Summary
engrailed is required to establish and maintain developmental compartments within each segment of the fly. To understand the role of the engrailed protein in this process, we have raised antibodies against engrailed and have visualized an engrailed protein in embryos by indirect immunofluorescence. The protein accumulates in the nucleus, supporting the notion that engrailed is a regulatory factor. The first pattern of expression is in alternating segments followed by expression in every segment, suggesting that engrailed may be responding to pair-rule segmentation gene products. Overall, engrailed protein levels peak in areas undergoing morphogenesis. Finally, the complex final form of the head and terminalia derive from earlier simple subdivision of these areas into developmental fields by engrailed.
Introduction
The formation of the segmental body plan of Drosophila provides a model system for analysis of the mechanisms involved in embryonic pattern formation. A 2 hr period of rapid nuclear divisions converts the zygote into a shell of about 6000 nuclei at the periphery of a syncytial cytoplasm. Cellularization of each of these nuclei produces the cellular blastoderm. Although the embryo shows no morphologically evident pattern at this stage, a molecular prepattern is somehow encoded within individual cells because the approximate position to which their descendents will contribute is already specified (Wieschaus and Gehring, 1976; Szabad et al., 1979; Simcox and Sang, 1983). During a 7 hr period of morphogenetic movements of gastrulation this prepattern is interpreted and becomes visibly manifest as the basic segmental divisions. A genetic and molecular analysis of the genes involved in the creation of this prepattern will lead us toward an understanding of the molecular mechanisms involved.
Mutations in a number of genes produce discrete defects in the segmental pattern (Nüsslein-Volhard and Wieschaus, 1980). The patterns of defects seen in these segmentation mutants suggest that at least two repeat periods characterize the embryonic pattern. Some mutations (e.g., engrailed) produce defects in every segment, others (e.g., fushi tarazu) produce defects with a periodicity that has a paired-segment repeat and others (e.g., Kruppel) give distinctive gaps in the pattern (Kornberg, 1981a; Wakimoto et al., 1984; Wieschaus et al., 1984). The genes affected by these mutations are thought to encode products that regulate segmental pattern formation.
In situ hybridization of cloned DNA probes to RNA immobilized in tissue sections has been used to examine the spatial distribution of transcripts from three segmentation genes, engrailed, fushi tarazu, and Kruppel (Kornberg et al., 1985; Hafen et al., 1984; Knipple, Rosenberg, and Jackle, submitted). For each of these genes, the spatial pattern of early accumulation of transcripts anticipates the position of defects that would become evident in the corresponding mutants. These observations have suggested that it is the spatial pattern of expression of these segmentation genes that gives rise to morphological pattern by limiting the action of these developmental regulators to the appropriate positions.
engrailed gene function is required for embryonic and adult pattern formation. The function of engrailed was examined by classical studies that focused on the requirement for engrailed in the imaginal discs that give rise to adult structures. These studies showed that the adult segments are subdivided into anterior and posterior compartments (Garcia-Bellido et al., 1973; Lawrence et al., 1979) that the cells contributing to these compartments are clonally distinct throughout most of development, and that this distinction requires engrailed function in the cells of the posterior compartment (Lawrence and Morata, 1976; Kornberg, 1981b; Lawrence and Struhl, 1982). This led to the proposal that engrailed encodes a regulator that acts, wherever it is expressed, to direct cells along a pathway of posterior compartment development. Analyses of engrailed mutant phenotypes and spatial patterns of gene expression strongly support this proposed function and suggest that embryonic and adult functions are similar (Kornberg et al., 1985).
The capacity to analyze the spatial pattern of engrailed distribution in embryos can offer new insights into the basis of embryonic pattern formation. The mechanisms that determine the spatial distribution of developmental regulators such as engrailed must constitute a fundamental step in the process of pattern formation. Here we present a detailed analysis of how the pattern of engrailed expression first becomes established. These observations are discussed in terms of the possible mechanisms responsible for its restricted pattern of expression. Additionally, because the engrailed product acts as a determinant of developmental fate, molecular probes for its expression offer us a way to visualize a “developmental field” early in development. We can thereby observe how a simple, segmental molecular prepattern is transformed to produce more complex structures during morphogenesis.
Results
Expression of engrailed Fusion Proteins
We isolated recombinant clones that express segments of the engrailed coding sequence as tribrid fusion proteins (ompF-engrailed-lacZ) in E. coli (see Experimental Procedures). Two of these recombinant plasmids, penORF1 and penORF2, were selected for detailed analysis. Restriction mapping, Southern and Northern blot analysis, and DNA sequencing indicated that the engrailed inserts are present in the same orientation and reading frame as they are in the major exon of an engrailed cDNA (Poole et al., 1985). The inserts are 391 and 505 bp long and do not overlap (Figure 1C). They do not contain homeo box sequences, but do exhibit some homology to sequences present at several developmentally regulated loci (Wharton et al., 1985).
Figure 1. Expression of enORF Fusion Proteins and Analysis of Insert-Specific Antibodies.
(A) Extracts run on 7.5% PAGE-SDS gel stained with Coomassie Blue. Lanes 1 and 2, TK1046 with penORF1 and penORF2, respectively. Lane H, TK1046 with an ompF-lacZ fusion as a control. Lane D, strain deleted for lacZ. Markers: myosin (200 kd), β-galactosidase (116 kd), phosphorylase B (93 kd), and bovine serum albumin (69 kd). Bracket: position of engrailed fusion proteins. In lane 1, we attribute the presence of two novel bands and the presence of a band the size of β-galactosidase to partial degradation of the fusion protein.
(B) Autoradiogram of immunoblotted gel similar to that in A. Left, probed with anti-enORF1. Right, anti-enORF2. The pattern was detected with 125I-labeled donkey anti-rabbit secondary antibody (Amersham). M, 14C-labeled molecular weight markers: myosin, phosphorylase B, bovine serum albumin, ovalbumin (46 kd), and carbonic anhydrase (30 kd). Left, lane 1 shows that this affinity-purified antisera detects a band the size of the fusion protein and additional bands presumed to be degradation products. Negligible cross-reaction is seen to β-galactosidase or the penORF2 fusion. Similarly, as shown on the right, the anti-enORF2 serum is specific for the penORF2 fusion.
(C) Diagram of the engrailed cDNA (Poole et al., 1985). Filled boxes numbered 1 and 2, enORFs characterized in this study. Open boxes mark position of the homeo box sequence.
