Abstract
The Drosophila brain is a highly complex structure composed of thousands of neurons that are interconnected in numerous exquisitely organized neuropile structures such as the mushroom bodies, central complex, antennal lobes, and other specialized neuropiles. While the neurons of the insect brain are known to derive in a lineage-specific fashion from a stereotyped set of segmentally organized neuroblasts, the developmental origin and neuromeric organization of the neuropile formed by these neurons is still unclear. In this report, we use genetic labeling techniques to characterize the neuropile innervation pattern of engrailed-expressing brain lineages of known neuromeric origin. We show that the neurons of these lineages project to and form most arborizations, in particular all of their proximal branches, in the same brain neuropile compartments in embryonic, larval and adult stages. Moreover, we show that engrailed-positive neurons of differing neuromeric origin respect boundaries between neuromere-specific compartments in the brain. This is confirmed by an analysis of the arborization pattern of empty spiracles-expressing lineages. These findings indicate that arborizations of lineages deriving from different brain neuromeres innervate a non-overlapping set of neuropile compartments. This supports a model for neuromere-specific brain neuropile, in which a given lineage forms its proximal arborizations predominantly in the compartments that correspond to its neuromere of origin.
Keywords: engrailed, neuropile, compartment, neuromere, arborization, lineage
INTRODUCTION
The insect CNS is composed of two spatially separated parts, the ventral nerve cord (VNC) which is located in the trunk region, and the brain which is located within the head. The overall composition of the VNC consists of a chain of relatively uniform segmental units referred to as neuromeres; each neuromere corresponding to one body segment of the trunk (thorax and abdomen. In the head, segments have fused and become strongly modified, and as a result, neuromere boundaries are difficult to define in the brain. The insect brain can be divided into two parts, namely a supraesophageal ganglion and a subesophageal ganglion (Hanström, 1928; Holmgren, 1928; Bullock and Horridge, 1965). The subesophageal ganglion can be further subdivided into three fused neuromeres called the mandibular, maxillary and labial neuromeres, which correspond to the neuromeres of the three fused gnathal segments of the head posterior to the esophagus. The supraesophageal ganglion is also classically subdivided into three parts, namely the protocerebrum, deutocerebrum and tritocerebrum. Tritocerebrum and deutocerebrum are generally considered to be the segmental neuromeres of two strongly modified segments, the intercalary segment and antennal segment respectively. The neuromeric nature of the protocerebrum, by far the largest part of the insect brain, is unclear; it may be correspond to a single, unsegmented acron (called ocular segment in some recent papers), or have multiple segmental (ocular, labral) and non-segmental (acron) components (Diederich and Kaufman, 1991; Schmidt-Ott and Technau, 1992; Urbach and Technau, 2003a, b).
This structural complexity of the supraesophageal ganglion contrasts with the relative simplicity of the thoracic and abdominal neuromeres. Each of these manifests the same, basic bilaterally symmetric structure consisting of a cortex of cell bodies surrounding a series of longitudinal tracts, transverse commissures and regionalized domains of interspersed neuropile areas (Tyrer and Gregory, 1982; Burrows, 1996). Many aspects of this structural organization are clearly serially reiterated throughout the VNC such that segmentally homologous tracts, neuropile regions and even individual neurons can be identified in each of the neuromeric units. Due to their relative structural simplicity, the ganglionic neuromeres of the VNC have been intensively studied, both anatomically and physiologically. In consequence, much is known about the organization of these neuromeres as well as the structure of a large number of their component neurons. One general neuroanatomical feature of neurons in the VNC that has emerged from these studies is the fact that most of the neurons in a given neuromere restrict their proximal (often dendritic) arborizations to the neuropile of that neuromere. This is obviously the case for the numerous local neurons, which by definition have no aborizations outside of the neuromere. Over half of the neurons in a thoracic or abdominal ganglion are local neurons of this type (see Burrows, 1996). However, it is also true for the vast majority of the motoneurons and intersegmental interneurons in the VNC. Although the processes of large intersegmental interneurons by definition spread over several ganglia, most of these interneurons have their proximal arborizations located in the same neuromere as their cell body, while their distal processes project to other neuromeres and ganglia. Most sensory neurons of a given segment project their axons into the neuromere of that segment and also form their terminal arborizations there, and motoneurons typically form dendrites in the neuromere that contains their somata (Merritt and Whitington, 1995; Schrader and Merritt, 2000; Landgraf et al., 2003; Zlatic et al., 2003).
To a significant extent, the internal structure and neuronal organization of the three neuromeres of the subesophageal ganglion in terms of tracts commissures and neuropiles is similar to that of the VNC neuromeres. In contrast, the larger supraesophageal ganglion manifests highly organized neuropile structures such as the mushroom bodies, central complex and antennal lobes which have no obvious equivalents in other neuromeres of the CNS. Moreover, due to its complex and hidden segmental organization, it is difficult to determine the neuromere boundaries within the neuropile of the supraesophageal ganglion. The antennal and intercalary segments possess afferent and efferent axons, and the neuropile compartments that contain arborizations of these fibers (in particular the afferent, sensory axons) are usually assigned “with some confidence” to the corresponding neuromeres. Thus, the morphologically well delineated antennal lobe and antenno-mechanosensory-motor center (AMMC), which receive input from the antenna, are considered part of the deutocerebrum (Homberg et al., 1989). Similarly, the domain of arborization of the pharyngeal nerve, belonging to the intercalary segment, delineates a small neuropile domain usually referred to as tritocerebrum (Rajashekhar and Singh, 1994). However, these sensory compartments most likely represent only parts of the deutocerebrum and tritocerebrum, given that in a prototypical trunk segment, the arborizations of sensory afferents fill only part of the volume of the corresponding segmental neuromere. Thus, the lack of information about neuromeric boundaries together with the highly fused nature of the brain neuropile confound attempts to understand the neuroanatomical construction principles of the supraesophageal ganglion. It also makes it difficult to relate the serially homologous organization of the prototypical ganglionic pattern elements in the VNC to the structural organization of the brain.
The serially homologous organization of the insect CNS arises during development. From an early stage onward, the insect embryo (germ band) becomes subdivided into a series of reiterated metameres. Each segmental metamere contains a relatively stereotyped set of progenitor cells, called neuroblasts, which generate the neurons and glia of the mature nervous system. These neuroblasts as well as the cellular and molecular mechanisms by which they generate the neurons of the CNS have been most throughly studied in Drosophila (Technau et al., 2006; Hartenstein et al., 2008a). All of the neuroblasts in the developing brain and VNC of Drosophila have been individually identified based on their position in the neuroectoderm and on their specific combination of marker gene expression. The supraesophageal ganglion derives from approximately 100 bilaterally symmetrical neuroblast pairs (Younossi-Hartenstein et al., 1996; Urbach and Technau, 2003a), each of which generates a characteristic lineage of neural progeny (neurons and glia). Based on the expression of segment polarity genes, it is possible to assign a defined neuromere of origin to each of the embryonic brain neuroblasts (Urbach and Technau, 2003a). Moreover, through clonal labeling techniques, it is also possible to follow the development of the neurons generated by individual brain neuroblasts through embyronic, larval and pupal stages and into the adult brain. Thus, the neuropile domains of the brain, in which neurons of a known neuromeric origin form their arborizations; can now in principle be determined. This, in turn, should make it possible to delineate the specific brain neuropile domains that correspond to specific brain neuromeres.
In this report, we reconstruct the projection pattern of the Drosophila neuroblast lineages expressing the segment polarity gene engrailed (en) from embryonic to adult stages in order to contribute to our understanding of neuromere boundaries in the brain. The en gene is expressed in neuroblasts located at the posterior boundary of each CNS neuromere (Bossing et al., 1996; Younossi-Hartenstein et al., 1996; Schmid et al., 1999; Urbach and Technau, 2003a) and, hence, the neurons that derive from these neuroblasts are of known neuromeric origin. We identify en-expressing neural lineages in protocerebrum, deuterocereberum and tritocerebrum, and determine the trajectory of the axon tracts as well as the innervated brain neuropile compartments for these lineages in protocerebrum and deutocerebrum. Moreover, we show that the neurons of the en-expressing lineages project to and innervate the same brain neuropile compartments in embryonic, larval and adult stages. An analysis of the arborization domains of en-expressing neurons in these stages reveals boundaries between neuromere-specific compartments in the brain that are respected by neurons of differing neuromeric origin. This is confirmed by analysis of the arborizations made by deutocerebral ems-expressing neurons, which in part also restrict their arborizations to the neuropile region delimited by deutocerebral en-expressing neurons.
MATERIAL AND METHODS
Fly strains and genetics
For this study, stock containing a recombinant chromosome was constructed: P{en2.4-Gal4}e16E, UAS mCD8::GFPLL5, UAS-nlslacZ20b on chromosomal arm 2R and FRT 82B, tubP-GAL80LL3 on chromosome 3. To generate MARCM clones (Lee and Luo, 1999) +; UAS mCD8::GFPLL5, UAS-nlslacZ20b; FRT82B (Bello et al., 2003) males were crossed to females of the MARCM driver stock hs-FLP; en-GAL4, UAS mCD8::GFPLL5, UAS-nlslacZ20b; FRT82B, tubP-GAL80LL3 resulting in wild-type clones of en expressing cells. For MARCM experiments, embryos of the appropriate genotype were collected on standard cornmeal/yeast/agar medium supplemented with live yeast over a 4 hour time window and raised at 25°C for 21 to 25 hours before heat-shock treatment. Heat-shock induction of FLP was done at 37°C for 60 minutes. MARCM clones were examined in brains dissected at wandering third instar stage and 0 – 10 days after adult fly eclosion. Other transgenic fly lines used were rhx25lacZ (expressed in the engrailed domains; Hama et al., 1990), and a151 Gal4 (expressed in subsets of sensory neurons in the adult fly; J. Simpson, unpublished).
Immunohistochemistry
Larval and adult brains were dissected in PBS, fixed in 2% paraformaldehyde in PBL (75mM lysine HCL in sodium phosphate buffer PH 7.4) for 1 hour at room temperature (RT), washed three times for 10 minutes in PBS containing 0.5% Triton X-100 (PBT), blocked for 1 hour at RT in PBT containing 10 % normal goat serum, and incubated with primary antibodies in blocking solution overnight at 4°C. Samples were washed three times for 10 minutes in PBT at RT, and secondary antibodies were applied in blocking solution for 3 hours at RT. After washing three times for 15 minutes in PBS, samples were mounted in Vectashield (Vector Labs).
Antibodies used
The following is a list of antibodies used in the course of this study. Immunogens used for immunization and their specificity are discussed.
