhibited s.c. growth of NSCLC cells in SCID mice. dnFGFR1-positive tumors were characterized by enhanced apoptosis rates, diminished mitotic index, and reduced invasion of surrounding tissue. The latter observation corresponds to a distinct blockade of tumor cell migration by inhibition of FGFR. In agreement with our data in NSCLC, expression of dnFGFR1 and dnFGFR2 in glioma cells inhibited in vivo tumor growth and led to increased apoptosis and decreased vessel formation in the tumor (33, 34). Our observations implicate that FGF/FGFR-mediated signals promote NSCLC progression by stimulating growth, survival, and migration as well as invasion.
In the clinical setting, inhibition of RTK-mediated signals is predominantly done by using small-molecule inhibitors or monoclonal antibodies. Thus, in addition to the dnFGFR1 construct, we have investigated the small-molecule FGFR inhibitors SU5402 and PD166866. Both substances led to a dose-dependent repression of NSCLC cell proliferation in the majority of cell lines, whereas nonmalignant bronchial epithelial cells BEAS-2B turned out to be rather insensitive against the FGFR inhibitors. Comparable growth-inhibitory effects as observed in NSCLC cells were also observed in other tumor entities such as multiple myeloma (35, 36), prostate carcinoma (37), and synovial sarcoma (38). Considering the difference in their in vitro kinase inhibiting activities (11, 12), the pan-FGFR inhibitor SU5402 proofed to possess higher growth-inhibiting potency than the FGFR1-specific PD166866. This is not surprising considering the constitutive expression of several FGFR in NSCLC cells (8, 27). Moreover, the pharmaceutical inhibitors might be differentially influenced by the presence of drug transporter molecules frequently expressed in NSCLC (9, 39). A second explanation might lie in the fact that, in addition to the transmission of growth/survival signals, FGFR are also involved in cell adhesion in a ligand-independent fashion (40).
Successful application of targeted agents has been frequently based on combinations with chemotherapeutic drugs. For example, the tyrosine kinase inhibitor SU6668 targeting FGFR1, VEGF receptor 2, and platelet-derived growth factor receptor displayed synergistic activities with Taxol against ovarian carcinoma xenografts (41). Likewise, soluble FGFR gene therapy combined with either paclitaxel or γ-irradiation exerted synergistic effects on DU145 prostate cancer cells (42). Thus, we analyzed whether a combination of FGFR inhibitors with diverse chemotherapeutics would enhance activity against NSCLC cells. Interestingly, the interactions of FGFR inhibitors and cytotoxic agents were frequently sequence dependent, a fact that has also been reported for combinations of EGFR inhibitors and chemotherapy as well as new targeted drugs in NSCLC cells (43– 45). Although the application of the FGFR inhibitor before chemotherapy rendered NSCLC cells sensitive to chemotherapy with drugs like cisplatin and vinblastine, the opposite sequence or coadministration resulted in only additive or antagonistic activities. This suggests that the FGFR inhibitors induce hypersensitivity to induction of cell death possibly by down-modulation of survival signals. Accordingly, previous studies have implicated FGF/FGFR-mediated signals in the acquisition of broad-spectrum resistance against anticancer drugs (46).
The relatively moderate effect of FGFR inhibition by small-molecule inhibitors resembles the one observed with EGFR inhibitors and might be explained by activation of multiple RTK molecules in NSCLC (27). Thus, we set out to investigate a combined inhibition of FGFR- and EGFR-mediated signals in NSCLC cells. Indeed, we found an almost general synergistic anticancer activity when combining inhibitors of these two RTK systems, which was obvious in terms of both growth inhibition and blockade of downstream signal pathways. These observations indicate that EGFR and FGFR cooperate in promoting growth and survival of NSCLC cells. In additional experiments, we have observed that, at least in some cases, EGFR inhibition or serum starvation leads to up-regulation of FGF ligand expression.6 Accordingly, the FGFR inhibitor PD166866 further attenuates ERK phosphorylation in serum-starved NSCLC cells (Fig. 1C). This might indicate that a feedback loop exists between these two mitogen-activated protein kinase activator signals leading to up-regulation of FGFR-mediated survival signals following EGFR blockade and/or serum starvation. To the best of our knowledge, this is the first study showing synergistic activity of FGFR and EGFR inhibitors in any type of cancer. However, several recent reports indicated that a combined inhibition of several RTK molecules and/or downstream effectors might be a feasible strategy for therapy even of drug-resistant solid tumors including NSCLC (47, 48).
