Abstract
We demonstrated a convenient, flexible and modular synthetic approach for preparation of a small library of DNA encapsulated supramolecular nanoparticles SNPs⊃DNA and RGD-SNPs⊃DNA with different sizes and RGD target ligand coverage for targeted gene delivery.
Gene therapy generally requires delivery vehicles that are capable of (i) carrying/protecting genetic materials, e.g., DNA and siRNA, and (ii) target-specific delivery to desired tissues or subsets of cells.1 Over the past decades, significant endeavors have been devoted to develop non-viral gene delivery vehicles2,3 as alternatives to their viral counterparts, whose applications are restricted due to the potential safety issues and complex processes of preparing. Among the existing non-viral gene delivery systems,4–8 nanoparticle-based gene delivery vehicles9–12 have received extensive attention.
Recently, we developed a novel assembly approach13 for the preparation of size-controllable supramolecular nanoparticles (SNPs) via multivalent molecular recognition based on β-cyclodextrin (CD) and adamantane (Ad) motifs. A collection of SNPs with sizes ranging from 30 to 450 nm were prepared by mixing three molecular building blocks, including (i) cationic Ad-grafted polyamidoamine dendrimer (Ad-PAMAM), (ii) cationic CD-grafted branched polyethylenimine (CD-PEI) and (iii) Ad-grafted polyethylene glycol (Ad-PEG), all at different concentrations. Given the fact that the interior of SNPs is composed of a cationic Ad-PAMAM/CD-PEI hydrogel network, it is conceivable that SNPs can encapsulate anionic plasmid DNA via electrostatic interactions. This new type of gene delivery system can provide significant protection of the encapsulated DNA from degradation in an extracellular context.
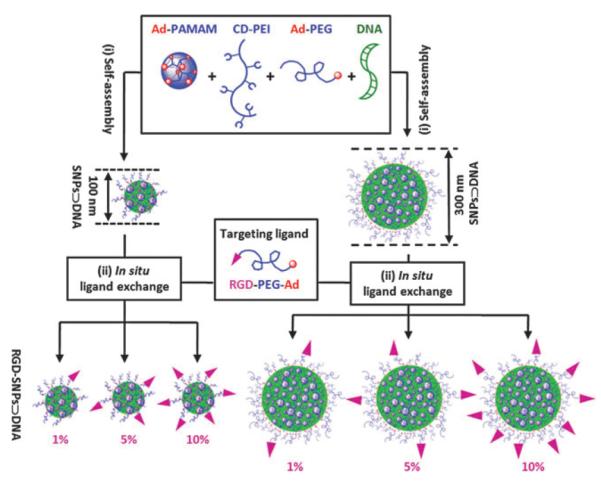
Here, we adopted this supramolecular assembly approach to prepare a small library of DNA-encapsulated SNPs (SNPs⊃DNA and RGD-SNPs⊃DNA, Scheme 1) with controllable sizes and tunable surface coverage of a targeting ligand, i.e., arginine-glycine-aspartic (RGD) peptide. A two-step preparation process has been developed to first generate both 100 and 300 nm SNPs⊃DNA from Ad-PAMAM, CD-PEI, Ad-PEG and DNA, followed by in situ RGD ligand exchange of SNPs⊃DNA to give six different RGD-SNPs⊃DNA with ligand coverage of 1, 5 and 10 mol% (based on Ad-PEG). In this proof-of-concept study, a plasmid DNA encoded with an enhanced green fluorescent protein (EGFP) driven by a CMV promoter was used as a reporter system, and the RGD ligand14 was employed to recognize the αvβ3 integrin receptor on the membranes of certain types of tumor cells. To characterize the sizes, morphologies and surface charges of the resulting SNPs⊃DNA and RGD-SNPs⊃DNA, we carried out dynamic light scattering (DLS), transmission electron microscope (TEM) and zeta potential measurements, respectively. Finally, the gene transfection effciency and specificity of each SNPs⊃DNA and RGD-SNPs⊃DNA in the small library were examined using αvβ3 high-expressed and low-expressed cells, along with the control delivery systems.