Monoclonal mouse anti-Engrailed/Invected antibody [Developmental Studies Hybridoma bank (DSHB), catalog ID: 4D9, 1:10]
Antigen: Raised against the C-terminal two thirds of the Invected protein and the epitope has been localized to residues 38–58 of the homeodomain (Patel et al., 1989). Specificity: The antibody recognizes both the engrailed and invected gene products of Drosophila. 4D9 binds on Western blots to the intact protein but not to the protein lacking the homeodomain. In addition, it also binds to Engrailed protein in chicken, leech, zebrafish and grasshopper embryos but not to recombinant mouse and human engrailed proteins. It also detects the patterning of segmentation from the cellular blastoderm stage onward. Staining is seen in a large number of CNS neurons in Drosophila including most of the median neuroblast progeny as well as a small number of PNS neurons.
Monoclonal mouse anti-Nc82 antibody (DSHB, 1:20)
Antigen: Raised against adult Drosophila head homogenates. Recently, the specific immunogen was identified as Bruchpilot (Wagh et al., 2006). Specificity: In Western blots of homogenized Drosophila heads, the antibody recognized two proteins of 190 and 170 kDa apparent size which were later found to be part of the same transcription unit of the bruchpilot gene. In vivo, the antibody recognizes brain neuropil as well as synaptic active zones during most stages of brain development in Drosophila. In addition, the antibody is also known to cross react with several insect species.
Polyclonal rat anti DN-cadherin antibody (DN-cad, DSHB, DN-EX#8, 1:25) (Iwai et al., 1997)
Antigen: Peptide encoded by exon 8, amino acid residues 1210–1272. DN-Cadherin fusion protein was produced in E.coli by subcloning a cDNA fragment covering an extracellular domain (exon 8 or CR8 in this case) into pGEX-1λT. The inclusion body of this fusion protein, mixed with adjuvant was then used to generate rat anti-sera. Specificity: This antibody detected two major bands, 300 kDa and 200 kDa molecular weights on Western blot of S2 cells only after transfection with a cDNA encoding the DN-cadherin protein (Iwai et al., 1997). In addition, the specificity of this antibody was tested with immunostaining of Drosophila Df(2L)TW119 embryos. Signal was hardly detectable in homozygous mutant, l(2)36DaM19 with nonsense mutation causes premature termination of protein translation. In contrast, this antibody gave a signal in mutant embryos with N-cadherin transgene.
Monoclonal mouse anti-fasciclin II (FasII) antibody (DSHB; catalog ID: 1D4, 1:10)
Antigen: 103 amino acids of the C-terminal transmembrane domain of FasII, beginning at amino acid 771. The cDNA fragment encoding this portion of the protein was fused to a bacterial vector and cloned. The purified inclusion bodies were used to raise antibodies in mice (Grenningloh et al., 1991). Specificity: On Western blot, 1D4 identifies a 97-kDa molecular-weight protein whose mobility is similar to that of a polyclonal antibody directed against the extracellular portion of FasII protein (Mathew et al., 2003).
Monoclonal mouse anti-Neurotactin (Nrt) antibody (DSHB; catalog ID: BP106, 1:10)
Antigen: A neurotactin CDNA fragment coding for the 419 carboxyterminal amino acid residues was subcloned into a protein A fusion protein expression vector pRIT31. This fusion protein was then expressed in pop2136 cells and isolated fusion protein was used as an immunogen (Hortsch et al., 1990). Specificity: Monoclonal BP106 generated using membrane proteins from cultured embryonic Drosophila neuronal cells detected the same Drosophila embryonic pattern as that of a polyclonal antisera raised against a fusion protein using part of the Neurotactin cDNA (Hortsch et al., 1990). In addition, another monoclonal antibody, Mab E1C, against Neurotactin gave a similar expression pattern in Drosophila embryos to that of BP106 (Piovant and Lena, 1988).
Other Antibodies used
To visualize reporter constructs, anti-β-galactosidase (in rabbit; Cappel MP Biomedicals, catalog#. 55976) was used after diluting 1:1000. The antibody was raised against the C-terminal end of E.coli beta-galactosidase protein. The specificity of the antibody is shown from its failure to label tissues that do not express the bacterial transgene. To highlight the arbors of MARCM clones, anti-GFP (in rabbit; Torrey Pines Biolabs Inc, #TP401) was used after diluting 1:450. Anti-Ems (gift of U. Walldorf, University of Saarland, Homburg, Germany) was used to visualize Ems positive MARCM clones. The antibody was generated in rat and diluted 1:200 before use on larval brains. Antigen: Antibody raised against a fusion protein containing amino acid positions 40–342 of the Ems homeodomain (Walldorf and Gehring, 1992). Specificity: In Drosophila, the Ems antibody detects a single circumferential stripe at the cellular blastoderm stage at the anterior end of the embryo. Later in embryogenesis, the antibody can be detected in the developing cephalic region. In addition, Ems immunostaining also extends over one-half of the embryonic head and includes much of the preantennal, antennal and intercalary segments (Hartmann et al., 2000).
Microscopy, image processing and generation of three dimensional models
All fluorescent images were recorded using a Leica TCS SP scanning confocal microscope. Complete series of optical sections were taken at 1 μm intervals in line average mode with picture size of 512 × 512 pixels. Digital image stacks were processed using ImageJ (http://rsb.info.nih.gov/ij/). All images including enGal4 driven MARCM clones were recorded in the above mentioned manner. Digital 3D-models were generated using the AMIRA software. Surface-rendered digital atlas models were created by manually labeling each lineage and neuropile compartment. For clonal analysis, the surface rendered neuropile compartments were brought into registration with the dataset of the clone by warping to the standard master template. The Amira program also allows one to adjust virtual lighting, camera angle, transparency, reflection, and other parameters in a straightforward manner.
RESULTS
The engrailed-expressing primary neurons in embryonic brain development
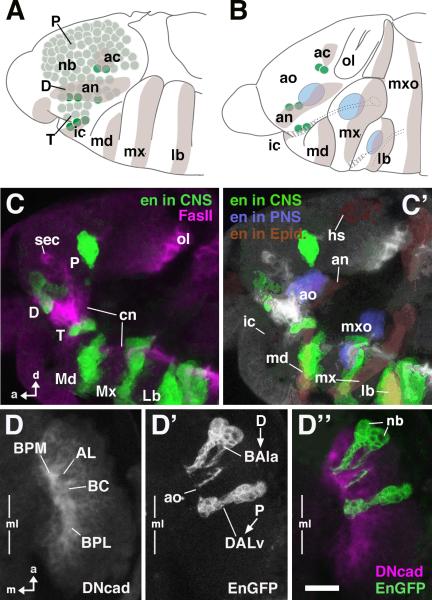
The engrailed (en) gene is expressed in metamerically reiterated stripes in the embryonic neuroectoderm and the neuroblasts that delaminate from these neuroectodermal domains. The stripes of en expression define the posterior segmental compartments and, correspondingly, the en-expressing neuroblasts define the posterior boundary of each neuromere (Bossing et al., 1996; Younossi-Hartenstein et al., 1996; Urbach and Technau, 2003b). In the preoral procephalic neuroectoderm that gives rise to the supraesophageal ganglion of the brain, we can distinguish, in all, nine en-positive neuroblasts; three belong to the intercalary segment, four to the antennal segment, and two to the more anterior ocular segment. As is the case for the neuromeres of the ventral nerve cord, these neuroblasts define the posterior boundaries of the preoral neuromeres; the en-expressing neuroblasts of the intercalary segment define the posterior boundary of the tritocerebrum, those of the antennal segment the boundary of the deutocerebrum, and those of the ocular segment the boundary of the protocerebrum (Fig. 1A).
Fig.1. Metameric engrailed expression in the embryonic head.
A: Schematic of early (stage 11) embryonic head, lateral view. Ectodermal engrailed (en) stripes corresponding to posterior domains of head segments are shaded [lb labium; mx maxilla; md mandible; ic intercalary segment; an antennal segment; ac acron (“head spot”)]. Brain neuroblasts (nb) are shaded green. Dark green indicates sets of en-positive neuroblasts. It is notable that these neuroblasts spatially overlap with en expression domains. B: Schematic of mid-stage (stage 13) embryonic head, lateral view. Spatial relationship of en-positive neuroblasts and ectodermal en strips is maintained. Blue areas indicate primordia of head sensory complexes (labium, maxilla, antenna) which partially overlap with en stripes. C: Z-projections of confocal sections of stage 14 embryonic head labeled with antibody against En (green) and Fasciclin II (magenta); lateral view. Note metamerical clusters of neurons (lineages) derived from en-positive neuroblasts. The three posterior clusters (Md, Mx, Lb) demarcate the three neuromeres of the gnathal segments, which will later form the subesophageal ganglion. The anterior clusters correspond to the neuromeres of the supraesophageal ganglion (P protocerebrum, derived from acron; D deutocerebrum, derived from antennal segment; T tritocerebrum, derived from intercalary segment). C': Same Z-projection as in C. The different tissues contributing to en expression domains are shown in different colors. en-positive clusters of neurons are in green. en in the ectoderm (by that stage: epidermal primordium) is shaded brown; parts of sensory primordia expressing en (ao antennal organ; mxo maxillary organ) are in blue. Note that the en-positive clusters and the corresponding en stripes from which they derive are still in close proximity, except for the protocerebral cluster of neurons that, due to morphogenetic movements in the head, has moved away from the epidermal head spot (hs). D, D', D": Z-projection of confocal sections of stage 15 embryonic brain labeled with antibodies against DN-cadherin (D; magenta in D") and GFP expressed by an en-Gal4 driver (D'; green in D"); dorsal view; only right brain hemisphere is shown; vertical line indicates midline (ml). Note pattern of en-positive lineages (deutocerebral BAla3; protocerebral DALv) and their relationship to the primordia of brain compartments (AL Antennal lobe; BC basocentral; BPL baso-posterior lateral; BPM baso-posterior medial). Each lineage consists of the superficial neuroblast (white line and arrowhead indication, D') and a chain of primary neurons. Other abbreviations: cn cervical connective; ol optic lobe; sec supraesophageal commissure. Bar: 10μm (for C–D)
Following their delamination from the neuroectoderm at embryonic stages 9–11, procephalic neuroblasts begin to proliferate and generate the primary neurons of the larval brain. During this phase, en expression, as assayed by anti-En immunoreactivity, is maintained in the neuroblasts and the neurons generated by several of these neuroblasts. Subsequently, at embryonic stages 13–15 (Campos-Ortega and Hartenstein, 1997), neurons initiate neuronal differentiation and axonal outgrowth. At these stages, the en-positive neurons in the supraesophageal (procephalic) neuromeres are observed in three clusters grouped along the neuraxis anterior and dorsal to the three clusters of the en-positive neurons of the subesophageal (gnathal) neuromeres (Fig.1C). The protocerebral cluster, which appears to be composed of 2–3 closely apposed neuroblast lineages, emits a single, short primary axon tract that extends straight medially towards the center of the nascent brain neuropile. This position marks the point where the baso-central (BC; nomenclature of larval brain compartments according to Younossi-Hartenstein et al., 2003; Younossi-Hartenstein et al., 2006) neuropile compartment will appear several hours later (see Fig. 2). The deutocerebral cluster of en-expressing neurons, which may comprise 1 or 2 neuroblast lineages, emits axon tracts that converge posteriorly and defines the position where the baso-posterior medial compartment (BPM) will appear (see Fig. 2). (In addition, en-positive sensory neurons of the antennal organ, the larval equivalent of the antenna, project towards the central neuropile laterally adjacent to the deutocerebral cluster, a position that defines the nascent antennal lobe). The tritocerebral cluster of en-positive cells encircles the base of the embryonic tritocerebral primordium in the shape of a horizontal crescent. No axons can be discerned emanating from this tritocerebral cluster, and at late embryonic stages, and most of the en-expressing cells in the cluster have adopted the shape and position of glial cells; these cells were not characterized further in this study.