Extracts from cells expressing these fusion proteins are displayed in Figure 1A. Gel-purified fusion protein was used to raise antisera, which was subsequently affinity-purified to isolate antibodies directed against the engrailed determinants. Figure 1B demonstrates the specificity of the affinity-purified sera on immunoblots of bacterial extracts. Each serum recognizes the fusion protein against which it was raised, in addition to some break-down or premature termination products. Though these sera were independently raised against nonoverlapping portions of the engrailed coding sequences, they gave indistinguishable results when used to localize the engrailed protein by indirect immunofluorescence.
Synopsis of Embryogenesis
A short description of D. melanogaster embryogenesis will assist in the presentation of our results below. Detailed treatments have been presented by Turner and Mahowald (1976; 1977; 1979) and by Hartenstein and Campos-Ortega (1984; 1985).
Within a few minutes following cellularization (2.5 hr after egg laying, AEL) the complex morphogenetic movements of gastrulation begin with a set of invaginations that establish the primary germ layers, the ectoderm, mesoderm, and endoderm. In the first of these movements the presumptive mesoderm invaginates along the ventral midline forming the ventral furrow (VF). About 5 min later, the transverse cephalic furrow begins to form laterally near the anterior edge of the ventral furrow. Posterior to the developing cephalic furrow, movements and changes in cell shape begin to compact the ectoderm on the ventral surface of the embryo (3 hr AEL). The ectoderm and underlying mesoderm constitute the germ band, which elongates posteriorly around the tip of the embryo and, folding back over itself, extends along the dorsal surface to a position adjacent to the presumptive head segments (5–6 hr AEL). It is only after full germ-band elongation that the first outward signs of segmentation appear transiently, a series of lateral grooves in the ectoderm spaced at segmental repeat intervals.
Subsequently, the germ band shortens (8–9 hr AEL). During this time, the final segmental appearance of the embryo becomes evident. Additionally, there are complicated morphogenetic movements in the head and terminalia of the embryo. In the head, movements produce the oral (gnathal) lobes, that is, the labial, maxillary, and mandibular oral appendages; while in the terminal abdominal segments (A8–A10), reorganization leads to eventual segment fusion and the elaboration of specializations such as posterior spiracles, sense organs, and analia. Shortly after germ-band shortening, the embryonic head structures involute (about 12 hr AEL). An acephalic larva hatches with three thoracic segments, seven abdominal segments, and a complex eighth abdominal-terminal segment externally visible (22 hr AEL).
Many of these dramatic movements of gastrulation are highlighted by visualizing the temporal and spatial patterns of accumulation of the engrailed protein.
The engrailed Protein Is Localized to the Nucleus
Figure 2 shows a lateral view of an embryo (3.5 hr AEL) stained with affinity-purified anti-engrailed antibodies and the DNA-specific fluorochrome, DAPI. Both treatments highlight the nuclei of cells, indicating that the engrailed immunofluorescent signal is localized to the nucleus. All nuclei are lightly stained with the antibody, but as shown below, the more intense signal is caused by the engrailed protein. Thus, the engrailed antigen is confined to particular nuclei forming a series of stripes having segmental periodicity.
Figure 2. Nuclear Localization of engrailed and Its Dispersal during Mitosis.
Detail of embryo shown in Figure 6a (3.5 hr AEL). Left, indirect immunofluorescence with anti-engrailed antibody. Arrow marks position of two mitotic cells exhibiting diffuse fluorescence. Center and right, nuclei revealed by DAPI fluorochrome. Right shows engrailed expressing nuclei blackened out for ease of alignment. Arrows mark mitotic chromosomes in cells with diffuse engrailed signal. Vertical hash-marks delimit engrailed stripe. Objective 63×.
To determine whether the engrailed sera were specific for the engrailed gene product, we examined an engrailed deficiency mutant (en28lenSF31). The results (Figure 3) demonstrate that the strong immunofluorescent signal seen in wild-type embryos is from a protein encoded by the engrailed region (or, less likely, a protein the expression of which is controlled by the engrailed region). Aside from these results with engrailed deficiencies, we are convinced that the intense, localized expression is due to the engrailed gene product for three reasons. First, two antisera, each raised against different portions of the engrailed coding sequence, give the same patterns. Second, at least for some of these patterns, the results are as expected from classical genetic analysis of the engrailed locus. Finally, the patterns seen by immunofluorescence are consistent with the patterns detected by in situ hybridization of cloned sequences to RNA in tissue sections (Kornberg et al., 1985).
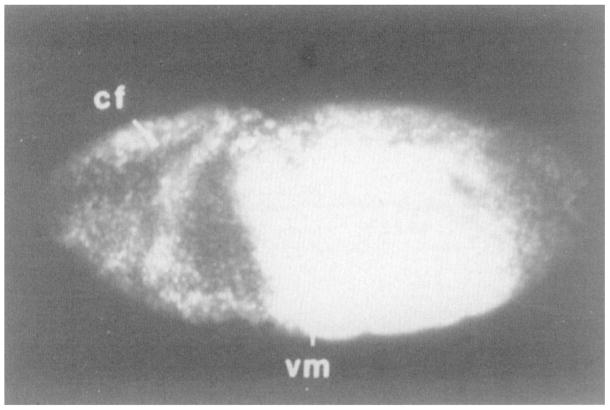
Figure 3. Stripes Are a Product of the engrailed Locus.
A ventro-lateral view of an anti-engrailed stained en28lenSF31 deficiency embryo during germ-band elongation. No striped immunofluorescent signal is evident. Photograph is overexposed to highlight low levels of signal due to cross-reacting non-engrailed nuclear antigens. Additionally, nonspecific staining of the central yolk region is seen. Anterior is to the left in all figures. cf, cephalic furrow; vm, ventral midline. Objective 25×, in this and remaining micrographs.
In addition to the intense, localized engrailed signal, the antisera give a weak signal in all nuclei. Controls demonstrated that all of the nuclear staining is due to the engrailed antisera. In the deficiency embryos, though there is no hint of intense staining whether localized into stripes or not, we see a weak, general nuclear staining. Thus, the engrailed sera cross-react with non-engrailed nuclear antigens, but only weakly. Consequently, we are uncertain whether the weak immunofluorescence seen between engrailed stripes in wild-type embryos reflects low level engrailed expression or simply cross-reactivity. This uncertainty does not affect our analysis of the pattern of intense, engrailed immunofluorescence.
Since the engrailed locus encodes a sequence-specific DNA binding protein (Desplan, Theis, and O’Farrell, submitted), we anticipated that the immunofluorescent signal would be localized to chromatin because of interactions with DNA, and therefore would condense with mitotic chromosomes. However, in mitotic cells, the location of the immunofluorescent staining did not parallel the location of DAPI staining. Rather, a dispersed engrailed signal is detected throughout the cell (Figure 2). The lack of immunofluorescence of mitotic chromosomes could result if antigenic determinants recognized by our sera are not accessible in mitotic chromosomes.