Scheme 1.
A two-step modular assembly approach for preparation of a small library of DNA-encapsulated supramolecular nanoparticles (SNPs⊃DNA and RGD-SNPs⊃DNA) with controllable sizes and tunable RGD ligand coverage.
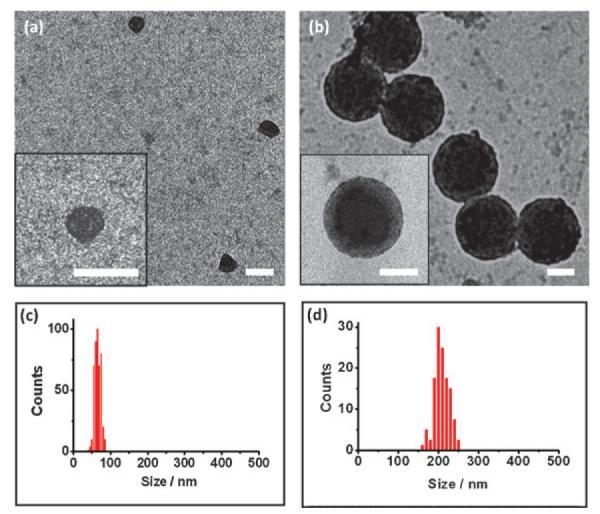
We first determined the DNA loading capacity to be used for preparation of SNPs⊃DNA and RGD-SNPs⊃DNA. Similar to cationic polymer based gene delivery systems,15,16 the DNA loading capacity of SNPs depends on the net cationic charges embedded in the interior Ad-PAMAM/CD-PEI hydrogel network. We utilized both electrophoresis analysis17 and ethidium bromide exclusion assay18 to measure the DNA loading capacity of the Ad-PAMAM/CD-PEI hydrogel (Fig. S1 and S2, ESI†), resulting in the respective nitrogen/ phosphate (N/P) ratios of 2.6 and 5.0. The N/P ratio of 5.0 was chosen to ensure complete DNA encapsulation in our studies. Next, SNPs⊃DNA with 100 and 300 nm diameters were prepared separately by slowly adding a PBS solution (pH = 7.2) of CD-PEI (600 nM) into PBS solution containing Ad-PAMAM (300 nM for 100 nm SNPs⊃DNA and 600 nM for 300 nm SNPs⊃DNA), Ad-PEG (3 μM) and DNA (2.2 nM), followed by incubation at room temperature for 20 min. The DLS measurements indicated that the hydro-dynamic sizes of the 100 and 300 nm SNPs⊃DNA were 106 ± 14 and 312 ±47 nm, respectively. Subsequently, the samples of each size of SNPs⊃DNA were split into four aliquots, and three of them were subjected to the in situ ligand exchange by adding 30, 150 or 300 nM of RGD-PEG-Ad (Scheme S1, ESI†). A collection of RGD-SNPs⊃DNA with different RGD coverage,19 namely 100-1%, 100-5%, 100-10%, 300-1%, 300-5% and 300-10%, were obtained accordingly. After in situ ligand exchange, the hydrodynamic sizes of RGD-SNPs⊃DNA exhibited negligible changes (o5%, Fig. S4, ESI†). The morphologies of SNPs⊃DNA and RGD-SNPs⊃DNA were then examined by using TEM. The TEM images (Fig. 1) showed smaller sizes (62 8 for 100 nm SNPs⊃DNA and 210 ± 24 nm for 300 nm SNPs⊃DNA), spherical shapes and narrow size distributions of SNPs⊃DNA and RGD-SNPs⊃DNA. Zeta potential measurements indicated that the surface-charge densities of 100 and 300 nm SNPs⊃DNA were 3.7 ± 0.4 and 6.8 ± 0.5 mV, respectively. After ligand exchange, small increases (3–11%)in zeta potentials of RGD-SNPs⊃DNA were observed (Fig. S5, ESI†).
Fig. 1.