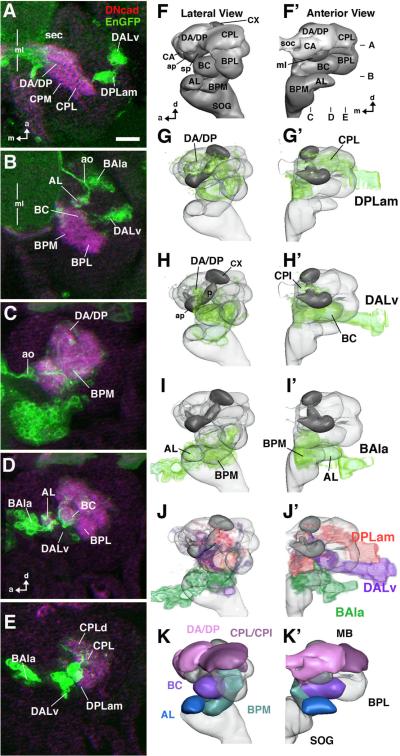
Fig.2. engrailed lineages in the late embryonic brain.
A, B: Z-projection of confocal sections of stage 17 embryonic brain labeled with antibodies against DN-cadherin (magenta) and GFP expressed by an en-Gal4 driver (green); dorsal view; only right brain hemisphere is shown; vertical line indicates midline (ml). Dorso-ventral focal plane of A and B is indicated in panel F' to the right. C–E: Z-projections of parasagittal confocal sections of stage 17 embryonic brain prepared as the one shown in A/B. Anterior is to the left, dorsal up. Medio-lateral focal planes of C–E are indicated in panel F'. F, F': 3D digital models of stage 17 embryonic brain hemisphere in lateral view (F) and anterior view (F'), showing neuropile compartments (AL larval antennal lobe; ap anterior appendix of larval mushroom body; BC baso-central; BPL baso-posterior lateral; BPM baso-posterior medial; CA centro-anterior; CPL centro-posterior lateral; CX calyx; DA dorso-anterior; DP dorso-posterior; sec supraesophageal commissure; ml medial lobe of mushroom body; SOG subesophageal ganglion (=anterior ventral nerve cord); sp spur of mushroom body). G–I, G'–I': Digital brain models as in F/F', with volume renderings of the en lineages visible in stage 17 embryo (DPLam, DALv, BAla3; shaded green). J, J': Montage of the en lineages in one model (DPLam red; DALv purple; BAla3 green). K, K': Digital brain models as in F/F', with neuropile compartments innervated by en lineages shown in different colors. En expression in late embryo appears in three main clusters. The dorsal-most cluster corresponds to the DPLam lineage (A, E, G/G'), which arborizes in CPL, CPI, DP and DA (A, C–E, G/G'). The medial cluster (DALv; A, B, E, H/H') consists of two adjacent lineages, DALv2 and DALv3, with indistinguishable projections in the embryo and early larva. Proximal projections are in the BC compartment (B, D, H/H'); more distally, projections overlap with those of DPLam in CPI (see G' and H') and DA and CA (see G, H, F). In the deutocerebrum, one lineage (BAla3) is distinguished (B, E, I/I'). It projects to the BPM compartment (C, I/I'). The larval AL compartment (antennal lobe) is labeled by sensory afferents of en-positive antennal organ (ao; BD, I/I'). Bar: 10μm
Compartment-specific arborizations of engrailed-expressing primary neurons
In the late embryo (stage 16 to hatching), the primary axon tracts of the neurons in all primary lineages, including those that are en-positive, begin to form extensive arborizations, and as a result the developing brain neuropile, subdivided into several compartments, emerges (Pereanu and Hartenstein, 2006; Younossi-Hartenstein et al., 2006). These neuropile compartments can be individually identified and followed throughout larval and pupal development into the adult brain (see Hartenstein et al., 2008b). The entire morphology of the developing En-immunoreactive neurons, including their cell bodies, neurites, and arborizations, can be revealed by an en-Gal4 driver coupled to a UAS-mCD8::GFP reporter and then related to nascent brain neuropile structures revealed by anti-DNcadherin immunolabeling (Fig. 1D). Moreover, by relating the compartments innervated by en-expressing neuronal clusters to the compartments innervated by identified neuroblast lineages (see Pereanu and Hartenstein, 2006), one can assign the neurons of a given en cluster to identified neuroblast lineages.
In the protocerebrum of late embryonic stages, two clusters of en-expressing neurons become apparent (Fig. 2A). The protocerebral cluster of neurons that had already been en-positive at earlier stages is located next to the nascent BC compartment (Fig. 2B, H). The primary axon tract that derives from these neurons projects to the BC compartment. The neurites from this tract form arborizations throughout the compartment and also continue medially and anteriorly and form arborizations in the CPI, CPL and CA compartments, where they intermingle with branches from other procephalic lineages, as well as with en-positive fibers that ascend from the ventral nerve cord (Fig. 2). Based on their close association with the BC compartment we can identify the en-expressing neurons in this protocerebral cluster as members of the two DALv2/3 neuroblast lineages of the larva (see below). The second en-positive protocerebral cluster is located anterior and dorsal to the DALv2/3 cluster (Fig.2A, E, G). (At the earlier embryonic stages described above, this group of neurons might have been nested in the observed protocerebral en-cluster; alternatively, en-expression may appear de novo in this cell cluster between embryonic stage 15 and late16.) The primary axon tract from this second cluster enters the neuropile more dorsally than the DALv2/3 axons, and arborizes in the CPL, posterior CPI, and DA compartments. Based on this arborization pattern, one can identify the neurons in this second cluster as members of a third protocerebral en-positive lineage namely the DPLam lineage of the larva (see below). The deutocerebral cluster of en-positive neurons projects its axon tracts from anterior into the BPM compartment (Fig. 2 D, E, I). There they form arborizations which are intermingled with unidentified en-positive ascending fibers. The BPM compartment (like the antennal lobe) is also filled with en-positive sensory endings from the antennal organ. Their compartment-specific arborization pattern identifies the neurons in the deutocerebral cluster as members of the BAla3 lineages as defined for the larva (see below).
This general pattern of neurite projections and arborizations of the three groups of en-positive primary neurons in the late embryo is maintained throughout the larval period (Fig. 3B, C). Based on this, one can assign distinct larval neuropile compartments to at least part of the arborizations made by the neurons of any of these given lineages. Thus, the proximal arborization of the larval DPLam lineage, as described above for the embryo, is found in the CPL compartment of the larval brain, close to where the cell bodies are located. Similarly, the proximal arborizations of the larval DALv lineages, as described above for the embryo, are in the BC compartment of the larval brain that is adjacent to the DALv cell bodies. The more distal branches of DPLam and DALv are found in the larval DA, DP, CPI and CPM compartments, where they appear to intermingle (Fig. 3B, C, D). Finally, the en-positive neurons of the larval BAla3 lineage, as described above for the embryo, form a long tract that passes the antennal lobe and reaches the more posteriorly located larval BPM compartment, where it arborizes.
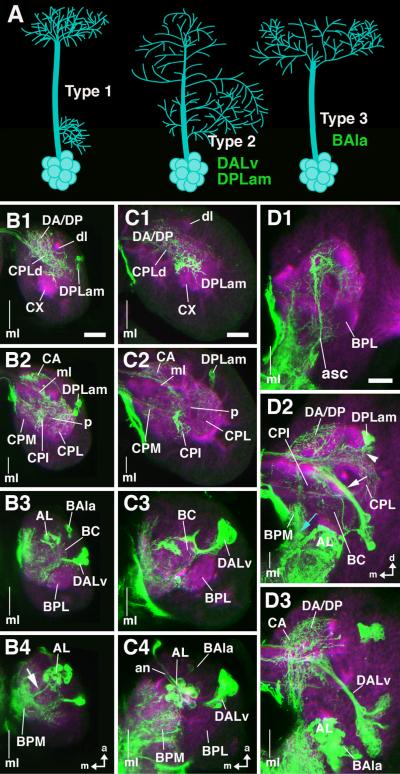
Fig.3. engrailed lineages during larval development.
A: Schematic representation of different types of lineages encountered in brain (type 1: separate proximal and distal arborization; type 2: continuous arborization; type 3: distal arborization; Larsen et al., personal communication). B1–B4, C1–C4, D1–D3: Z-projection of confocal sections of larval brains labeled with antibodies against DN-cadherin (magenta) and GFP expressed by an en-Gal4 driver (green); only right brain hemisphere is shown; vertical line indicates midline (ml). B1– B4: first larval instar, dorsal view, focal plane goes from dorsal (B1) to ventral (B4). C1–C4: second larval instar, dorsal view, focal plane goes from dorsal (C1) to ventral (C4). D1–D3: late third larval instar; anterior view; focal plane goes from anterior (D1) to posterior (D3). The arborization pattern of en lineages is similar as shown for late embryo in Fig.2. The protocerebral DPLam lineage (cell bodies shown in B2, C2, D2) arborizes widely in CPL, CPI, DP/DA, and CA. DALv (comprising two neighboring lineages; cell bodies in B3, C3, D3) has arborizations in BC, as well as CPI and DP/DA where they overlap with fibers of DPLam. BAla3 (cell bodies in B3, C4, D3) arborizes in BPM. en-Gal4 driven GFP labeling in larval antennal lobe (AL; B4, C4, D3) is due to en-positive afferents from antennal organ. At third instar, secondary neurons have been added to en lineages. They form distinctive secondary axon tracts (SATs) that project into territory innervated by primary neurons (white arrow head in D2: SAT of DPLam; arrow in D2: SAT of DALv2/3; blue arrow in D2: SAT of BAla3). Note that there are en–positive fibers ascending from the ventral cord (asc), as well as en-positive surface glia (sg). Other abbreviations: BPL baso-posterior lateral compartment; CPM centro-posterior medial compartment; CX calyx; dl dorsal lobe; ml medial lobe; p peduncle). Bars: 10μm (B1–B4; C1–C4); 25μm (D1–D3)
In summary, most of the neuropile compartments of the late embryonic as well as the larval brain can be assigned to at least one of the en-lineages; CPL and BC to the proximal arborizations of the DPLam lineage and the DALv lineages, respectively; CPI, CPM, DP and DA to the distal arborizations of the DPLam lineage and the DALv lineages; BPM to arborizations of the BAla3 lineage (Fig. 2 F, J, K). The only major brain compartments that appear to lack arborizations from neurons of en-expressing lineages are the mushroom body, and the BPL.
