Abstract
Prostaglandin endoperoxide H synthases (PGHS)-1 and -2, also called cyclooxygenases, convert arachidonic acid (AA) to prostaglandin H2 (PGH2) in the committed step of prostaglandin biosynthesis. Both enzymes are homodimers, but the monomers often behave asymmetrically as conformational heterodimers during catalysis and inhibition. Here we report that aspirin maximally acetylates one monomer of human (hu) PGHS-2. The acetylated monomer of aspirin-treated huPGHS-2 forms 15-hydroperoxyeicosatetraenoic acid from AA, whereas the nonacetylated partner monomer forms mainly PGH2 but only at 15 to 20% of the rate of native huPGHS-2. These latter conclusions are based on the findings that the nonsteroidal anti-inflammatory drug diclofenac binds a single monomer of native huPGHS-2, having an unmodified Ser530 to inhibit the enzyme, and that diclofenac inhibits PGH2 but not 15-hydroperoxyeicosatraenoic acid formation by acetylated huPGHS-2. The 18R- and 17R-resolvins putatively involved in resolution of inflammation are reportedly formed via aspirin-acetylated PGHS-2 from eicosapentaenoic acid and docosahexaenoic acid, respectively, so we also characterized the oxygenation of these omega-3 fatty acids by aspirin-treated huPGHS-2. Our in vitro studies suggest that 18R- and 17R-resolvins could be formed only at low rates corresponding to less than 1 and 5%, respectively, of the rates of formation of PGH2 by native PGHS-2.
Prostaglandin endoperoxide H synthases (PGHS)-1 and -2, also called cyclooxygenases (COXs), catalyze the conversion of arachidonic acid (AA) to prostaglandin (PG) H2 in the committed step in prostaglandin and thromboxane biosynthesis (van der Donk et al., 2002; Rouzer and Marnett, 2003, 2008; Schneider et al., 2007; Smith, 2008). The enzymes have two catalytic activities: 1) a COX activity responsible for oxygenating AA to prostaglandin G2; and 2) a peroxidase activity that catalyzes a two-electron reduction of prostaglandin G2 to PGH2. PGHS are homodimers that exhibit half of sites with COX activity with AA as the substrate (Yuan et al., 2006). That is, only one monomer is able to catalyze a reaction at a given time. The noncatalytic monomer functions as an allosteric regulator of the catalytic monomer (Kulmacz and Lands, 1985; Yuan et al., 2006, 2009). This is of potential importance in vivo, where certain fatty acids can function as allosteric regulators of PGHS inhibiting PGHS-1 and stimulating PGHS-2 (Yuan et al., 2009).
Nonspecific nonsteroidal anti-inflammatory drugs (nsNSAIDs) inhibit the COX activities of both PGHS-1 and PGHS-2, whereas COX-2 inhibitors called coxibs are more selective for huPGHS-2 (Grosser et al., 2006). nsNSAIDs and coxibs fall into three general categories based on their mechanisms of action, and their mechanisms are related in part to the way in which they interact with the dimeric structures of PGHS (DeWitt, 1999; Smith and DeWitt, 1999; Smith et al., 2000; Walker et al., 2001; Blobaum and Marnett, 2007). One category of inhibitors, which includes ibuprofen and mefenamic acid, are freely reversible competitive inhibitors. Binding of these inhibitors to the COX sites of both monomers comprising a dimer is required for inhibition of AA oxygenation by PGHS-2 (Prusakiewicz et al., 2009). This mechanism of inhibition may also underlie the actions of inhibitory fatty acids acting on PGHS-1 (Yuan et al., 2009). A second group of inhibitors is composed of time-dependent, noncovalent inhibitors, including the nsNSAIDs flurbiprofen, indomethacin, meclofenamate, and diclofenac. Flurbiprofen, meclofenamate, and indomethacin are allosteric inhibitors that bind to one monomer of PGHS to inhibit PGHS (Kulmacz and Lands, 1985; Yuan et al., 2006). We provide evidence in this report that inhibition by diclofenac also involves binding to one monomer of PGHS and that an intact Ser-530 is required in at least one of the monomers. Finally, aspirin is unique to a third group of inhibitors that cause a time-dependent, covalent inhibition. Binding of aspirin by PGHS-1 or PGHS-2 leads to an irreversible acetylation of a highly conserved Ser-530 (van der Donk et al., 2002; Rouzer and Marnett, 2003; Schneider et al., 2007; Smith, 2008). Aspirin acetylates one monomer of PGHS-1 to cause a temporally correlated loss of COX activity (Rimon et al., 2010). However, the situation with PGHS-2 is more complex. Treatment of PGHS-2 with aspirin converts the enzyme to a form that generates 15R-hydroxyeicosapentaenoic acid (15R-HETE) from AA (Holtzman et al., 1992; Lecomte et al., 1994). We report here an investigation of the stoichiometry of PGHS-2 acetylation by aspirin and the consequences of acetylation on product formation from arachidonic acid (AA), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA). We investigated the oxygenation of EPA and DHA because bioactive, trihydroxylated compounds called resolvin E1 (RvE1) and 17R-resolvin D1 (17R-RvD1) are reportedly formed at least in part via aspirin-acetylated PGHS-2 from the ω3 fatty acids EPA and DHA, respectively (Serhan et al., 2000; Serhan et al., 2008), and there is incomplete quantitative information on the oxygenation of EPA or DHA by purified PGHS-2. RvE1 is derived from 18R-hydroxy-eicosapentaenoic acid (18R-HEPE) formed from EPA and 17R-RvD1 is derived from 17R-hydroxy-docosahexaenoic acid (17R-HDHA) formed from DHA by aspirin-treated preparations containing PGHS-2.