Summarizing, our data show autocrine involvement of FGF/FGFR-mediated signals in growth and survival of NSCLC cells. Based on the synergistic anticancer activity of FGFR and EGFR inhibitors, we suggest that substances targeting the FGF/FGFR system should be considered as new tools for multimodality treatment of NSCLC.
Supplementary Material
Acknowledgments
We thank Vera Bachinger, Christian Balcarek, and Maria Eisenbauer for skillful technical assistance, Annemarie Losert for supplying dnEGFR-GFP, and StofflMayer and Hedwig Sutterlüty for fruitful discussion of the data.
Grant support: Fonds zur Förderung der Wissenschaftlichen Forschung projects P17630-B12 and P19920-B12; Forschungsentwicklungspreis des Fonds der Stadt Wien für innovative interdisziplinäre Krebsforschung 2007; Initiative Krebsforschung of the Medical University Vienna.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
In preparation.
Unpublished data.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Herbst RS, Dang NH, Skarin AT. Chemotherapy for advanced non-small cell lung cancer. Hematol Oncol Clin North Am. 1997;11:473–517. doi: 10.1016/s0889-8588(05)70445-7. [DOI] [PubMed] [Google Scholar]
- 2.Smith PW, Denlinger CE, Jones DR. Novel targeted therapies for non-small cell lung cancer. Thorac Surg Clin. 2006;16:353–66. doi: 10.1016/j.thorsurg.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Byers LA, Heymach JV. Dual targeting of the vascular endothelial growth factor and epidermalgrowth factor receptor pathways: rationale and clinical applications for non-small-cell lung cancer. Clin Lung Cancer. 2007;8(Suppl2):S79–85. doi: 10.3816/clc.2007.s.006. [DOI] [PubMed] [Google Scholar]
- 4.Jeffers M, LaRochelle WJ, Lichenstein HS. Fibroblast growth factors in cancer: therapeutic possibilities. Expert Opin Ther Targets. 2002;6:469–82. doi: 10.1517/14728222.6.4.469. [DOI] [PubMed] [Google Scholar]
- 5.Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7:165–97. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- 6.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:1–12. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronauer MV, Schulz WA, Seifert HH, Ackermann R, Burchardt M. Fibroblast growth factors and their receptors in urological cancers: basic research and clinical implications. Eur Urol. 2003;43:309–19. doi: 10.1016/s0302-2838(03)00005-8. [DOI] [PubMed] [Google Scholar]
- 8.Berger W, Setinek U, Mohr T, et al. Evidence for a role of FGF-2 and FGF receptors in the proliferation of non-small cell lung cancer cells. Int J Cancer. 1999;83:415–23. doi: 10.1002/(sici)1097-0215(19991029)83:3<415::aid-ijc19>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 9.Berger W, Elbling L, Hauptmann E, Micksche M. Expression of the multidrug resistance-associated protein (MRP) and chemoresistance of human non-small-cell lung cancer cells. Int J Cancer. 1997;73:84–93. doi: 10.1002/(sici)1097-0215(19970926)73:1<84::aid-ijc14>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Sutterluty H, Mayer CE, Setinek U, et al. Down-regulation of Sprouty2 in non-small cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and independent mechanisms. Mol Cancer Res. 2007;5:509–20. doi: 10.1158/1541-7786.MCR-06-0273. [DOI] [PubMed] [Google Scholar]
- 11.Panek RL, Lu GH, Dahring TK, et al. In vitro biological characterization and antiangiogenic effects of PD 166866, a selective inhibitor of the FGF-1 receptor tyrosine kinase. J Pharmacol Exp Ther. 1998;286:569–77. [PubMed] [Google Scholar]
- 12.Mohammadi M, McMahon G, Sun L, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–60. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Kharbanda S, Hanfelt J, Kern FG. Both autocrine and paracrine effects of transfected acidic fibroblast growth factor are involved in the estrogen-independent and antiestrogen-resistant growth of MCF-7 breast cancer cells. Cancer Res. 1998;58:352–61. [PubMed] [Google Scholar]
- 14.Steiner E, Holzmann K, Pirker C, et al. The major vault protein is responsive to and interferes with interferon-g-mediated STAT1 signals. J Cell Sci. 2006;119:459–69. doi: 10.1242/jcs.02773. [DOI] [PubMed] [Google Scholar]