TEM micrographs of (a) 100 nm SNPs⊃DNA and (b) 300 nm SNPs⊃DNA. Insets: the respective higher magnification TEM images. Scale bars: 100 nm. (c) and (d) Histograms summarize the size distributions of 100 nm SNPs⊃DNA and 300 nm SNPs⊃DNA in dry states.
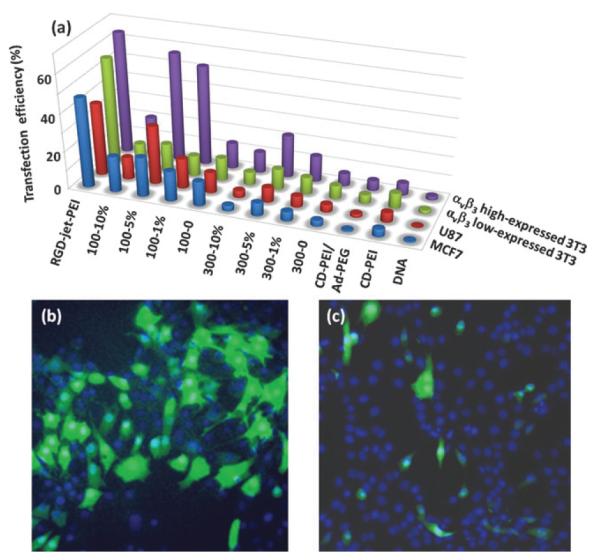
We carried out an in vitro EGFP transfection study of a collection of SNPs⊃DNA and RGD-SNPs⊃DNA along with the controls, i.e., DNA, DNA complexes of CD-PEI, CD-PEI/ Ad-PEG and RGD-jet-PEI, in 8-well chamber slides containing two αvβ3 high-expressed cells (i.e., U87 and scraping-collected 3T3 cells)20 and two αvβ3 low-expressed cells (i.e., MCF7 and 0.25% trypsin-treated 3T3 cells).21
For the purpose of comparison, an equal amount of EGFP-encoded plasmid DNA (100 ng) was added to individual cell culture chambers in this transfection study. The resulting 48 individual EGFP transfection experiments were incubated at 37 °C (5% CO2) for 24 h. After para-formaldehyde fixation and DAPI nuclear staining, a fluorescence microscope was used to quantify the EGFP expression levels in individual cells. These levels were then used to determine the transfection effciency for each vehicle. The transfection study was repeated three times, and the results of average transfection effciency of gene delivery vehicles for different cell lines were summarized in Fig. 2. First, DNA complexes based on each of the molecular building blocks (CD-PEI and CD-PEI/Ad-PEG) gave very poor transfection performance similar to free plasmid DNA, indicating that the formation of supramolecular nanoparticles is crucial for achieving enhanced transfection effciency. Second, it is apparent that 100-nm RGD-SNPs⊃DNA exhibited higher transfection effciency than those of 300-nm analogues. This observation is consistent with the results from the reported polymer-based gene delivery systems,22–24 in which vehicles with 10–100 nm size range display better gene transfection effciency.13 Third, 100-5% RGD-SNPs⊃DNA gave the highest transfection effciency compared to those observed for SNPs⊃DNA and other targeted RGD-SNPs⊃DNA. The reduced transfection effciency observed for 100-10% RGD-SNPs⊃DNA can be attributed to an excess amount of free RGD ligand in the culture medium, which compromised the targeted binding of RGD-SNPs⊃DNA as a result of a competition effect.11 Overall, 100-5% RGD-SNPs⊃DNA demonstrated the best transfection effciencies (57 ± 11% and 31 ± 8% for αvβ3 high-expressed 3T3 and U87, respectively). These results are comparable to those observed for the commercially available RGD-jet-PEI (64 ± 15% and 38 ± 9% for αvβ3 high expressed 3T3 and U87, respectively), which is a well-known selective and effcient transfection reagent for integrin-expressing cell lines.25 Fourth, in addition to high transfection effciency, 100-5% RGD-SNPs⊃DNA also exhibited outstanding delivery specificity to the αvβ3 high expressed cells, U87 (31 ± 8%) and 3T3 (57 ± 11%), over the α3β3 low expressed cells, MCF7 (21 ± 6%) and trypsin-treated 3T3 (15 ± 4%). Four-fold difference in transfection effciencies were observed for 100-5% RGD-SNPs⊃DNA between αvβ3 high-expressed and αvβ3 low expressed 3T3 cells, while only 1.2-fold difference was observed for RGD-jet-PEI. In contrast to non-target-specific transfection performance of RGD-jet-PEI, 100-5% RGD-SNPs⊃DNA had higher transfection effciency for the U87 cell line with respect to the MCF7 cell line, which indicated good transfection specificity of RGD-SNPs⊃DNA for the αvβ3 high expressed cell lines. Moreover, we tested the toxicity of SNPs⊃DNA and RGD-SNPs⊃DNA by using the cell viability assay. The cells transfected by SNPs⊃DNA and RGD-SNPs⊃DNA were compared with the cells cultured in the normal medium. There were no significant differences in viability (97 ± 2%), which suggested that the toxicity of SNPs⊃DNA and RGD-SNPs⊃DNA is negligible for in vitro transfection studies (Fig. S6, ESI†).