Secondary neurons in engrailed lineages innervate the same brain compartments as their primary neuron siblings
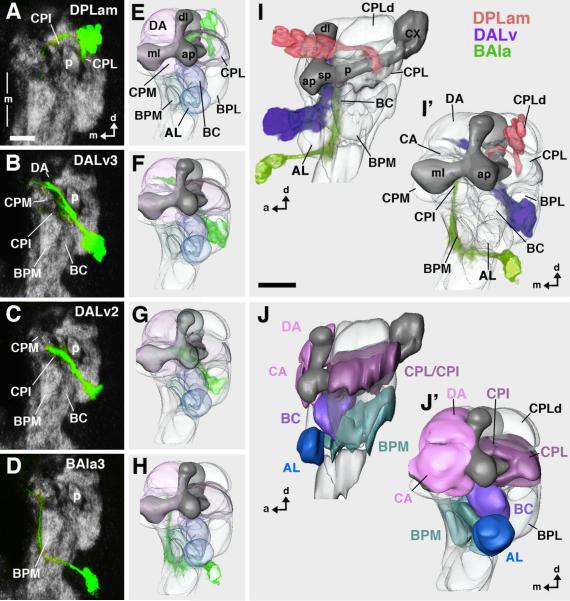
After a period of mitotic quiescence during the early larval period, neuroblasts reactivate proliferation and produce adult-specific secondary neurons. Secondary neurons belonging to one and the same neuroblast lineage form axons that fasciculate in a coherent bundle referred to as a secondary axon tract (Dumstrei et al., 2003). To investigate the lineages of secondary en-expressing neurons in more detail, we carried out a MARCM-based clonal analysis with a faithful en-Gal4 driving UAS-mCD8::GFP and therefore, we recovered positively labeled clones for analysis that comprised en-expressing neurons. (Clones were induced at larval hatching and therefore only adult-specific secondary cells were labeled.) In this analysis, we consistently recovered, in the late larval brain, en-positive clones in the supraesophageal ganglion that correspond to 4 different neuroblast lineages, and all en-expressing secondary neurons in the supraesophageal ganglion belong to one of these four lineages. (However, not all of the secondary neurons in these lineages are en-expressing.) Location of cell bodies, projection of secondary axon tract, and the innervated set of compartments positively identify these four lineages as the DPLam, DALv2, DALv3, and BAla3 lineages (Fig. 4).
Fig.4. Secondary neurons of engrailed lineages visualized in late third instar larva by en-Gal4 driven MARCM clones.
Heat shock to induce Flippase mediated recombination was given shortly after larval hatching, leading to labeling of secondary neurons. A–D: Z-projection of confocal sections of late larval brains labeled with antibodies against Nc82 (grey) and GFP expressed by an en-Gal4 driver (green); anterior view; only right brain hemisphere is shown; vertical line indicates midline (ml). E–H: Digital 3D models of third instar brains (anterior view) with volume renderings of secondary en lineages (green). The lineages correspond to that shown in A–D. In models, mushroom body is shown in dark gray for better orientation. Compartments are rendered transparent. I, I': Digital 3D model of third instar brain with montage of all four en lineages; lateral view (I) and anterior view (I'). J, J': Digital models showing neuropile compartments in lateral view (J) and anterior view (J'). Lineages depicted in I/I' are shown in different colors: DPLam red, DALv2/3 purple; BAla3 green. Compartments in E–J' are shaded in colors that reflect the lineages they are innervated by (dark blue: antennal organ; light blue: BAla3; purple: DALv2/3; burgundy/pink: DPLam plus DALv. Note that axon tracts of secondary lineages are associated with the same compartments that were innervated by primary neurons of corresponding lineages (see Fig.2/3). For abbreviations of compartments see legend of Fig.2. Bar: 25μm
Secondary axon tracts of most, if not all, neuroblast lineages in the brain are thought to grow along the neuronal processes of primary neurons, possibly using them as guidance structures (Larsen et al., personal communication). This suggests that the secondary neurons of the four en-expressing lineages might innervate the same brain compartments as do their lineage-related en-expressing primary neurons. MARCM clonal analysis indicates that this is indeed the case (Fig. 4I, J). The protocerebral DPLam lineage projects its secondary axon tract postero-medially into the CPL compartment, thus innervating the same neuropile domain densely innervated by the primary DPLam neurons (Fig.4A, E). Approaching the peduncle of the mushroom body from an antero-dorsal direction, this secondary axon tract branches and sends one branch medially across the peduncle, the other one ventrally. The secondary axon tracts of the two protocerebral DALv2/3 lineages form one fascicle that grows along the BC compartment, passes underneath the medial lobe of the mushroom body, and reaches the midline. The DALv3 lineage has a secondary axon tract that is split, forming a dorsal and a ventral branch (Fig. 4B, F). These represent the larval forerunners of the two commissures that flank the ellipsoid body in the adult brain (see below). The DALv2 has a single unbranched secondary axon tract that stops short of the midline (Figs. 4C, G). The deutocerebral BAla3 secondary axon tract, similar to the primary BAla3 neurons, passes ventro-medially of the antennal lobe and grows backward into the BPM compartment (Fig. 4D, H). (Although one or two deutocerebral BAla3 neuroblast lineages may express en in the late embryo, only a single clone of secondary neurons can be recovered in larval stages suggesting that there is indeed only one lineage or, alternatively, that one of the two neuroblasts does not generate en-expressing secondary neurons since anti-En antibody immunostaining reveals only a single deutocerebral cluster of secondary neurons; data not shown). No en-positive secondary clones were recovered for the en-positive tritocerebral neuroblasts implying that they do not generate en-positive secondary cells.
In a recent clonal analysis of Drosophila neuronal lineages, three main morphological classes of lineages were distinguished (Larsen et al., personal communication; Fig.3A). Lineages belonging to the first class (type 1) elaborate distinct, spatially separate proximal and distal arborizations. For example, the lineages of olfactory projection neurons between antennal lobe and protocerebrum form proximal, dendritic arborizations in one or more glomeruli of the antennal lobe, and have their terminal, axonal arborizations in the calyx or lateral horn of the protocerebrum (Rodrigues and Hummel, 2008). The second class of lineages (type 2) forms branches that are distributed more or less evenly along the entire length of the axon tracts. One cannot distinguish, in these lineages, between a defined proximal input and a distal output domain. Type 3 lineages form a relatively long tract without proximal arborizations (i.e., branches that are formed in the neuropile adjacent to the cell bodies); arborizations are restricted to the distal part of fibers. Our MARCM-based clonal analysis shows that all three of the protocerebral en-positive lineages (DPLam, DALv2/3) are type 2 lineages. In contrast, the deutocerebral BAla3 neurons show characteristics of type 3 lineages.
The arborization pattern of secondary engrailed-lineages may delimit boundaries between adult brain neuropile compartments
Secondary neurons remain undifferentiated during the larval stage; aside from the single undivided fibers that are assembled into the secondary axon tracts, no terminal branches or synaptic connections are formed (see Hartenstein et al., 2008a). During the subsequent pupal phase, secondary neurons undergo substantial maturation processes involving generation of terminal and proximal arborizations as well as formation of synaptic interconnections and, thus, attain the mature neuron morphology that characterizes the adult brain. To determine the neuroanatomical features of the en-expressing neuroblast lineages in the mature brain (supraesophageal ganglion), we generated MARCM clones at larval hatching and recovered labeled, en-positive clones in the adult. In order to reveal the spatial relationship between the neuroblast clones and the mature neuropile compartments of the adult brain, these preparations were also labeled with the Nc82 antibody, which stains synaptic neuropile.
Figure 5 shows the MARCM-labeled arborizations of the BAla3, DALv3 and DPLam lineages in different frontal sections of the adult brain as well as 3D digital models of the lineages relative to the neuropile compartments. It is notable that the DALv2 lineage can no longer be recovered in the adult brain, suggesting that the neuroblast or the neurons of this lineage no longer express en. In all cases, the adult brain compartments occupied by the arborizations of the secondary lineages largely conform to the larval compartments innervated by the primary lineages. Moreover, in most cases the arborizations of the different en-positive lineages largely respect each others' territories and do not intermingle (Fig. 6A–C). In doing so, these lineages appear to delimit boundaries that may correspond to the borders of the protocerebrum, deutocerebrum and tritocerebrum in the adult brain neuropile.
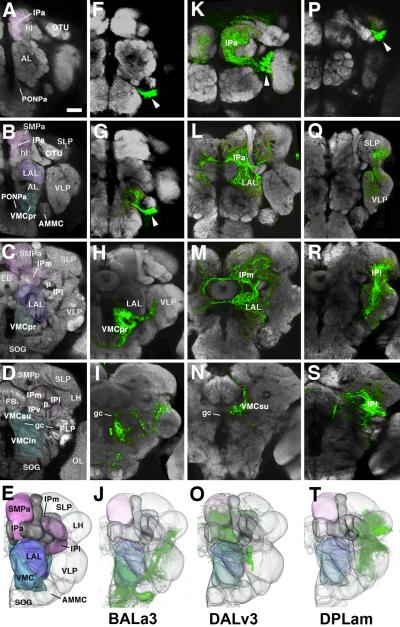
Fig.5. Arborization of secondary engrailed lineages in adult brain.