Onset of engrailed Protein Accumulation in Stripes
Among the many changes in the pattern of engrailed expression that occur throughout development, the most dramatic feature of engrailed protein accumulation is its localization in a series of stripes that transect the anterior–posterior axis of the embryo. These highlight the segment anlagen before they become morphologically identifiable. Because we are particularly interested in processes that define the spatially restricted patterns of engrailed expression, we have examined the earliest stages of development of this discrete pattern. The basic periodic pattern that characterizes the oral, thoracic, and abdominal segments is established during a 30 min period between ventral furrow formation and the start of germ-band elongation. Within this short time, the pattern of engrailed appearance is quite complex (Figure 4).
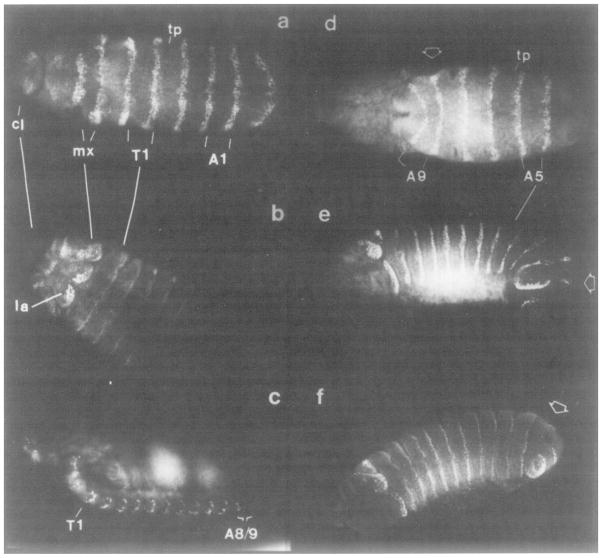
Figure 4. Onset of engrailed Protein Accumulation in Early Embryos.
(a and b) Ventral surface down. (c and d) Ventral surface toward viewer. Embryo in d is approximately 30 min older than embryo in a. (a) Embryo just beginning gastrulation, ventral furrow has formed (not visible here). engrailed antigen is first visible in the posterior regions of the maxillary (mx) segment primordium. (b) Cephalic furrow is just forming anterior to maxillary stripe. Accumulation of engrailed protein is evident in more posterior segment primordia. This first pattern is in alternating segments. Antigen is visible in more ventrally located nuclei first. By this stage the maxillary stripe is two cells wide in some areas. (c) Cephalic furrow has deepened, almost drawing the maxillary stripe into its folds. The alternating pattern is still obvious, but the posterior region of each segment anlage is beginning to accumulate engrailed. The area posterior to A2 is delayed in engrailed accumulation (cf b and c with d). vf, ventral furrow. (d) Onset of germ-band elongation, the cephalic furrow is quite deep with much of the maxillary stripe having been pulled in. The maxillary through A2 signals are equalizing in intensity. Posterior to A2, each alternate stripe is slightly more intense than the one in between. The signals have progressed laterally over the dorsal ectoderm. A2, second abdominal segment.
When they first become apparent, the stripes of engrailed accumulation are a single cell wide in most areas. The stripes are usually separated by about three cells in which nuclear staining is less intense (Figures 4c and 4d). The cells are not precisely aligned but rather seem to be primarily in a hexagonal close-packed array. As a result, counting rows of cells is somewhat imprecise. Nonetheless, it is clear that in many stripes, each positive cell usually has only two positive neighbors positioned to produce an almost straight line transecting the anterior–posterior axis of the embryo. Specification of engrailed expression must require very precise regulatory interactions that reiterate along the anterior–posterior axis.
The engrailed signal does not appear simultaneously in all presumptive segments. Rather, there are four superimposed patterns of accumulation.
First, a single stripe of engrailed-positive nuclei appears on the lateral ectoderm just posterior to the region where the cephalic furrow will form (Figure 4a). This first stripe of engrailed accumulation is unusual because shortly after its appearance it is two cells wide through some of its length. As the cephalic furrow appears, but before the germ band begins to elongate, engrailed antigen accumulates in more posterior segment primordia.
Second, although there is an overall gradient of engrailed accumulation posteriorly from the cephalic furrow, it is not a smooth one. Surprisingly, engrailed-positive nuclei are initially spaced at alternating segment intervals (Figures 4b and 4c). Within the next few minutes, engrailed antigen does appear in the lagging segment primordia, although it is still at higher levels in the alternate segments corresponding to maxillary, first thoracic, third thoracic, and second, fourth, sixth, and eighth abdominal segments (Figure 4d). The signal comprising the posterior primordia of the mandibular segment develops within the cephalic furrow and is not visible here.
Third, there is also a ventral–dorsal discordancy in accumulation. As engrailed antigen appears, it is more evident in ventral and ventro-lateral nuclei (Figure 4b). As development proceeds, the stripes do progress over most of the dorsal ectoderm (Figure 4d and Figure 6a).
Figure 6. Reorganization of Segmented Developmental Fields.
(a and b) Morphological transformation of maxillary and labial segments into lobed appendages. Ventral view. cl, clypeolabrum; la, labial segment; tp, tracheal pit. In b, note that the signal is decaying in the thoracic and abdominal segments, but there is a strong signal in the maxillary and labial lobes. (c) Embryo after head involution (11 hr AEL). Ventral down. Nuclei in posterior regions of the segmented ventral nervous system accumulate engrailed protein. The large, diffuse signal is nonspecific yolk staining. (d, e, and f) Reorganization of segments A8, A9, and rudimentary A10 into the fused terminalia. In the dorsal view shown in d, the germ band is still fully extended over the dorsal surface so that the posterior tip of the embryo is just to the left of A9. Open arrow head points to the posterior compartment of A8. This region and a similar area in A9 are shown reorganizing in e and f. (e) Dorso-lateral view, late germ-band shortening. The central yolky mass stains nonspecifically. Note the break in engrailed stripes in A6 and A9. (f) Fully germ-band shortened, during dorsal closure. Lateral view, dorsal down. A8 and A9 ectodermal signals have shifted quite far dorsally. The engrailed signal remains high in the terminalia, and in the labial and maxillary lobes toward the anterior end (see b).
Finally, engrailed protein accumulates first in the regions from the maxillary through A2 segments, and only then in areas posterior to A2 (compare Figures 4b and 4c with 4d).
Early in germ-band elongation (3.5 hr AEL), the engrailed antigen accumulates to an equivalent extent in all presumptive segments in the trunk of the embryo (not shown, but see Figure 5a, 4 hr AEL). Thus specification of compartment boundaries over the length of the embryo occurs in a complex but ordered sequence. The progression is anterior to posterior, ventral to dorsal, and alternating segment to every segment.