- 15.Johnson DE, Lu J, Chen H, Werner S, Williams LT. The human fibroblast growth factor receptor genes: a common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol Cell Biol. 1991;11:4627–34. doi: 10.1128/mcb.11.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korynevska A, Heffeter P, Matselyukh B, et al. Mechanisms underlying the anticancer activities of the angucycline landomycin E. Biochem Pharmacol. 2007;74:1713–26. doi: 10.1016/j.bcp.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Allerstorfer S, Sonvilla G, Fischer H, et al. FGF5 as an oncogenic factor in human glioblastoma multiforme: autocrine and paracrine activities. Oncogene. 2008;27:4180–90. doi: 10.1038/onc.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonvilla G, Allerstorfer S, Stattner S, et al. FGF18 in colorectal tumour cells: autocrine and paracrine effects. Carcinogenesis. 2008;29:15–24. doi: 10.1093/carcin/bgm202. [DOI] [PubMed] [Google Scholar]
- 19.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 20.Berger W, Setinek U, Hollaus P, et al. Multidrug resistance markers P-glycoprotein, multidrug resistance protein 1, and lung resistance protein in non-small cell lung cancer: prognostic implications. J Cancer Res Clin Oncol. 2005;131:355–63. doi: 10.1007/s00432-004-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornmann M, Ishiwata T, Matsuda K, et al. IIIc isoform of fibroblast growth factor receptor 1 is overexpressed in human pancreatic cancer and enhances tumorigenicity of hamster ductal cells. Gastroenterology. 2002;123:301–13. doi: 10.1053/gast.2002.34174. [DOI] [PubMed] [Google Scholar]
- 22.Kwabi-Addo B, Ropiquet F, Giri D, Ittmann M. Alternative splicing of fibroblast growth factor receptors in human prostate cancer. Prostate. 2001;46:163–72. doi: 10.1002/1097-0045(20010201)46:2<163::aid-pros1020>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 23.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Giaccone G, Rodriguez JA. EGFR inhibitors: what have we learned from the treatment of lung cancer? Nat Clin Pract Oncol. 2005;2:554–61. doi: 10.1038/ncponc0341. [DOI] [PubMed] [Google Scholar]
- 25.Arora A, Scholar EM. Role of tyrosine kinase inhibitors in cancer therapy. J Pharmacol Exp Ther. 2005;315:971–9. doi: 10.1124/jpet.105.084145. [DOI] [PubMed] [Google Scholar]
- 26.Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556–68. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 27.Muller-Tidow C, Diederichs S, Bulk E, et al. Identification of metastasis-associated receptor tyrosine kinases in non-small cell lung cancer. Cancer Res. 2005;65:1778–82. doi: 10.1158/0008-5472.CAN-04-3388. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Weir BA, LaFramboise T, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–70. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 29.McKeehan WL, Wang F, Kan M. The heparan sulfate-fibroblast growth factor family: diversity of structure and function. Prog Nucleic Acid Res Mol Biol. 1998;59:135–76. doi: 10.1016/s0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- 30.Ozen M, Giri D, Ropiquet F, Mansukhani A, Ittmann M. Role of fibroblast growth factor receptor signaling in prostate cancer cell survival. J Natl Cancer Inst. 2001;93:1783–90. doi: 10.1093/jnci/93.23.1783. [DOI] [PubMed] [Google Scholar]
- 31.Aoki T, Kato S, Fox JC, et al. Inhibition of autocrine fibroblast growth factor signaling by the adenovirus-mediated expression of an antisense transgene or a dominant negative receptor in human glioma cells in vitro. Int J Oncol. 2002;21:629–36. [PubMed] [Google Scholar]
- 32.Ozen M, Medrano EE, Ittmann M. Inhibition of proliferation and survival of melanoma cells by adenoviral-mediated expression of dominant negative fibroblast growth factor receptor. Melanoma Res. 2004;14:13–21. doi: 10.1097/00008390-200402000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Auguste P, Gursel DB, Lemiere S, et al. Inhibition of fibroblast growth factor/fibroblast growth factor receptor activity in glioma cells impedes tumor growth by both angiogenesis-dependent and -independent mechanisms. Cancer Res. 2001;61:1717–26. [PubMed] [Google Scholar]