A–S: Upper four panels of each column show Z-projections of frontal confocal sections of adult brains labeled with Nc82 antibody (synapses, grey) which delineates neuropile compartment. Each panel represents a Z projection of optical sections (“brain slice”) of approximately 20 micron thickness. Rows of panels start anteriorly in the brain (top) and move posteriorly (second to bottom). Panels of the first row (A, F, K, P) show the neuropile right in front of the horizontal lobe of the mushroom body (hl); the second row (B, G, L, Q) right behind the horizontal lobe; third row (C, H, M, R) at the level of the ellipsoid body (EB); fourth row (D, I, N, S) at the level of the fan-shaped body. E–T: The bottom row presents 3D digital models of adult brain neuropile in anterior view. The first column identifies the compartments (AL antennal lobe; AMMC antennomechanosensory and motor center; CCX central complex; CX calyx of mushroom body; EB ellipsoid body; FB fan-shaped body; gc great commissure; hl horizontal lobes of mushroom body; IP inferior protocerebrum; IPa anterior domain of IP; IPl lateral domain of IP; IPm medial domain of IP; IPv ventral domain of IP; LAL lateral accessory lobe; LH lateral horn; OL optic lobe; OTU optic tubercle; p peduncle of mushroom body; PLP postero-lateral protocerebrum; PONPa anterior perioesophageal neuropile; PSi inferior posterior slope (VMCpo); PSs superior domain of posterior slope (=VMCpo); SLP superior lateral protocerebrum; SMP superior medial protocerebrum; SMPa anterior domain of SMP; SMPp posterior domain of SMP; SOG suboesophageal ganglion; vl vertical lobes of mushroom body; sp spur of mushroom body; VLP ventrolateral protocerebrum; VMC ventromedial cerebrum; VMCin infracommissural domain of VMC; VMCpo postcommissural domain of VMC; VMCpr precommissural domain of VMC; VMCsu supracommissural domain of VMC). Color scheme corresponds to that one of Figure 4: the VMC, whose larval forerunner (BPM) is innervated by en lineage BAla3, is shaded blue; LAL (larval precursor: BC), innervated by DALv2/3, is purple; IPa (larval precursor: CA), SMPa (larval precursor: DA) and IPm (larval precursor: CPI), all innervated by DALv and DPLam, are pink; IPl (larval precursor: CPL), innervated mostly by DPLam, is burgundy. The second, third and fourth column each shows the arborization pattern of a single en lineage (BAla3 (F–J), DALv3 (K–O), and DPLam (P–T), respectively). Clusters of cell bodies of the lineages are identified by arrowheads. Note that the compartments that contain arborizations of the secondary neurons largely correspond to those that had input from primary neurons of the corresponding lineage. An exception is DPLam, where secondary neurons contribute substantially to the VLP, which had no primary arborizations; moreover, secondary DPLam neurons do not project to the SMP and IPa, both of which did receive primary neuronal input. Bar: 25μm
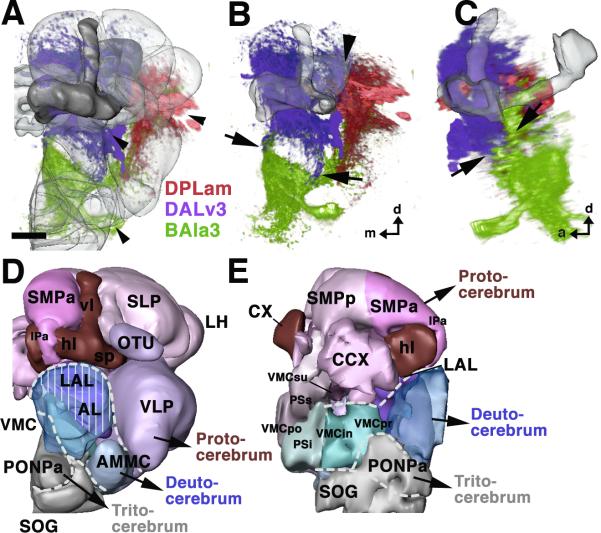
Fig.6. Secondary engrailed lineages in the adult brain and suggested neuromere boundaries.
A–C: montages of volume renderings of the three en lineages shown in different colors (DPLam: red; DALv3: purple; BAla3: green). A and B show anterior views; in A, neuropile compartments are shaded. C presents a lateral view. Arrowheads in A point at the cell body clusters of the three lineages. Arrows in B and C outline the sharp boundary between arborizations of DALv3 (above) and BAla3 (below). This boundary corresponds to the structurally distinct boundary between LAL compartment (above) and VMC (below). Large arrowhead in B demarcates another, less sharply demarcated boundary between the IPm and IPl (figure 5); DPLam neurites are largely confined to the IPl, DALv3 neurites to the IPm. D, E: 3D digital models of the neuropile compartments of the adult brain, presenting suggested neuromere boundaries as white hatched lines. D shows anterior view, E medial view. Color coding is similar to that used in Figures 4 and 5. Compartments of the protocerebrum are shown in shades of red and purple, deutocerebral compartments in blue, and the tritocerebrum in grey. Note that in D, the hatched line indicating the proto-deutocerebral boundary arches dorsally over the deutocerebral antennal compartment (AL), which is rendered transparent to let one see through to the VMC and LAL located behind. The LAL (vertical lines) belongs to the protocerebrum, while the VMC (no vertical lines) to the deutocerebrum. Also, note that the volume renderings in A–C do not reveal the fact that BAla3 terminal arbors are excluded from the PONPa; this is visible in Z-Projections shown in Fig.5F, G. For abbreviations of neuropile compartments, see Fig.5. Bar: 25μm
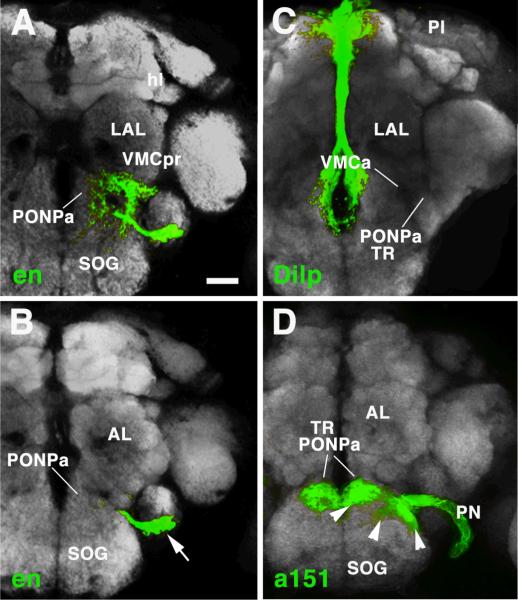
The BAla3 lineage forms dense terminal arborizations in the anterior part of the ventro-medial cerebrum (VMC), the compartment that develops from the larval BPM (Pereanu et al., personal communication; Fig. 5H). Some longer branches are directed laterally into the ventro-lateral protocerebrum (VLP), the descendant of the larval BPL, as well as ventrally into the subesophageal ganglion (Fig. 5H, I). The BAla3 arborization defines a relatively sharp boundary medially and anteriorly towards the anterior perioesophageal neuropile (PONPa) (Fig. 7A, B). This BAla3 arborization-negative domain corresponds to the anterior perioesophageal domain that harbors the sensory terminals of pharyngeal nerve axons, which originate in the intercalary segment (Fig. 7D). Furthermore, overlapping with this sensory neuropile, but continuing further posteriorly, is the compartment called flange (Strausfeld, 1976; Rajashekhar and Singh, 1994), which receives input from the pars intercerebralis (PI) (Fig. 7C). Both pharyngeal sensory afferents and PI input define the tritocerebrum in Drosophila (Rajashekhar and Singh, 1994) and other insects (Bullock and Horridge, 1965; Zaretsky and Loher, 1983). Based on this, we suggest that this anterior perioesophageal domain, which is not innervated by the en-expressing BAla3 lineage, corresponds to the tritocerebrum (Fig. 6D, E).
Fig.7. Tentative delineation of the tritocerebrum.
A, B: Z-projections of frontal confocal sections of adult brain labeled with Nc82 antibody (grey; labels neuropile compartments). The en-positive BAla3 lineage is shown in green; arrow in B points at cell bodies. Panel A represents neuropile at level of mushroom body horizontal lobe (hl) and lateral accessory lobe (LAL); focal plane shown in B represents neuropile more anteriorly, at the level of the antennal lobe (AL). C, D: Z-projections of confocal sections at levels corresponding to those shown in A and B, respectively. In C, projection of Dilp-positive neurons of the Pars intercerebralis (PI) to the anterior perioesophageal neuropile (PONPa) is visualized by Dilp-Gal4, UAS-mcd8GFP (Rulifson et al., 2002). In D, the sensory afferents of the pharyngeal nerve (PN, labeled by Gal4-driver line a151; kindly provided by Dr.Julie Simpson, JFRC, USA), which carries axons of the sensory neurons that originate in the intercalary segment, are shown in the anterior-most tip of the perioesophageal compartment. Note that the anterior perioesophageal neuropile is devoid of arborizations of the deutocerebral BAla3 lineage (A, B). We take this finding to support the notion that the PONPa represents the tritocerebrum (TR). Bar: 25μm
The DALv3 lineage fills the lateral accessory lobe (LAL, descendant of the BC compartment) and the medial inferior protocerebrum (IPm, descendant of the larval CPI; Fig. 5L, M). In addition, the anterior part of the inferior protocerebrum (IPa) that wraps around the horizontal lobes of the mushroom body like a cuff contains dense terminal arborizations of DALv3. Commissural fibers that cross dorsal and ventral of the ellipsoid body carry input of DALv3 to the contralateral IPm and LAL (Fig.5M). Noteworthy is the sharp boundary between DALv3 and BAla3 that coincides with the morphologically distinct boundary of LAL (DALv3) and anterior (precommissural) VMC (BAla3). More posteriorly, at the level of the great commissure (gc), the boundary coincides with this commissure; BAla3 fibers are restricted to the territory below the commissure (the infracommissural VMC; Fig.5I), and DALv3 to the one above the commissure (supracommissural VMC; Fig.5N). We propose that this boundary can be used to delineate the protocerebral-deutocerebral borderline within the neuropile (Fig. 6D, E).
The domain of arborization of DPLam is in the inferior lateral protocerebrum (IPl; Fig.5R) which develops from the larval CPL, and, more posteriorly, reaches into the inferior medial protocerebrum (Fig. 5S). In addition, branches extend into the ventrolateral protocerebrum (VPL) and superior lateral protocerebrum (SLP) adjacent to the IPl (Fig. 5Q, R). Projecting both DPLam and DALv3 into the same 3D model makes it clear that the two lineages divide the inferior protocerebrum in such a way that DALv3 innervates mostly medial and anterior domains while DPLam innervates mostly posterior and lateral domains (Fig. 6D, E).
Secondary lineages expressing the empty spiracles gene arborize in neuromere-specific neuropile compartments
To what extent do other neuroblast lineages respect the proposed boundaries between neuromere-specific compartments in the brain? To address this, we focused on the secondary neuron lineages in the larval brain that comprise empty spiracles (ems)-positive neurons (Fig. 8B); the development of these lineages was analysed by Lichtneckert et al. 2007; Lichtneckert et al., 2008. In the embryo, two lineages are known to derive from protocerebral neuroblasts that co-express ems and en, and are thus identfied as the DALv2/3 neuroblasts (Fig.8A). However, in the post embryonic larval brain, only one of the two neuroblast lineages, DALv3, co- expresses ems and en. (We show above that the dendritic arbors of the DALv2/3 secondary neuron lineages are restricted to protocerebral-specific compartments, hence they will not be considered further here.)