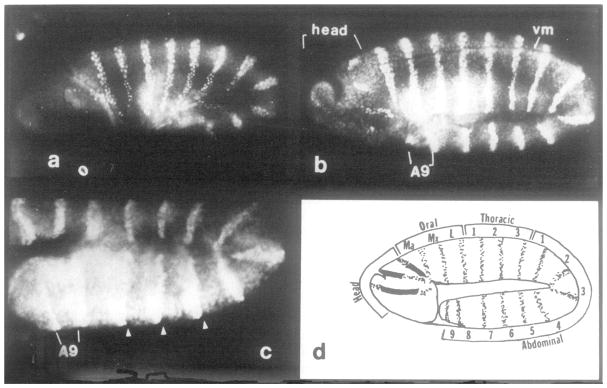
Figure 5. Number and Position of Segments and Parasegments as Revealed by engrailed Protein Localization.
(a) Embryo during germ-band elongation (4 hr AEL), ventral surface up. Germ band has extended partway around the end of animal (right side) and back over 25% of dorsal surface. The maxillary and mandibular stripes are within the cephalic furrow (out of focal plane). engrailed staining is evident in labial, three thoracic, and seven abdominal primordia. (b) Embryo at full germ-band elongation (5.5 hr AEL), same orientation as in a. The deep stomadeal pocket is visible near the anterior end of the head region on the ventral surface. The mandibular and maxiliary signals are evident, since the cephalic furrow has become shallow. Three oral (gnathal), three thoracic, and nine abdominal signals are visible. engrailed protein first appears in the head region in four discrete areas on each lateral face. A9, ninth abdominal segment. (c) Detail of an embryo at full germ-band extension where the transient lateral grooves are evident on the ectoderm. Same orientation and approximate stage as that shown in b. Grooves within segments A5, AS, and A7 are marked by arrows. The grooves are anterior to the engrailed signal and are therefore at the anterior–posterior compartment boundary. Groove to groove marks a parasegment and not a segment. (d) Schematic representation of the segmental divisions in germ-band elongated embryos, as suggested by the pattern of engrailed protein accumulation. The stippling indicates positions where nuclei stain with engrailed antisera (e.g., b). The four positions of staining in the head might be derived from three prepattern stripes as illustrated by the blackened-in regions. Segmental identity can be unambiguously assigned by examining a succession of older embryos. Note that there are nine abdominal stripes, indicating nine complete abdominal segmental divisions, and a rudimentary tenth that lacks a posterior compartment. Thus, the number of parasegments is 15 at this stage in development, if counting is initiated with posterior mandibular (Martinez-Arias and Lawrence, 1985) and extended to anterior tenth abdominal (this work).
When a stripe first becomes evident, not all of the nuclei within the stripe stain equally. Even neighboring nuclei can behave quite differently. It appears as if the induction of engrailed expression in each cell is a stochastic process. A consequence of this is heterogeneity of engrailed appearance within a given stripe.
Within a short time, as the germ band elongates, the engrailed stripe increases in width from one to two or three cells (cf. Figure 4d with Figure 5a). Given this short time interval, it is unlikely that respecification of engrailed expression is taking place. Two processes are responsible for movement of the germ band, oriented cell division along the dorsal ectoderm and rearrangement or interdigitation of cells along the ventral ectoderm (Hartenstein and Campos-Ortega, 1985). Throughout germ-band extension there is no obvious change in the fraction of engrailed positive nuclei, although we have not yet rigorously scored all nuclei in these embryos. To maintain the same proportion of engrailed positive cells, the commitment to express engrailed must be transmitted to both daughter cells during cell divisions (an element of determination). To maintain the integrity of the engrailed-positive stripes requires that any cell movements are governed such that engrailed-expressing cells remain adjacent to one another.
engrailed Protein Localization Marks the Position and Number of Segments Along the Extended Germ Band
Assuming that engrailed expression defines the posterior compartment of each segment (see below), then the position of engrailed staining illuminates the compartment and segment boundaries along the anterior–posterior axis. The total number of stripes of engrailed protein expression is most evident when the germ band is fully extended (Figure 5b). At this stage, the cephalic furrow has shallowed so that the signals corresponding to the labial (la), maxillary (mx), and mandibular primordia are visible on the surface of the embryo. In addition to these three oral stripes, three thoracic and nine abdominal stripes are visible. These 15 signals representing segmental divisions develop relatively early during germ-band elongation (4 hr AEL Figure 5a, or earlier, Figure 4). Their segmental identity can be unambiguously defined by examining a series of stages up to the time of obvious morphological segmentation (Figure 6). Later in embryogenesis, the region posterior to the seventh abdominal segment, along with the oral segment primordia, undergoes extensive reorganization (see below).
It is toward the end of germ-band elongation that the earliest morphological signs of segmentation appear in the embryo. A series of lateral grooves appear transiently along the extended germ band. The engrailed protein accumulates just posterior to each of these grooves in TZ through A8, indicating that the lateral grooves are not at the segmental boundaries but rather are at the anterior–posterior compartment boundary within each segment (Figure 5c). A second segmental marker, tracheal pits, becomes evident at this stage. As shown in Figures 6a and 6d, the pits are located in an unstained region and correspond to neither segmental nor compartmental borders.
The number and position of segments in the head is debated. While the final morphology of head structures is not obviously segmental, these structures are thought to develop from three segmental units that undergo highly specialized patterns of development (for the larval head, see Jurgens et al., submitted; for the adult head, see Struhl, 1981). Our observations fit this interpretation. At late germ-band extension, engrailed staining first appears in the head region in four discrete areas (Figure 5b). Since fate maps show that invaginations and rotational distortions have occurred in the head by this time, it is possible that engrailed expression was specified by an earlier, more simple three-stripe prepattern that was then distorted by cell movements.
Figure 5d summarizes our current interpretation of the total number and position of segment primordia at germ-band elongation.
Germ-Band Shortening: Dynamics of engrailed Localization during Formation of Head and Terminal Structures
At germ-band shortening, when segmentation becomes quite apparent morphologically, it is clear that engrailed protein is indeed restricted to the posterior portion of each thoracic and abdominal segment. Figure 6f shows that a positive signal is seen in a strip of nuclei at the posterior edge of the groove at each segment border. The signal is narrow because a region of the posterior compartment is apparently drawn into the grooves at the segment boundaries.