- 34.Miraux S, Lemiere S, Pineau R, et al. Inhibition of FGF receptor activity in glioma implanted into the mouse brain using the tetracycline-regulated expression system. Angiogenesis. 2004;7:105–13. doi: 10.1007/s10456-004-1037-0. [DOI] [PubMed] [Google Scholar]
- 35.Grand EK, Chase AJ, Heath C, Rahemtulla A, Cross NC. Targeting FGFR3 in multiple myeloma: inhibition of t(4;14)-positive cells by SU5402 and PD173074. Leukemia. 2004;18:962–6. doi: 10.1038/sj.leu.2403347. [DOI] [PubMed] [Google Scholar]
- 36.Paterson JL, Li Z, Wen XY, et al. Preclinical studies of fibroblast growth factor receptor 3 as a therapeutic target in multiple myeloma. Br J Haematol. 2004;124:595–603. doi: 10.1111/j.1365-2141.2004.04814.x. [DOI] [PubMed] [Google Scholar]
- 37.Udayakumar TS, Bair EL, Nagle RB, Bowden GT. Pharmacological inhibition of FGF receptor signaling inhibits LNCaP prostate tumor growth, promatrilysin, and PSA expression. Mol Carcinog. 2003;38:70–7. doi: 10.1002/mc.10146. [DOI] [PubMed] [Google Scholar]
- 38.Ishibe T, Nakayama T, Okamoto T, et al. Disruption of fibroblast growth factor signalpathway inhibits the growth of synovialsarcomas: potential application of signal inhibitors to molecular target therapy. Clin Cancer Res. 2005;11:2702–12. doi: 10.1158/1078-0432.CCR-04-2057. [DOI] [PubMed] [Google Scholar]
- 39.Young LC, Campling BG, Cole SP, Deeley RG, Gerlach JH. Multidrug resistance proteins MRP3, MRP1, and MRP2 in lung cancer: correlation of protein levels with drug response and messenger RNA levels. Clin Cancer Res. 2001;7:1798–804. [PubMed] [Google Scholar]
- 40.Cavallaro U, Niedermeyer J, Fuxa M, Christofori G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat Cell Biol. 2001;3:650–7. doi: 10.1038/35083041. [DOI] [PubMed] [Google Scholar]
- 41.Garofalo A, Naumova E, Manenti L, et al. The combination of the tyrosine kinase receptor inhibitor SU6668 with paclitaxel affects ascites formation and tumor spread in ovarian carcinoma xenografts growing orthotopically. Clin Cancer Res. 2003;9:3476–85. [PubMed] [Google Scholar]
- 42.Gowardhan B, Douglas DA, Mathers ME, et al. Evaluation of the fibroblast growth factor system as a potential target for therapy in human prostate cancer. Br J Cancer. 2005;92:320–7. doi: 10.1038/sj.bjc.6602274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li T, Ling YH, Goldman ID, Perez-Soler R. Schedule-dependent cytotoxic synergism of pemetrexed and erlotinib in human non-small cell lung cancer cells. Clin Cancer Res. 2007;13:3413–22. doi: 10.1158/1078-0432.CCR-06-2923. [DOI] [PubMed] [Google Scholar]
- 44.Piperdi B, Ling YH, Perez-Soler R. Schedule-dependent interaction between the proteosome inhibitor bortezomib and the EGFR-TK inhibitor erlotinib in human non-small cell lung cancer cell lines. J Thorac Oncol. 2007;2:715–21. doi: 10.1097/JTO.0b013e3180f60bb3. [DOI] [PubMed] [Google Scholar]
- 45.Shimoyama T, Koizumi F, Fukumoto H, et al. Effects of different combinations of gefitinib and irinotecan in lung cancer cell lines expressing wild or deletional EGFR. Lung Cancer. 2006;53:13–21. doi: 10.1016/j.lungcan.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 46.Song S, Wientjes MG, Gan Y, Au JL. Fibroblast growth factors: an epigenetic mechanism of broad spectrum resistance to anticancer drugs. Proc Natl Acad Sci U S A. 2000;97:8658–63. doi: 10.1073/pnas.140210697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cascone T, Gridelli C, Ciardiello F. Combined targeted therapies in non-small cell lung cancer: a winner strategy? Curr Opin Oncol. 2007;19:98–102. doi: 10.1097/CCO.0b013e328011beec. [DOI] [PubMed] [Google Scholar]
- 48.Gridelli C. Targeted therapies and non-small-cell lung cancer: new developments. Curr Opin Oncol. 2007;19:75–7. doi: 10.1097/CCO.0b013e328012a05b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.