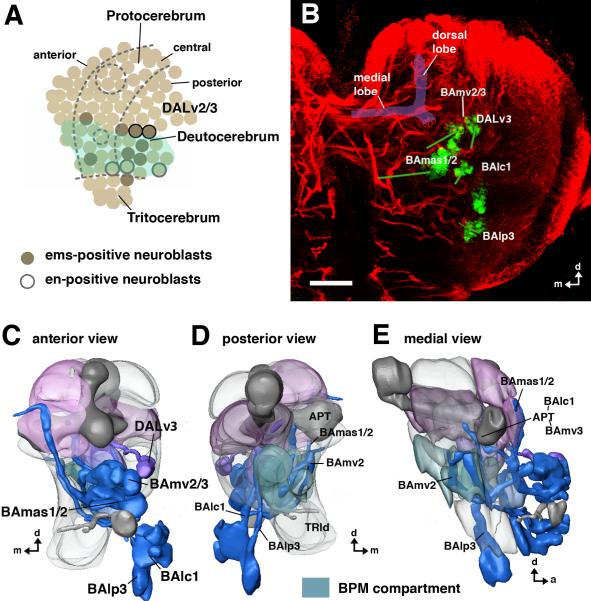
Fig.8. Projection of ems-positive lineages in the late larval brain.
A: Schematic map of embryonic brain neuroblasts (lateral view; after Urbach and Technau, 2003a, b; Sprecher et al., 2007). Neuroblasts that were classified as deutocerebral based on their spatial relationship to en and other markers are shaded green. ems (shaded in brown) is expressed in one tritocerebral neuroblast, four deutocerebral neuroblasts, and four neuroblasts along the posterior border of the protocerebrum. Two of these are the DALv2/3 neuroblasts that also turn on en (shown as black-frame neuroblasts). B: Z-projection of confocal section of a late larval brain labeled with anti-Neurotactin (red; visualizes secondary lineages), and anti-Ems (green). The Ems-positive lineages of interest are pointed out by labels; they include the protocerebral DALv3, and the deutocerebral BAmv2/3, BAlc1/2 and BAlp3. Green lines connect the clusters of somata to the belonging, BP106-positive axon tracts. C–E: Digital 3D models of late larval brains with neuropile compartments rendered semi-transparent and ems-positive lineages rendered blue (deutocerebral) and purple (protocerebral). Three deutocerebral lineages, BAmv3, BAlc1, and BAlp3 connect the deutocerebral antennal lobe with the protocerebrum; two deutocerebral lineages, BAmv2/3, project to the BPM compartment (D, E). BAmas1/2 are most likely tritocerebral, since they have proximal arbors in this compartment (Lichtneckert et al., 2007). Bar: 25μm.
Among the remaining lineages that express ems in the larval brain, five are deutocerebral (Younossi-Hartenstein et al., 1997; Urbach and Technau, 2003a). Analysis of the secondary axon tracts of these lineages relative to major brain neuropile structures using clonal MARCM labeling combined with anti-Ems and anti-Neurotactin co-immunostaining identifies them as BAlc1/2, BAmv2/3 and BAlp3 (Fig. 8C–E). Moreover, characterization of their domains of arborization in the adult brain indicates that arbors are indeed found in deutocerebral compartments as proposed above. Thus, BAlc1/2 and BAmv3 form projection neurons which have dendritic arbors in the antennal lobe (of the deutocerebrum) and which connect the antennal lobe with the protocerebrum (Pereanu and Hartenstein, 2006; Lichtneckert et al., 2008; VH, unpublished). Also, the BAlp3 and BAmv2 lineages project neurites to the VMC and form arbors there in the adult brain, providing further support for the idea that this compartment forms part of the deutocerebrum.
The remaining ems-positive lineage can be identified as BAmas2 based on its neuroanatomical features (Fig. 8B–E). The somata of the secondary neurons in this lineage are located most ventrally of all the ems-positive lineages in the larval brain; these neurons most likely represent the progeny of the unique tritocerebral neuroblast that expresses ems (Urbach and Technau, 2003a). The characteristic axon tract of BAmas2 projects up in the median bundle towards the protocerebrum (Pereanu and Hartenstein, 2006; Lichtneckert et al., 2007). In the adult, BAmas2 produces dendritic arborizations in the small neuropile domain that receives sensory afferents from the pharyngeal nerve, and thereby probably represents part of the tritocerebrum (see Fig.7).
Taken together, these findings indicate that the adult-specific neurons of both the en-expressing lineages and the ems-expressing lineages form their domain of arborization in discrete sets of compartments in the brain neuropile. Moreover, they suggest that lineages deriving from different brain neuromeres innervate a non-overlapping set of neuropile compartments. These observations support a model for neuromere-specific brain neuropile, in which a given lineage forms its arborizations predominantly in the compartments that correspond to its neuromere of origin.
DISCUSSION
engrailed expression and neuromere boundaries in the Drosophila brain
In this paper we have analyzed the pattern of neurite projections and arborizations of the en-positive lineages in the brain of Drosophila embryos, larvae and adults. Our data demonstrate that arborizations of individual lineages are restricted to discrete neuropile compartments, or parts thereof. Furthermore, they show that the arborization pattern seen at the larval stage (formed by primary neurons) is remarkably similar to the pattern of the adult (formed by secondary neurons).
Based on the characteristic trajectory of axon tracts and their terminal arborizations, we can follow the en-positive lineages unambiguously from late embryo to adult. But the question arises whether the en-positive neuroblasts, which demarcate the posterior neuromere boundary within the early embryonic (stage 8–11) neural primordium, are the same cells that we recognize at later embryonic stages. It could be argued that the expression pattern of transcription factors, generally speaking, is dynamic; in the neural primordium, neuroblasts expressing en at stage 11 could lose expression during stage 12/13, and at the same time, other neuroblasts gain expression, so that the neuroblasts visible at stage 15 are different ones from those at stage 11. We argue that this scenario is very unlikely, and even if correct, the neuroblasts gaining en-expression would have to be the immediate neighbors of the neuroblasts that lose expression. The basis for this argument is the fact that the time interval between stage 11 and stage 15 (when tracts are clearly distinguishable) lasts only about 5 hours, and observations of embryos at very close increments (15min) can be, and were, made. In such a sequence of observations it is evident that initially, small groups of en-positive neuroblasts demarcate the posterior neuromere boundary. Some of these neuroblasts generate en-positive clusters of GMCs and neurons, which stay in contact with their parent neuroblasts. As time passes, the clusters grow larger, and finally start producing axon tracts. Given this continuity of observations we can be quite certain that the metameric groups of lineages we define in the late (stage 15) embryo stem from within the metameric groups of neuroblasts defined at stage 11.
The arborization patterns of en-expressing lineages provide support for the existence of neuromere boundaries that coincide with morphologically defined neuropile compartment boundaries. According to our data, the boundary between protocerebrum and deutocerebrum corresponds to the interface between LAL/CC and ventromedial cerebrum (VMC). In view of the domains of the fly brain neuropile that are classically referred to as protocerebral (see, for example, Strausfeld, 1976), the findings reported here are entirely confirmatory; these domains include the superior and inferior protocerebrum, lateral horn, mushroom body, central and accessory complex, ventrolateral protocerebrum, and optic lobe. In regard to deutocerebral domains, we add the VMC to the two traditionally accepted sensory domains of the deutocerebrum, namely the antennal lobe and AMMC. Specifically, the precommissural VMC and the infracommissural VMC that flank the great commissure receive dense innervation from the en-positive BAla3 lineage, as well as from other deutocerebrally derived lineages expressing ems (e.g., BAmv2). For the tritocerebral neuropile, we tentatively assign the anterior perioesophageal neuropile domain, which lies medial and anterior of the VMC and is excluded from the BAla3 arborization, to the tritocerebrum. This coincides well with previous anatomical studies (Rajashekhar and Singh, 1994) that designated the region receiving input from the pars intercerebralis (via median bundle) and from the pharyngeal nerves as tritocerebral.
Our argument for assigning the VMC to the deuterocerebrum is based on the unity of neuronal cell bodies and their (proximal) arborizations. Thus, in the clearly metameric segmental ganglia of the ventral nerve cord the majority of neurons have most of their terminal arbors (i.e., arbors of all local neurons, dendrites of motorneurons, dendrites of many intersegmental interneurons, axon terminals of sensory neurons) in the neuropile of the same neuromere that produces their cell bodies. In other words, neurons whose cell bodies derive from segment A have neurites that largely arborize in the segmental neuropile of segment A. If this situation also applies to the neuromeres of the brain, then we would predict, for example, that the VMC should be considered part of the deutocerebral neuropile. This is because the BAla3 neurons clearly derive from the deutocerebrum and the large majority (more than 90%) of their terminal arborizations fall within the realm of the VMCpr and VMCi. To test this notion, future analyses of the arborization pattern of other lineages, as well as of connectivity patterns within the VMC, will be needed. Some insight may also be gained by identifying the serial homologs of the brain lineages in the thoracic ganglia of the ventral nerve cord, and then comparing key neuroanatomical as well as other features between homologous lineages in brain and ventral ganglion neuromeres. We are optimistic that the numerous genetic markers (e.g. Pfeiffer et al., 2008) that are becoming available at an increasing rate will soon provide more tools to study the development of anatomical modules such as compartments and neuromeres in the Drosophila brain.
VMC: features of a novel deutocerebral neuropile compartment
Currently, very little is known about the structure and function of the VMC. Classical neuroanatomical accounts dealing with the deutocerebrum in arthropods make no mention of this region (Bullock and Horridge, 1965; Strausfeld, 1976; Homberg et al., 1989). In Strausfeld's atlas of the Musca brain (1976), arguably the most detailed account of insect brain anatomy, the only named compartment posterior to the lateral accessory complex (= ventral body) is the posterior slope. An anterior boundary of the slope is not indicated, leaving open the question whether or not a separate, distinct compartment is located between the slope and the LAL. Thus, based on published data it is not currently possible to relate the posterior slope to parts of the VMC; the posterior slope could correspond to only the posterior-most VMC layer or even to the entire VMC compartment. Irrespective of this uncertainty regarding boundaries, it has been found that the posterior slope of Musca and other insects receives ascending fibers from the subesophageal and thoracic ganglia (Strausfeld, 1976). Similarly, collateral axons of descending neurons from the ventrolateral protocerebrum are known to branch in the posterior slope. In a recent study of neurons connecting the brain and ventral nerve cord of Drosophila (Hartenstein et al., 2008a) we found that the larval BPM compartment, which corresponds to the adult VMC, also receives a large amount of ascending axons from the ventral nerve cord; furthermore, one of the groups of descending neurons (BP-DN) has cell bodies located in the posterior cortex adjacent to the BPM. These findings suggest that the BPM/VMC, similar to other basal neuropile compartments like the LAL (Homberg, 1994; Homberg et al., 2004) or ventrolateral protocerebrum (Strausfeld and Gronenberg, 1990), may represent an important output center where multimodal sensory input, integrated upstream or within the VMC neuropile, is used to drive descending “command neurons” that influence motor behavior.