Materials and Methods
Materials.
Flag-affinity resin, hemin, and acetylsalicylic acid (aspirin, ASA) were purchased from Sigma-Aldrich (St. Louis, MO). Arachidonic acid (AA), 11-hydroxyeicosatetraenoic acid (11-HETE), 15-hydroxyeicosatetraenoic acid (15-HETE), 15-hydroxyeicosatetraenoic-d8 acid, 11-hydroxyeicosapentaenoic acid (11-HEPE), 12-hydroxyeicosapentaenoic acid, 15-hydroxyeicosapentaenoic acid (15-HEPE), 18-hydroxyeicosapentaenoic acid (18-HEPE), 13-hydroxydocosahexaenoic acid (13-HDHA), and 17-hydroxydocosahexaenoic acid (17-HDHA) were from Cayman Chemical (Ann Arbor, MI). [1-14C]AA (55 mCi/mmol), [1-14C]EPA (55 mCi/mmol), and [1-14C]acetylsalicylic acid (55 mCi/mmol) were from American Radiolabeled Chemicals (St. Louis, MO). The nonionic detergents C10E6 and β-octylglucopyranoside were from Anatrace (Santa Clara, CA). BCA protein reagent was from Pierce Chemical (Rockford, IL). Complete EDTA-free protease inhibitor was from Roche Applied Science (Indianapolis, IN). Restriction enzymes were from New England Biolabs (Ipswich, MA). Nickel-nitrilotriacetic acid was from QIAGEN (Valencia, CA). Methanol and water were HPLC-grade and were purchased from Honeywell Burdick and Jackson (Muskegon, MI). Ammonium acetate was analytical reagent grade from Mallinckrodt Baker, Inc. (Paris, KY). Trichloroacetic acid (TCA) was purchased from Thermo Fisher Scientific (Waltham, MA). Methanol and water were HPLC-grade and were purchased from Honeywell Burdick and Jackson. All other materials were purchased from Thermo Fisher Scientific.
Mutagenesis, Protein Expression, and Purification.
Protocols for expressing and purifying native and S530A huPGHS-2 were similar to those reported previously (Yuan et al., 2006, 2009; Liu et al., 2007; Wada et al., 2007). A cDNA for huPGHS-2 containing a hexahistidine (His6) tag at the N terminus was subcloned into pFastBac plasmid (Invitrogen, Carlsbad, CA). S530A mutations of huPGHS-2 were prepared by site-directed mutagenesis, starting with His6-tagged huPGHS-2 in pFastBac vector and by using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The presence of the mutations was confirmed by DNA sequencing. The primers used to prepare the S530A mutation were ggAgCACCATTCgCCTTgAAAggACTTATg and CATAAgTCCTTTCAAggCgAATggTgCTCC.
Using the Invitrogen Bac-to-Bac expression system protocol, bacmid DNA was generated and was used to transfect Sf21 insect cells. Virus was harvested and used to infect cultures of sf21 cells. After 72 to 96 h of infection, the cells were collected and used to purify protein.
Sf21 cell pellets were resuspended in nickel-solubilization buffer (20 mM Tris-HCl, pH 7.4, and 100 mM NaCl) containing Complete EDTA-Free Protease Inhibitor (Roche Applied Sciences). The enzyme was solubilized with C10E6 [0.8% (v/v)], and insoluble material was removed by centrifugation. Enzymes were purified from the solubilized supernatant using a combination of chromatography on fast-flow nickel-nitrilotriacetic acid resin (QIAGEN) and FLAG-affinity resin (Sigma) essentially as reported previously (Liu et al., 2007; Yuan et al., 2009). Purified protein at a concentration of approximately 100 μg/ml was concentrated to 0.5 to 4 mg/ml using a centrifugal filter (Millipore Corporation, Billerica, MA) with a cutoff molecular weight of 50,000.
Radio-Thin Layer Chromatography of Prostaglandin Products.