Fig. 2.
(a) EGFP transfection effciency of a collection of SNPs⊃DNA and RGD-SNPs⊃DNA along with control delivery systems for two αvβ3 high-expressed cells (U87 and scraping-collected 3T3 cells) and two αvβ3 low-expressed cells (MCF7 and 0.25% trypsin-treated 3T3 cells). The representative fluorescence micrographs of 57 ± 11% and 9 ± 4% transfection effciencies observed for (b) 5 mol% RGD-grafted 100 nm RGD-SNPs⊃DNA (100-5%)-treated αvβ3 high-expressed 3T3 cells and (c) 1 mol% RGD-grafted 300 nm RGD-SNPs⊃DNA (300-1%)-treated αvβ3 low-expressed 3T3 cells.
In conclusion, we demonstrated a convenient, flexible and modular synthetic approach for preparation of a small library of SNPs⊃DNA and RGD-SNPs⊃DNA with different sizes and RGD ligand coverage. Gene transfection studies of SNPs⊃DNA and RGD-SNPs⊃DNA library for αvβ3 high-expressed cells and αvβ3 low-expressed cells were performed. The results revealed that the size and target ligand coverage of RGD-SNPs⊃DNA played a critical role in the target-specific gene delivery. In conjunction with the use of a miniaturized high throughput screening platform26 and molecular imaging technology,13 we will dramatically accelerate the discovery processes of SNPs-based gene delivery vehicles toward in vivo application.
Supplementary Material
Acknowledgments
This research was supported by NIH-NCI NanoSystems Biology Cancer Center (U54CA119347) and NIH R21 grant (EB008419–01). We appreciate the reviewers suggestive comments to help us improve our manuscript.
Footnotes
Electronic supplementary information (ESI) available: Preparation and characterization of SNPs⊃DNA and RGD-SNPs⊃DNA, electrophoresis analysis, ethidium bromide exclusion assay, dynamic light scattering experiments, and gene transfection protocol. See DOI: 10.1039/b923711a
Notes and references
- 1.Kim DH, Rossi JJ. Nat. Rev. Genet. 2007;8:173–84. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 2.Glover DJ, Lipps HJ, Jans DA. Nat. Rev. Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- 3.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 4.Niidome T, Huang L. Gene Ther. 2002;9:1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 5.Prata CAH, Li Y, Luo D, McIntosh TJ, Barthelemy P, Grinstaff MW. Chem. Commun. 2008:1566–1568. doi: 10.1039/b716247b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Saltzman WM. Nat. Mater. 2009;8:526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Yu L, Feng Z, Hou S, Liu Y. Chem. Commun. 2009:4106–4108. doi: 10.1039/b906794a. [DOI] [PubMed] [Google Scholar]
- 8.Torchilin VP, Levchenko TS, Rammohan R, Volodina N, Papahadjopoulos-Sternberg B, D’Souza GG. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1972–1977. doi: 10.1073/pnas.0435906100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang H, Harries D, Wong GC. Proc.Natl.Acad.Sci.U.S.A. 2005;102:11173–11178. doi: 10.1073/pnas.0502416102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, Kumar V. Chem. Commun. 2009:5433–5435. doi: 10.1039/b907283g. [DOI] [PubMed] [Google Scholar]