Some of its neuroanatomical features suggest that the VMC might correspond to the dorsal “motor neuropile” of the deutocerebrum. Given the 90 degree upward turn of the neuraxis that occurs in the head between subesophageal ganglion and brain, positions that are ventral in the subesophageal (or thoracic) ganglia are anterior in the (basal) brain, and positions that are dorsal in the gnathal and ventral ganglia (like the motor neuropiles of the thoracic segments) are posterior in the brain. Are there indications that the VMC, a posterior (deutocerebral) compartment, receives dendrites of motorneurons, like the dorsal neuropile in the thoracic segments? Although strongly modified from early developmental stages onward, the antennal segment, to which the deutocerebrum belongs, has all the hallmarks of a segment, including a myomere that gives rise to a number of mesodermal tissues, including somatic muscles and part of the vascular system (Ullman, 1964; DeVelasco et al., 2006). In the antennal segment, the myomere-derived muscles are the extrinsic muscles that move the antenna. These muscles are innervated by deutocerebral motoneurons. The central and peripheral projection of these antennal motor neurons has been studied in several insect taxa (ant: Ehmer and Gronenberg, 1997; moth: Kloppenburg et al., 1997; cockroach: Baba and Comer, 2008), but not in Drosophila. Dendrites of the antennal motorneurons are reported to branch in the AMMC, which is called the “dorsal lobe” in certain species by some authors (e.g., Ehmer and Gronenberg, 1997). From the description given and markers shown, it is not clear whether the AMMC/dorsal lobe of other insects corresponds to what is defined (strictly by input from the antennal nerve) as the AMMC in Drosophila, or whether it might also include a more posterior domain corresponding to the VMC in Drosophila (Peterson et al., 1998). Careful anatomical studies of the antennal motor neurons in Drosophila should clarify this issue.
Expression and function of engrailed at neuromere boundaries in other animals
Detailed studies of early en expression in the head neurectoderm have been performed for a variety of arthropods, including many insects (Diederich et al., 1991; Schmidt-Ott and Technau, 1992; Younossi-Hartenstein et al., 1993; Rogers and Kaufman, 1996; Peterson et al., 1998; Boyan and Williams, 2002), as well as a few crustaceans (Sintoni et al., 2007). Outside the arthropods, we know of en expression in several annelids (Wedeen and Weisblat, 1991; Shain et al., 2000; Seaver et al., 2001; Prud'homme et al., 2003; Seaver and Kaneshige, 2006), mollusks (Moshel et al., 1998; Jacobs et al., 2000; Wanninger and Haszprunar, 2001; Nederbragt et al., 2002; Iijima et al., 2008), ascidians (Imai et al., 2002; Canestro et al., 2005), Cephalochordates (Holland et al., 1997; Beaster-Jones et al., 2008) and, of course, vertebrates (Joyner, 1996; Simon et al., 2004; 2005). In arthropods and most annelids, en expression is seen in the early embryo in a metameric striped pattern that coincides with segment boundaries. In addition, as shown for Drosophila, en is typically expressed in the progeny (or derivatives) of cells in the early en stripes (e.g. segmental clusters of neurons). Some annelids seem to have lost the early phase of en expression and maintain the gene only during a later phase in neurons and other metamerically organized cell types (Seaver et al., 2001). Metameric expression of en is not seen in vertebrates and invertebrate deuterostomians, except Amphioxus (Holland et al., 1997). In vertebrates, en appears in the neurectoderm in a single broad stripe that demarcates the mid-hindbrain boundary. However, as for invertebrates, some of the derivatives of the en-positive neurectoderm (e.g. parts of the cerebellum) maintain en expression during subsequent developmental phases.
In view of the fact that en is a transcription factor, its function very much depends on the type of genes activated or repressed by it. It is sometimes assumed that these downstream factors (in case of en as well as many other early expressed patterning genes) form part of a defined molecular developmental pathway. This assumption is often misleading, or at least unproductive. All one can reliably generalize about the role of en is that is required for both early (morphogenetic) and late (differentiative) aspects of the cells it is expressed in. With respect to its early function, en appears to be involved in setting up segmental boundaries, such that adhesion molecules or other structural proteins that form part of the cellular mechanism for generating or maintaining segmental boundaries (invagination of cells to form furrow; differential adhesion of cells to stabilize boundaries) are under the control of en (Dahmann and Basler, 2000). As far as the late function of en is concerned, loss of function studies of en in Drosophila and vertebrates show that cell types expressing en, such as the adrenergic and serotonergic neurons of the vertebrate brainstem, are often absent or reduced in en mutants (Simon et al., 2004; Simon et al., 2005), or show pathfinding defects (Joly et al., 2007). In most of these studies, antibodies raised against the En protein were used to study en expression, and, since En is a nuclear protein, the morphology of en-expressing neurons (i.e., neurite projections, terminal arborizations) as well as the neuropile compartments they contribute to could not be analyzed in detail. It will therefore be important to develop and extend the type of genetic labeling techniques available in the Drosophila model system and as utilized in this report, to investigate the precise anatomical features of wildtype and mutant en neurons and neuronal compartments in the developing brains of other vertebrate and invertebrate animals.
ACKNOWLEDGEMENTS
We thank Bruno Bello and Susanne Flister for helpful discussions and technical assistance. The project was funded by NIH Grant NS054814A to VH and Swiss NSF Grant 3100A0-112024 to HR.
FUNDED BY: NIH Grant R01 NS054814 to VH and the Swiss NSF Grant 3100A0-112024 to HR.
REFERENCES
- Baba Y, Comer CM. Antennal motor system of the cockroach, Periplaneta americana. Cell Tissue Res. 2008;331:751–62. doi: 10.1007/s00441-007-0545-9. [DOI] [PubMed] [Google Scholar]
- Beaster-Jones L, Kaltenbach SL, Koop D, Yuan S, Chastain R, Holland LZ. Expression of somite segmentation genes in amphioxus: a clock without a wavefront? Dev Genes Evol. 2008;218:599–611. doi: 10.1007/s00427-008-0257-5. [DOI] [PubMed] [Google Scholar]
- Bello BC, H. F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–19. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Boyan G, Williams L. A single cell analysis of engrailed expression in the early embryonic brain of the grasshopper Schistocerca gregaria: ontogeny and identity of the secondary headspot cells. Arthropod Struct Dev. 2002;30:207–18. doi: 10.1016/s1467-8039(01)00034-2. [DOI] [PubMed] [Google Scholar]
- Bullock TH, Horridge GA. Structure and function in the nervous system of invertebrates. Two volumes. Freeman; San Francisco: 1965. [Google Scholar]
- Burrows M. The Neurobiology of the Insect Brain. Oxford University Press; Oxford: 1996. [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. Springer; Berlin: 1997. [Google Scholar]
- Cañestro C, Bassham S, Postlethwait J. Development of the central nervous system in the larvacean Oikopleura dioica and the evolution of the chordate brain. Dev Biol. 2005;285:298–315. doi: 10.1016/j.ydbio.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Basler K. Opposing transcriptional outputs of Hedgehog signaling and engrailed control compartmental cell sorting at the Drosophila A/P boundary. Cell. 2000;100:411–22. doi: 10.1016/s0092-8674(00)80677-7. [DOI] [PubMed] [Google Scholar]
- De Velasco B, Mandal L, Mkrtchyan M, Hartenstein V. Subdivision and developmental fate of the head mesoderm in Drosophila melanogaster. Dev. Genes Evol. 2006;216:39–51. doi: 10.1007/s00427-005-0029-4. [DOI] [PubMed] [Google Scholar]
- Diederich RJ, Pattatucci AM, Kaufman TC. Developmental and evolutionary implications of labial, Deformed and engrailed expression in the Drosophila head. Development. 1991;113:273–81. doi: 10.1242/dev.113.1.273. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Nassif C, Hartenstein V. Early development of the Drosophila brain: V. Pattern of postembryonic neuronal lineages expressing DE- cadherin. J Comp Neurol. 2003;455:451–62. doi: 10.1002/cne.10484. [DOI] [PubMed] [Google Scholar]
- Ehmer B, Gronenberg W. Proprioceptors and fast antennal reflexes in the ant Odontomachus (Formicidae, Ponerinae) Cell Tissue Res. 1997;290:153–65. doi: 10.1007/s004410050917. [DOI] [PubMed] [Google Scholar]
- Grenningloh G, Rehm EJ, Goodman CS. Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell. 1991;67:45–57. doi: 10.1016/0092-8674(91)90571-f. [DOI] [PubMed] [Google Scholar]
- Hama C, Ali Z, Kornberg TB. Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev. 1990;4:1079–93. doi: 10.1101/gad.4.7.1079. [DOI] [PubMed] [Google Scholar]
- Hanström B. Vergleichende Anatomie des Nervensystems der Wirbellosen Tiere. Springer; Berlin: 1928. [Google Scholar]
- Hartenstein V, Spindler S, Pereanu W, Fung S. The development of the Drosophila larval brain. Adv Exp Med Biol. 2008a;628:1–31. doi: 10.1007/978-0-387-78261-4_1. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Cardona A, Pereanu W, Hartenstein AY. Modeling the developing Drosophila brain: Rationale, Technique and Application. BioOne. 2008b;58:823–36. [Google Scholar]
- Hartmann B, Hirth F, Walldorf U, Reichert H. Expression, regulation and function of the homeobox gene empty spiracles in brain and ventral nerve cord development of Drosophila. Mech Dev. 2000;90:143–53. doi: 10.1016/s0925-4773(99)00237-3. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Kene M, Williams NA, Holland ND. Sequence and embryonic expression of the amphioxus engrailed gene (AmphiEn): the metameric pattern of transcription resembles that of its segment-polarity homolog in Drosophila. Development. 1997;124:1723–32. doi: 10.1242/dev.124.9.1723. [DOI] [PubMed] [Google Scholar]
- Holmgren B. Vergleichende Anatomie des Nervensystems der Wirbellosen Tiere. Springer; Berlin: 1928. [Google Scholar]
- Homberg U. Flight-correlated activity changes in neurons of the lateral accessory lobes in the brain of the locust Schistocerca gregaria. J Comp Physiol A. 1994;175:597–610. [Google Scholar]
- Homberg U, Christensen TA, Hildebrand JG. Structure and function of the deuterocerebrum in insects. Annu Rev Entomol. 1989;34:477–501. doi: 10.1146/annurev.en.34.010189.002401. [DOI] [PubMed] [Google Scholar]