During and after germ-band shortening, striking patterns of engrailed expression in the anterior region (Figures 6a and 6b) and terminal segments (Figures 6d, 6e, and 6f) parallel the complex morphological transformations of these regions into the oral appendages and the terminalia, respectively. During these movements, a higher level of engrailed antigen accumulation can be seen in posterior portions of the labial lobe, maxillary lobe (Figure 6b), A8 and A9 (Figure 6f), as compared with the thoracic or other abdominal segments.
As an example of the dramatic restructuring of simple segmental divisions into complex three dimensional patterns, we describe the transformations in A8, A9, and A10 that occur toward the end of germ-band shortening. The most terminal engrailed signal is at the posterior region of A9 (Figure 6d). Posterior to this stripe there is nonstaining tissue that is presumed to represent a rudimentary tenth abdominal segment lacking a posterior compartment. Segments A8 to A10 will fuse and will eventually differentiate into the terminalia. The position of engrailed-positive cells is dynamic in these segments during the time leading up to the segment fusion. Given that the reorganization occurs over a relatively short period, 30 min, we assume that changes in the pattern of immunofluorescence reflect relative cell movements rather than respecification of engrailed-expressing cells to nonexpressing cells and vice versa. Initially the signal flanking the ventral midline in A8 and A9 is about as intense as that in the other thoracic and abdominal segments (arrow, Figure 6d). Then engrailed-expressing cells move dorsally to contribute to the developing spiracles and other specializations (Figure 6e). As a result of these movements, “anterior” A8, A9, and A10 cells abut each other along the ventral ectoderm (Figure 6f). The resulting juxtaposition of “anterior” cells, that is, those lacking engrailed antigen, may directly allow the segment fusion known to occur in A8–A10. Thus the developmental fields that started as a simple transection of the anterior–posterior axis have been morphologically transformed into the complex terminalia.
engrailed Protein in the Central Nervous System
Since a requirement for engrailed function in the adult nervous system has been described (Lawrence and Johnston, 1984) engrailed presence in the embryonic nervous system was anticipated. At 3.5 hr AEL (Figure 4) the presumptive neurogenic region of the ectoderm flanking the ventral midline is positive for engrailed protein. As development proceeds, neuroblasts bud inward from the neuroectoderm. Indeed we find that certain nuclei in the segmented ventral nervous system stain positively for engrailed protein at 10 hr of development. Not all nuclei extending laterally from the midline are positive. Staining is strong in the nervous system after the immunofluorescent signal has declined in the ectodermal nuclei of the trunk of the embryo (Figure 6c).
Discussion
The transcripts of several segmentation genes have been shown to be localized in spatially restricted patterns in the early embryo (Hafen et al., 1984; Kornberg et al., 1985). The positions of expression correspond to the pattern of defects caused by mutations at the corresponding loci. This suggests that spatially restricted patterns of expression give rise to morphological pattern by limiting the positions where regulatory gene products function. This perspective identifies two questions regarding the function of segmentation genes in development. How are the localized patterns of expression produced? How is the pattern of expression interpreted to produce morphological form?
The resolution afforded by immunofluorescent detection of the engrailed protein and the ease with which patterns of expression can be analyzed in whole mount preparations show that an extraordinarily elaborate spatial and temporal program of engrailed gene expression is superimposed on the fundamentally simple pattern of subdivision of the anterior–posterior axis of the embryo. Analyzing the way in which the early engrailed pattern is established gives indications as to how its spatially restricted expression is specified. Additionally, by examining the changes in the distribution of the engrailed protein throughout gastrulation, we see how complex morphological form is first generated by simple subdivision of the anterior–posterior axis into developmental fields.
The engrailed Protein as a Developmental Regulator
On the basis of the extraordinary conservation of the homeo box sequence, a number of different regulatory genes of Drosophila are thought to share a fundamental mechanism of action (McGinnis et al., 1984a; Scott and Weiner, 1984). Two of these genes, fushi tarazu and Utrabithorax, encode products that share other features with the engrailed protein, all are nuclear proteins (this report; White and Wilcox, 1984; Beachy et al., 1985; Carroll and Scott, 1985) and all have unusual stretches of homo-polymeric amino acid sequences (Laughon and Scott, 1984; Beachy et al., 1985; Poole et al., 1985). On the basis of these observations and a recent demonstration that engrailed encodes a sequence-specific DNA binding activity (Desplan, Theis, and O’Farrell, submitted), we favor the proposal that their shared mechanism of action is through regulation of transcription.
We note that engrailed protein levels peak in different regions of the embryo when these areas are undergoing morphogenesis (trunk, oral segments, terminal segments, and nervous system). Although the significance of the correlation and the mechanistic implications are unclear, the observation is consistent with the role of engrailed in maintaining segment integrity. engrailed is thought to mark groups of cells within each segment establishing them as members of the posterior compartment. We would view the assignment of compartment identity as a critical step preceeding any ordered movement of cells.
What Governs engrailed Expression
By the time nuclei have cellularized (the cellular blastoderm), individual cells must have received instruction as to their relative positions in the embryo (Simcox and Sang, 1983). Though largely speculative, it is often proposed that a chemical gradient across a spatial domain is produced and imparts this positional information to cells (Lawrence, 1973; Meinhardt, 1977; Kauffman et al., 1978; Russell, 1985). Such a mechanism cannot easily account for the extraordinarily complex pattern of initial expression of engrailed. We conclude that engrailed gene expression is specified by regulators distributed in a complex pattern rather than in a simple gradient.
The early pattern of engrailed expression in alternate segments is provocative. First, in most engrailed lethal genotypes, cuticular defects, though seen in every segment, are most severe in the alternate segments that also show early engrailed protein accumulation (Nüsslein-Volhard and Wieschaus, 1980; Kornberg, 1981a; Gubb, 1985). Perhaps this observation reflects a more stringent requirement for engrailed function at these paired-segment intervals. Second, engrailed expression appears to respond to a prepattern with paired-segment periodicity. Because segmentation genes in the pair-rule class are expressed with an alternate segment periodicity, we propose that some of these genes influence engrailed expression. Further, since expression of pair-rule genes is spatially restricted to alternate segment intervals, the gene product or combination of gene products that specify engrailed expression must differ in even, as opposed to odd, segments.
Could pair-rule gene products influence engrailed expression directly? There are two suggestions that this might be so. First, the earliest accumulation of the protein product of a particular pair-rule gene, fushi tarazu, is observed only about 15 min prior to the onset of engrailed accumulation into stripes (Carroll and Scott, 1985; and this report). Given that characterized examples of induction of gene expression indicate a lag time roughly comparable to this short time interval (Brown et al., 1984; Ucker and Yamamoto, 1984), this suggests that the route of pair-rule gene product action on engrailed expression would be relatively direct. The second argument is based on our current views regarding the activity of developmental genes containing a homeo box sequence. Based on homology arguments (Laughon and Scott, 1984; Sheperd et al., 1984) and the recent observation that engrailed sequences encode a sequence-specific DNA binding activity (Desplan, Theis, and O’Farrell, submitted), it seems likely that these regulatory proteins act directly to regulate transcription. This leads us to the testable hypothesis that some of the products of pair-rule loci will act directly on engrailed sequences to regulate transcription of the locus.