- 11.Bazin L, Gressier M, Taberna PL, Menu MJ, Simon P. Chem. Commun. 2008:5004–5006. doi: 10.1039/b807837h. [DOI] [PubMed] [Google Scholar]
- 12.Cheon J, Lee JH. Acc. Chem. Res. 2008;41:1630–1640. doi: 10.1021/ar800045c. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Wang ST, Su H, Chen KJ, Armijo AL, Lin WY, Wang YJ, Sun J, Kamei K, Czernin J, Radu CG, Tseng HR. Angew. Chem., Int. Ed. 2009;48:4344–4348. doi: 10.1002/anie.200900063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Cai WB, He LN, Nakayama N, Chen K, Sun XM, Chen XY, Dai HJ. Nat. Nanotechnol. 2007;2:47–52. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasachari S, Reineke TM. Biomaterials. 2009;30:928–938. doi: 10.1016/j.biomaterials.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Yang C, Li HZ, Wang X, Goh SH, Ding JL, Wang DY, Leong KW. Adv. Mater. 2006;18:2969–2970. [Google Scholar]
- 17.Zugates GT, Anderson DG, Little SR, Lawhorn IEB, Langer R. J. Am. Chem. Soc. 2006;128:12726–12734. doi: 10.1021/ja061570n. [DOI] [PubMed] [Google Scholar]
- 18.Meyer M, Philipp A, Oskuee R, Schmidt C, Wagner E. J. Am. Chem. Soc. 2008;130:3273–3273. doi: 10.1021/ja710344v. [DOI] [PubMed] [Google Scholar]
- 19.When mixing ratios of RGD to target ligand were 1, 5 and 10%, the actual RGD ligand coverages on the 100 nm RGD-SNPs⊃DNA were 0.8 ± 0.2%, 2.9 ± 0.5% and 6.7 ± 0.8%,respectively. For 300 nm RGD-SNPs⊃DNA, the RGD ligand coverages were 0.9 ± 0.2%, 2.2 ± 0.4% and 6.7 ± 0.6%,respectively. For details see ESI†.
- 20.Ng QKT, Sutton MK, Soonsawad P, Xing L, Cheng H, Segura T. Mol. Ther. 2009;17:828–836. doi: 10.1038/mt.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie J, Chen K, Lee HY, Xu CJ, Hsu AR, Peng S, Chen XY, Sun SH. J. Am.Chem. Soc. 2008;130:7542–7543. doi: 10.1021/ja802003h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis ME, Chen Z, Shin DM. Nat. Rev. Drug Discovery. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 23.Yu HJ, Wagner E. Curr. Opin. Mol. Ther. 2009;11:165–178. [PubMed] [Google Scholar]
- 24.Fukushima S, Miyata K, Nishiyama N, Kanayama N, Yamasaki Y, Kataoka K. J. Am. Chem. Soc. 2005;127:2810–2811. doi: 10.1021/ja0440506. [DOI] [PubMed] [Google Scholar]
- 25.Erbacher P, Remy JS, Behr JP. Gene Ther. 1999;6:138–45. doi: 10.1038/sj.gt.3300783. [DOI] [PubMed] [Google Scholar]
- 26 (a).Lin W-Y, Wang Y, Wang S, Tseng H-R. Nano Today. 2009;4:470–481. doi: 10.1016/j.nantod.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang YJ, Lin WY, Liu K, Lin RJ, Selke M, Kolb HC, Zhang NG, Zhao XZ, Phelps ME, Shen CKF, Faull KF, Tseng HR. Lab Chip. 2009;9:2281–2285. doi: 10.1039/b907430a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wang J, Sui G, Mocharla VP, Lin RJ, Phelps ME, Kolb HC, Tseng H-R. Angew. Chem., Int. Ed. 2006;45:5276–5281. doi: 10.1002/anie.200601677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