- Homberg U, Hofer S, Mappes M, Vitzthum H, Pfeiffer K, Gebhardt S, Müller M, Paech A. Neurobiology of polarization vision in the locust Schistocerca gregaria. Acta Biol Hung. 2004;55:81–9. doi: 10.1556/ABiol.55.2004.1-4.10. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Trquina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990;110:1327–40. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Iijima M, Takeuchi T, Sarashina I, Endo K. Expression patterns of engrailed and dpp in the gastropod Lymnaea stagnalis. Dev Genes Evol. 2008;218:237–51. doi: 10.1007/s00427-008-0217-0. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. Region specific gene expressions in the central nervous system of the ascidian embryo. Gene Expr Patterns. 2002;2:319–21. doi: 10.1016/s0925-4773(02)00383-0. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN- Cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Jacobs DK, Wray CG, Wedeen CJ, Kostriken R, DeSalle R, Staton JL, Gates RD, Lindberg DR. Molluscan engrailed expression, serial organization, and shell evolution. Evol Dev. 2000;2:340–7. doi: 10.1046/j.1525-142x.2000.00077.x. [DOI] [PubMed] [Google Scholar]
- Joly W, Mugat B, Maschat F. Engrailed controls the organization of the ventral nerve cord through frazzled regulation. Dev Biol. 2007;301:542–54. doi: 10.1016/j.ydbio.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Joyner AL. Engrailed, Wnt and Pax genes regulate midbrain--hindbrain development. Trends Genet. 1996;12:15–20. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- Kloppenburg P, Camazine SM, Sun XJ, Randolph P, Hildebrand JG. Organization of the antennal motor system in the sphinx moth Manduca sexta. Cell Tissue Res. 1997;287:425–33. doi: 10.1007/s004410050767. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Jeffrey V, Fujioka M, Jaynes JB, Bate M. Embryonic origins of a motor system: motor dendrites form a myotopic map in Drosophila. PLoS Biol. 2003;1:E41. doi: 10.1371/journal.pbio.0000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic Analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–61. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lichtneckert R, Bello B, Reichert H. Cell lineage-specific expression and function of the empty spiracles gene in the adult brain of Drosophila melanogaster. Development. 2007;134:1291–1300. doi: 10.1242/dev.02814. [DOI] [PubMed] [Google Scholar]
- Lichtneckert R, Nobs L, Reichert H. Empty Spiracles is required fort he development of olfactory projection neuron circuitry in Drosophila. Development. 2008;135:2415–24. doi: 10.1242/dev.022210. [DOI] [PubMed] [Google Scholar]
- Mathew D, Popescu A, Budnik V. Drosophila amphiphysin functions during synaptic Fasciclin II membrane cycling. J Neurosci. 2003;23:10710–6. doi: 10.1523/JNEUROSCI.23-33-10710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt DJ, Whitington PM. Central projections of sensory neurons in the Drosophila embryo correlate with sensory modality, soma position, and proneural gene function. J Neurosci. 1995;15:1755–67. doi: 10.1523/JNEUROSCI.15-03-01755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshel SM, Levine M, Collier JR. Shell differentiation and engrailed expression in the Ilyanassa embryo. Dev Genes Evol. 1998;208:135–41. doi: 10.1007/s004270050164. [DOI] [PubMed] [Google Scholar]
- Nederbragt AJ, van Loon AE, Dictus WJ. Expression of Patella vulgata orthologs of engrailed and dpp-BMP2/4 in adjacent domains during molluscan shell development suggests a conserved compartment boundary mechanism. Dev Biol. 2002;246:341–55. doi: 10.1006/dbio.2002.0653. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Elli MC, Kornberg TB, Goodman CS. Expression of engrailed proteins in arthropods, annelids and chordates. Cell. 1989;58:955–68. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages in the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J. Neurosci. 2006;26:5534–53. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MD, Popadić A, Kaufman TC. The expression of two engrailed-related genes in an apterygote insect and a phylogenetic analysis of insect engrailed-related genes. Dev Genes Evol. 1998;208:547–57. doi: 10.1007/s004270050214. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–20. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovant P, Lena P. Membrane glycoproteins immunologically related to the human insulin receptor are associated with presumptive neuronal territories and developing neurons in Drosophila melanogaster. Development. 1988;103:145–56. doi: 10.1242/dev.103.1.145. [DOI] [PubMed] [Google Scholar]
- Prud'homme B, de Rosa R, Arendt D, Julien JF, Pajaziti R, Dorresteijn AW, Adoutte A, Wittbrodt J, Balavoine G. Arthropod-like expression patterns of engrailed and wingless in the annelid Platynereis dumerilii suggest a role in segment formation. Curr Biol. 2003;13:1876–81. doi: 10.1016/j.cub.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Rajashekhar KP, Singh RN. Neuroarchitecture of the tritocerebrum of Drosophila melanogaster. J Comp Neurol. 1994;349:633–45. doi: 10.1002/cne.903490410. [DOI] [PubMed] [Google Scholar]
- Rodrigues V, Hummel T. Development of the Drosophila olfactory system. Adv Med Exp Biol. 2008;628:82–101. doi: 10.1007/978-0-387-78261-4_6. [DOI] [PubMed] [Google Scholar]
- Rogers BT, Kaufman TC. Structure of the insect head as revealed by the EN protein pattern in developing embryos. Development. 1996;122:3419–32. doi: 10.1242/dev.122.11.3419. [DOI] [PubMed] [Google Scholar]
- Rulifson E, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–89. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott U, Technau GM. Expression of en and wg in the embryonic head and brain of Drosophila indicates a refolded band of seven segment remnants. Development. 1992;116:111–25. doi: 10.1242/dev.116.1.111. [DOI] [PubMed] [Google Scholar]
- Schrader S, Merritt DJ. Central projections of Drosophila sensory neurons in the transition from embryo to larva. J Comp Neurol. 2000;425:34–44. [PubMed] [Google Scholar]
- Seaver EC, Paulson DA, Irvine SQ, Martindale MQ. The spatial and temporal expression of Ch-en, the engrailed gene in the polychaete Chaetopterus, does not support a role in body axis segmentation. Dev Biol. 2001;236:195–209. doi: 10.1006/dbio.2001.0309. [DOI] [PubMed] [Google Scholar]
- Seaver EC, Kaneshige LM. Expression of `segmentation' genes during larval and juvenile development in the polychaetes Capitella sp. I and H. elegans. Dev Biol. 2006;289:179–94. doi: 10.1016/j.ydbio.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Shain DH, Stuart DK, Huang FZ, Weisblat DA. Segmentation of the central nervous system in leech. Development. 2000;127:735–44. doi: 10.1242/dev.127.4.735. [DOI] [PubMed] [Google Scholar]
- Simon HH, Thuret S, Alberi L. Midbrain dopaminergic neurons: control of their cell fate by the engrailed transcription factors. Cell Tissue Res. 2004;318:53–61. doi: 10.1007/s00441-004-0973-8. [DOI] [PubMed] [Google Scholar]
- Simon HH, Scholz C, O'Leary DD. Engrailed genes control developmental fate of serotonergic and noradrenergic neurons in mid- and hindbrain in a gene dose-dependent manner. Mol Cell Neurosci. 2005;28:96–105. doi: 10.1016/j.mcn.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Sintoni S, Fabritius-Vilpoux K, Harzsch S. The engrailed-expressing secondary head spots in the embryonic crayfish brain: examples for a group of homologous neurons in Crustacea and Hexapoda? Dev Genes Evol. 2007;217:791–9. doi: 10.1007/s00427-007-0189-5. [DOI] [PubMed] [Google Scholar]
- Sprecher S, Reichert H, Hartenstein V. Gene expression patterns in primary neuronal clusters of the Drosophila embryonic brain. Gene Expr Patterns. 2007;7:584–95. doi: 10.1016/j.modgep.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld N. Atlas of an Insect Brain. Springer; Berlin: 1976. [Google Scholar]
- Strausfeld NJ, Gronenberg W. Descending neurons supplying the neck and flight motor of Diptera: organization and neuroanatomical relationships with visual pathways. J Comp Neurol. 1990;302:954–72. doi: 10.1002/cne.903020419. [DOI] [PubMed] [Google Scholar]
- Technau GM, Berger C, Urbach R. Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev Dyn. 2006;235(4):861–9. doi: 10.1002/dvdy.20566. [DOI] [PubMed] [Google Scholar]
- Tyrer NM, Gregory GE. A guide to the neuroanatomy of locust suboesophageal and thoracic ganglia. Phil Trans R Soc Lond B. 1982;297:91–123. [Google Scholar]
- Ullmann SL. The origin and structure of the mesoderm and the formation of the coelomic sacs in Tenebrio molitor L (Insecta, Coleoptera) Phil Trans R Soc Lond B. 1964;747:245–276. [Google Scholar]
- Urbach R, Technau GM. Segment polarity and DV patterning gene expression reveals segmental organization of the Drosophila brain. Development. 2003a;130:3607–20. doi: 10.1242/dev.00532. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003b;130:3621–37. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–44. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Walldorf U, Gehring WJ. Empty spiracles, a gap gene containing a homeobox involved in Drosophila head development. EMBO J. 1992;11:2247–59. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanninger A, Haszprunar G. The expression of an engrailed protein during embryonic shell formation of the tusk-shell, Antalis entalis (Mollusca, Scaphopoda) Evol Dev. 2001;3:312–21. doi: 10.1046/j.1525-142x.2001.01034.x. [DOI] [PubMed] [Google Scholar]
- Wedeen CJ, Weisblat DA. Segmental expression of an engrailed-class gene during early development and neurogenesis in an annelid. Development. 1991;113:805–14. doi: 10.1242/dev.113.3.805. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein AY, Tepass U, Hartenstein V. The embryonic origin of the primordia of the adult Drosophila head. Roux's Arch Dev Biol. 1993;203:60–73. doi: 10.1007/BF00539891. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370:313–29. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Green P, Liaw G, Rudolph K, Lengyel J, Hartenstein V. Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev Biol. 1997;182:270–283. doi: 10.1006/dbio.1996.8475. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra P, Hartenstein V. Early development of the Drosophila brain: IV. Larval neuropile compartments defined by glial septa. J Comp Neurol. 2003;455:435–50. doi: 10.1002/cne.10483. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nguyen B, Shy D, Hartenstein V. Embryonic origin of the Drosophila brain neuropile. J Comp Neurol. 2006;497:981–98. doi: 10.1002/cne.20884. [DOI] [PubMed] [Google Scholar]
- Zaretsky M, Loher W. Anatomy and Electrophysiology of individual neurosecretory cells of an insect brain. J Comp Neurol. 1983;216:253–63. doi: 10.1002/cne.902160304. [DOI] [PubMed] [Google Scholar]
- Zlatic M, Landgraf M, Bate M. Genetic specification of axonal arbors: atonal regulates robo3 to position terminal branches in the Drosophila nervous system. Neuron. 2003;37:41–51. doi: 10.1016/s0896-6273(02)01131-5. [DOI] [PubMed] [Google Scholar]