The precision of specification of engrailed gene expression is apparent from the observation that early expression is restricted to one cell in four, reiterated along the anterior–posterior axis. Since mutations at other segmentation loci cause cuticular pattern defects that are also reiterated along the body axis, we expect similarly precise specification of expression of these other loci. Additionally, this expectation implies that within a segment primordium, each band of nuclei around the cellular blastoderm is uniquely specified by one or a combination of segmentation gene products.
When the striped engrailed immunofluorescent signal first develops, the intensity of adjacent nuclei varies substantially. As a result, there are transient gaps in a stripe of positive nuclei. This pattern suggests that the precise time at which expression is induced within a stripe is a stochastic process. Perhaps this indeterminant, but short, delay before commitment reflects the interactions of positive and negative factors that together control engrailed expression. We can point to another example where a regulatory event is controlled by the interaction of a number of factors and therefore gives the appearance of a stochastic process. In the bacteriophage lambda lysislysogeny decision (reviewed in Herskowitz and Hagen, 1980) infecting lambda sits poised in a delicate balance for a short but indeterminant length of time before committing itself to a particular mode of growth.
Beyond the striking segmental and alternate segment pattern, we have detailed several other complexities in the establishment of engrailed expression (e.g., the lag in expression in dorsal ectoderm or the lag posterior to A2). In addition, we see later events (e.g., expression in the head and CNS) suggesting that the complex spatial and temporal program of engrailed expression continues.
All of these observations tend to support hierarchical and combinatorial models of interaction among the segmentation gene products.
engrailed Is a Marker of Anterior–Posterior Compartmental and Segmental Divisions
Classical experiments studying the behavior of clonally marked cells indicated that the adult epidermis is divided into anterior and posterior developmental compartments as well as segments (Garcia-Bellido et al., 1973). Further studies showed that engrailed function is required to specify cells as members of posterior compartments (Lawrence and Morata, 1976; Kornberg, 1981b; Lawrence and Struhl, 1982). Although analogous experiments cannot be done to show that the embryo is similarly subdivided, the embryonic phenotypes of engrailed mutations are consistent with the idea that this gene performs a similar function during embryonic segment formation (Nüsslein-Volhard and Wieschaus, 1980; Kornberg, 1981a). However, it is the pattern of expression (Kornberg et al., 1985; and this report) that dramatically supports this notion. Therefore, we consider that it is engrailed action that specifies cells as members of a posterior compartment and within this report we have referred to groups of engrailed-expressing cells as the posterior compartment of embryonic segments.
The visualization of the pattern of engrailed expression permits us to relate the molecular and morphological features of segmentation. For example, at the extended germ-band stage the reiterated grooves that appear on the ectoderm are anterior to the region of engrailed staining and are therefore, at the anterior–posterior compartment boundary within each segment and not at the segment boundary. This localization of the grooves is consistent with their recent description as parasegmental (Martinez-Arias and Lawrence, 1985). The tracheal pits, on the other hand, lie at neither the segment nor the compartment border. Their location suggests that additional positional information must exist within the segment intervals at these early stages. We expect that segmentation genes other than engrailed are spatially regulated and together the distribution of these gene products represents the molecular code for this positional information.
Complex Final Form May Derive from Initially Simple Subdivision
The final positions and interactions of the segmentation gene products are thought to establish the molecular code that is interpreted and elaborated upon during the morphogenetic events of gastrulation. While the early pattern of fushi tarazu and engrailed expression appears as a relatively simple subdivision along most of the anterior-posterior axis, the final structures, especially in the tail and head, have complex morphologies. In the abdominal region posterior to A7, the area is initially subdivided into simple developmental fields by engrailed just as in more anterior segments. Apparently, cell movement reorganizes A8, A9, and A10 into fused terminalia (Figures 6d, 6e, and 6f). In contrast to the transformation seen in the terminal segments, the pattern of engrailed expression in the head already looks complex when we first detect it. However, it appears here only after groups of cells in the head have changed their relative positions along the anterior–posterior axis (Struhl, 1981; Jurgens et al., submitted). Perhaps engrailed expression in the head is also following a fundamentally simple prepattern (Figure 5), but this pattern has been distorted by gastrulation movements.
Generalizations about Segment Specification and Final Form
The conservation of homeo box sequences has led to the supposition that the mechanisms involved in Drosophila development are fundamental and used almost universally in the specification of embryonic pattern (McGinnis et al., 1984b). These proposals concern conservation of molecular mechanisms and do not necessarily imply conservation of morphological form. Because the basic body plan of Drosophila is segmental, the action of segmentation genes might initially be seen inexorably connected to this developmental form. However, the involvement of engrailed in the development of the Drosophila head and terminalia suggest that the same mechanisms can be involved in pattern development where the final form is no longer obviously segmental. We suggest that herein lie generalities that may be directly applicable to vertebrate development.
Experimental Procedures
Expression Clones
Part of a major engrailed transcript is encoded within a 3.1 kb genomic Eco RI-Xho I restriction fragment (Drees et al., unpublished). We searched for open translational reading frames within this segment using an expression vector system (Weinstock et al., 1983). These vectors, pORF-1 and pORF-2, contain the ompF promoter and leader peptide coding sequence coupled out of frame, via a polylinker, to the 5′ end of the lacZ (β-galactosidase) gene. The Eco RI-Xho I fragment was sonicated to randomize ends and was blunt-ended by digestion with S1 nuclease followed by a fill-in reaction with T4 polymerase. The fragments were ligated into the blunt end Sma I site in the pORF-2 poly-linker. E. coli TK1046 was transformed and was plated on L plates containing 100 μg/ml Carbenicillin and 40 μg/ml Xgal and was incubated at 27°C. TK1046 carries a cold-sensitive mutation of the ompB product that is a positive regulator of ompF. At elevated temperature, transcription from the ompF promoter is increased, and the resultant over-production of the fusion protein is lethal to the cell (Weinstock et al., 1983). In a typical experiment, out of 750 transformants screened, 225 exhibited some blue color, with 48 showing dark blue color. When representatives of the different groups were tested for growth at 37°C, 14 of the 48 dark blue colonies showed lethality, indicating an open reading frame insert controlled from the ompF promoter. Five of these carried inserts ranging in size from about 200 bp to about 450 bp. The fusion proteins produced corresponded in size to that expected from the insert. Two of the five clones, designated penORFI and penORF2, detected the same transcript as the original 3.1 kb Eco I-Xho I fragment and were characterized in detail. The lighter blue colonies perhaps result from translational starts in the insert that match the outgoing frame of the β-galactosidase gene. A control fusion was constructed producing an ompF-lacZ hybrid fusion with no insert, by removing the short segment between the Bgl II site in the leader sequence and the down-stream Barn HI site in the polylinker.
Preparation of Fusion Proteins
Overnight cultures of Tk1046 carrying the fusion plasmids were diluted 100x into L broth and 100 μg/ml carbenicillin and grown at 27°C until an OD600 of 0.2–0.3. The cultures were then shifted to 37°C, grown for 2 hr, and harvested. Cells were chilled, precipitated by centrifugation, and frozen at −70°C for storage. The frozen cells from 500 ml of culture were resuspended in 5 ml lysis buffer (100 mM NaCl; 1 mM Na2EDTA; 50 mM Tris-HCl, pH 7.9; 1 mM dithiothreitol; 10 mM benzamidine; and 1 mM PMSF) and were sonicated 3 × 1 min at the maximum intensity possible without foaming. After centrifugation at 35,000 ×g for 60 min 50%–85% of the fusion protein would precipitate with the cell debris and membranes. Aliquots of the supernatant and pellet were processed and electrophoresed on a 3 mm thick 5% acrylamide (29 parts acrylamide: 1 part bisacrylamide) preparative slab gel, which provides a very efficient purification step. The proteins were visualized by blotting to nitrocellulose by capillary action for 1 hr. followed by staining with India ink (Hancock and Tsang. 1983). The fusion protein band was cut out and electroeluted.
lmmunlzation of Rabbits
Male New Zealand white rabbits were injected subcutaneously with 100–150 μg of fusion protein emulsified with an equal volume of Freund’s complete adjuvant (Sigma). Rabbits were boosted (75–100 μg protein) and bled on a 14 day schedule.
Affinity Purification of Anti-engrailed Antibodies
Fusion proteins electroeluted from gels were dialyzed overnight against 0.1 M Hepes, pH 7, at 4°C. The dialyzed protein was coupled to Affigel-10 (Bio-rad). The serum was exhaustively passed over a column containing the ompF-lacZ fusion. The flow-through was then passed through a column containing the specific tribrid fusion against which the serum was raised. The bound antibodies were eluted with 0.1 M glycine (pH 2.5) and were immediately neutralized with 2 M Tris (pH 7.6). Eluted antibodies were dialyzed into phosphate buffered saline, 50% glycerol, and 0.01% sodium azide, and were stored in −20°C.
Indirect Immunofluorescent Staining of Whole Fixed Embryos
Wild-type embryos were from Oregon R parents. The engrailed deficiencies, en28 and enSF31, have been described (Eberlein and Russell, 1984; Kornberg, 1981a). Embryos were permeabilized, fixed, and devitellinized en masse by the procedure of Mitchison and Sedat (1983) as modified by T. Karr (Karr and Alberts, submitted). Fixed embryos were placed in 1 ml test tubes and were incubated with a PBS solution containing 0.1% BSA and 0.1% Triton X-100 for 1–1.5 hr with gentle rotation to ensure mixing. All succeeding antibody incubations and rinses were done in this solution. Embryos were next incubated with a dilution of affinity-purified anti-engrailed serum for 1.5 hr. Primary antibody treatment was followed by rinsing for 1.5 hr. Embryos were stained for 1.5 hr with goat anti-rabbit IgG coupled to rhodamine (Cappel) followed by another 1.5 hr rinse. The embryos were transferred into PBS containing 1 μg/ml DAPI for 1 min, rinsed for 30 min, and mounted in Fluoromount G (Southern Biotechnology Associates).
Light Microscopy
Embryos were examined using a Zeiss standard microscope equipped with epi-fluorescence optics. Rhodamine and DAPI fluorescence were observed using Zeiss filter cassettes 01 and 15, respectively, and Kodak 2415 technical pan film was used to record the images obtained. A Zeiss Plan-Neofluor 25/0.8, a Zeiss Plan-Neofluor 100/1.3, or a Leitz Plan-Neofluor 63/.14 were used as the primary objective lenses.
Acknowledgments
We thank Gerd Jurgens, Sean Carroll, and Matt Scott for valuable discussions and communicating unpublished work; Marika Walter and Tim Karr for materials and advice on immunofluorescent procedures; Claude Desplan for contributions to the characterization of the antisera; members of the lab and especially Liz Sher, for critical reading of the manuscript; Judy Piccini for preparation of the manuscript; Mei Lie Wong for photography. The work was supported by N.S.F. grant DCB-8418016 to P O’F. S. D. is a fellow of the Helen Hay Whitney Foundation; J. M. K. was supported by A. C. S. and Giannini fellowships; J. T. by an N. I. H. training grant.
Footnotes
The work referred to throughout as Knipple, Rosenberg, and Jackle, submitted has been published: Knipple, D. C., Seifert, E., Rosenberg, U. B., Preiss, A., and Jäckle, H. (1985). Spatial and temporal patterns of Krüppel gene expression in early Drosophila embryos. Nature 317, 40–44.
References
- Beachy PA, Helfand SL, Hogness DS. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985;313:545–551. doi: 10.1038/313545a0. [DOI] [PubMed] [Google Scholar]
- Brown AMC, Jeltsch JM, Roberts M, Chambon P. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc Natl Acad Sci USA. 1984;81:6344–6348. doi: 10.1073/pnas.81.20.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Scott MP. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985;43 doi: 10.1016/0092-8674(85)90011-x. this issue. [DOI] [PubMed] [Google Scholar]
- Eberlein S, Russell M. Effects of deficiencies in the engrailed region of Drosophila melanogaster Dev. Biol. 1983;100:227–237. doi: 10.1016/0012-1606(83)90215-4. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalization of the wing disc of Drosophila. Nature New Biol. 1973;245:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- Gubb D. Further studies on engrailed mutants in Drosophila melanogaster Roux’s Arch. Dev Biol. 1985;194:236–246. [Google Scholar]
- Hafen E, Kuroiwa A, Gehring WJ. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984;37:833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- Hancock K, Tsang VCW. India ink staining of proteins on nitrocellulose paper. Anal Biochem. 1983;133:157–162. doi: 10.1016/0003-2697(83)90237-3. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Campos-Ortega JA. Early neurogenesis in wild-type Drosophila melanogaster. Roux’s Arch Dev Biol. 1984;193:308–325. doi: 10.1007/BF00848159. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Campos-Ortega JA. Fate-mapping in wild-type Drosophila melanogaster I. The spatio-temporal pattern of embryonic cell divisions. Roux’s Arch Dev Biol. 1985;194:181–195. [Google Scholar]
- Herskowitz I, Hagen D. The lysis-lysogeny decision of phage λ: explicit programming and responsiveness. Ann Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- Kauffman SA, Shymko RM, Trabert K. Control of Sequential compartment formation in Drosophila. Science. 1978;199:259–269. doi: 10.1126/science.413193. [DOI] [PubMed] [Google Scholar]
- Kornberg T. engrailed: a gene controlling compartment and segment formation in Drosophila. Proc Natl Acad Sci USA. 1981a;78:1095–1099. doi: 10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T. Compartments in the abdomen of Drosophila and the role of the engrailed locus. Dev Biol. 1981b;86:363–372. doi: 10.1016/0012-1606(81)90194-9. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Siden I, O’Farrell P, Simon M. The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment-specific expression. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- Laughon A, Scott MP. Sequence of a Drosophila segmentation gene: protein structure homology with DNA binding proteins. Nature. 1984;310:25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Lawrence PA. A clonal analysis of segment development in Oncopeltus (Hemiptera) J Embryol Exp Morph. 1973;30:681–699. [PubMed] [Google Scholar]
- Lawrence PA, Johnston P. On the role of the engrailed gene in internal organs of Drosophila. EMBO J. 1984;3:2839–2844. doi: 10.1002/j.1460-2075.1984.tb02217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Morata G. Compartments in the wing of Drosophila: a study of the engrailed gene. Dev Biol. 1976;50:321–337. doi: 10.1016/0012-1606(76)90155-x. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Struhl G. Further studies of the engrailed phenotype in Drosophila. EMBO J. 1982;1:827–833. doi: 10.1002/j.1460-2075.1982.tb01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Struhl G, Morata G. Bristle patterns and compartment boundaries in the tarsi of Drosophila. J Embryol Exp Morphol. 1979;51:195–208. [PubMed] [Google Scholar]
- Martinez-Arias A, Lawrence PA. Parasegments and compartments in the Drosophila embryo. Nature. 1985;313:639–642. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Garber RL, Wirz J, Kuroiwa A, Gehring WJ. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984a;37:403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Hart CP, Gehring WJ, Ruddle FH. Molecular cloning and chromosome mapping of a mouse DNA sequence homologous to homeotic genes of Drosophila. Cell. 1984b;38:675–680. doi: 10.1016/0092-8674(84)90262-9. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. A model of pattern formation in insect embryogenesis. J Cell Sci. 1977;23:117–139. [PubMed] [Google Scholar]
- Mitchison T, Sedat J. Localization of antigenic determinants in whole Drosophila embryos. Dev Biol. 1983;99:261–264. doi: 10.1016/0012-1606(83)90275-0. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Poole SJ, Kauvar LM, Drees B, Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985;40:37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Russell MA. Positional information in insect segments. Dev Biol. 1985;108:269–283. [Google Scholar]
- Shepherd JCW, McGinnis W, Carrasco AE, DeRobertis EM, Gehring WJ. Fly and frog homoeo domains show homologies with yeast mating type regulatory proteins. Nature. 1984;310:70–71. doi: 10.1038/310070a0. [DOI] [PubMed] [Google Scholar]
- Scott MP, Weiner AJ. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci USA. 1984;81:4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox AA, Sang JH. When does determination occur in Drosophila embryos? Dev Biol. 1983;97:212–221. doi: 10.1016/0012-1606(83)90078-7. [DOI] [PubMed] [Google Scholar]
- Struhl G. A blastoderm fate map of compartments and segments of the Drosophila head. Dev Biol. 1981;84:386–396. doi: 10.1016/0012-1606(81)90407-3. [DOI] [PubMed] [Google Scholar]
- Szabad J, Schupbach T, Wieschaus E. Cell lineage and development in the larval epidermis of Drosophila melanogaster Dev. Biol. 1979;73:256–271. doi: 10.1016/0012-1606(79)90066-6. [DOI] [PubMed] [Google Scholar]
- Turner FR, Mahowald AP. Scanning electron microscopy of Drosophila embryogenesis. I. The structure of the egg envelope and the formation of the cellular blastoderm. Dev Biol. 1976;50:95–108. doi: 10.1016/0012-1606(76)90070-1. [DOI] [PubMed] [Google Scholar]
- Turner FR, Mahowald AP. Scanning electron microscopy of Drosophila melanogaster embryogenesis. II. Gastrulation and segmentation. Dev Biol. 1977;57:403–416. doi: 10.1016/0012-1606(77)90225-1. [DOI] [PubMed] [Google Scholar]
- Turner FR, Mahowald AP. Scanning electron microscopy of Drosophila melanogaster embryogenesis. III. Formation of head and caudal segments. Dev Biol. 1979;68:96–109. doi: 10.1016/0012-1606(79)90246-x. [DOI] [PubMed] [Google Scholar]
- Ucker DS, Yamamoto KR. Early events in the stimulation of MTV RNA synthesis by glucocorticoids: novel assays of transcription rates. J Biol Chem. 1984;259:7416–7420. [PubMed] [Google Scholar]
- Wakimoto BT, Turner RF, Kaufman TC. Defects in embryogenesis in mutants associated with the Antennapedia gene complex of Drosophila melanogaster. Dev Biol. 1984;102:147–172. doi: 10.1016/0012-1606(84)90182-9. [DOI] [PubMed] [Google Scholar]
- Weinstock GM, Rhys G, Berman ML, Hampar B, Jackson D, Silhavy TJ, Weiseman J, Zweig M. Open reading frame expression vectors: a general method for antigen production in E. coli using protein fusion to beta galactosidase. Proc Natl Acad Sci USA. 1983;80:4432–4436. doi: 10.1073/pnas.80.14.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Gehring W. Clonal analysis of primordal disc cells in the early embryo of Drosophila. Dev Biol. 1976;50:249–265. doi: 10.1016/0012-1606(76)90150-0. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C, Kluding H. Kruppel, a gene whose activity is required early in the zygotic genome for normal embryonic segmentation. Dev Biol. 1984;104:172–186. doi: 10.1016/0012-1606(84)90046-0. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Yedvobnick B, Finnerty VG, Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985;40:55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- White RAH, Wilcox M. Protein products of the bithorax complex in Drosophila. Cell. 1984;39:163–171. doi: 10.1016/0092-8674(84)90202-2. [DOI] [PubMed] [Google Scholar]