Thin-layer chromatography was used to separate the products formed from [1-14C]AA or [1-14C]EPA by native huPGHS-2 or S530A/S530A huPGHS-2 before and after incubation with 500 μM aspirin for 1 h at 37°C. The reactions were performed in standard assay buffer containing 0.1 M Tris-HCl, pH 8.0, containing 1 mM phenol and 5 mM hematin and allowed to proceed for 40 s at 37°C with or without 12.5 mM diclofenac. The reactions were quenched with stop buffer consisting of ethyl ether/methanol/0.2 M citric acid (30:4:1). The resultant solutions were centrifuged at 4°C for 10 min at 1000g. An aliquot of the organic layer (100 μl) was subjected to thin-layer chromatography on a Silica Gel G plate in ethyl acetate/2,2,4-trimethylpentane/acetic acid/water (110:50:20:100). After exposing the thin-layer plates to X-ray film, the films were scanned, and the density of the radioactive bands was quantified with NIH Image J program (ver. 1.38x; http://rsbweb.nih.gov/ij/). The product compositions were calculated based on the percentage of the band intensity (e.g., measuring those bands that cochromatographed with AA, 15-HETE, or PGH2).
Cyclooxygenase Assays.
COX assays were performed as detailed previously (Liu et al., 2007). COX reaction mixtures typically contained 3 ml of 0.1 M Tris-HCl, pH 8.0, 100 μM arachidonic acid, 1 mM phenol, and 5 μM hematin equilibrated in a glass chamber at 37°C. Reactions were initiated by adding enzyme to the assay chamber. A Yellow Springs Instruments model 53 Oxygen monitor was used to monitor O2 consumption by native or mutant PGHS with kinetic traces recorded using software (Daisytec, Amherst, NH). The rates reported are the maximum rates occurring after a short lag phase. One unit of COX activity is defined as 1 μmol of O2 consumed per minute at 37°C in the assay mixture. IGOR Pro version 6.0 (WaveMetrics, Lake Oswego, OR) was used for graphing Km and Vmax. The errors are reported as standard deviations from multiple kinetic trials.
Quantitation of Aspirin Acetylation.
Native huPGHS-2 and S530A huPGHS-2 variants were incubated with 0.5, 1, or 2 mM [1-14C]acetylsalicylate for different times at either room temperature (approximately 24°C) or 37°C (Bala et al., 2008). The resulting mixture was treated with ice-cold 10% TCA and filtered through 1 μM pore-sized filters under vacuum. The filters were washed with 5 ml of ice-cold TCA containing 8 mM unlabeled ASA. After washing, the filters were transferred to scintillation vials and allowed to equilibrate with the scintillation fluid for at least 24 h. Radioactivity was quantified by liquid scintillation counting.
LC-MS/MS Analysis of EPA and DHA Reaction Products of PGHS-2.
Mass spectrometric analysis was performed with a Surveyor HPLC (Thermo Fisher Scientific) interfaced directly to the electrospray ionization source of an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific). Sample mixtures were separated using a Gemini C6-phenyl analytical column (150 × 2.00 mm, 3 μm particles; Phenomenex, Torrance, CA) with a binary gradient at a flow rate of 0.22 ml/min. Solvent A was 10 mM ammonium acetate, pH 8.5, and solvent B was methanol. A linear gradient from 50% solvent B to 100% solvent B over 50 min followed by a hold at 100% solvent B for 10 min was used to elute compounds of interest. The column was maintained at 50°C, and the sample compartment was cooled to 4°C. Negative ions were generated in the electrospray ionization source using nitrogen as the sheath and auxiliary gas under the following conditions: spray voltage, 3.0 kV; tube lens, −52 V; sheath gas, 30 psi; auxiliary gas, 5 units; capillary temperature, 300°C. For identification of the reaction products, the mass spectrometer was set up to collect survey data in full scan mode from m/z 150 to 1000 and four independent MS2 scans based on a mass list of precursor ions for the substrates and predicted monohydroxy reaction products. The normalized collision energy was set to 26%. An additional scan function was added to the method for semiquantitative analysis. Data were collected using selected ion monitoring of the [M-H] − ions of the substrates, the products, and the internal standard (15-hydroxyeicosatetraenoic-d8 acid). Xcalibur software (version 1.4; Thermo Fisher Scientific) was used for instrument control and data analysis.
Results
Properties of Native and S530A huPGHS-2.
We used preparations of the native huPGHS-2 homodimer and an S530A/S530A huPGHS-2 homodimer to characterize the interactions of aspirin and diclofenac with huPGHS-2 (Table 1). The Vmax value for S530A/S530A huPGHS-2 was approximately 35% of that observed with native enzyme, and there was only a small difference in the Km values with AA between the native and mutant enzymes. The native and mutant enzymes were both stimulated with 11 cis-eicosaenoic acid (20:1ω9) and by palmitic acid (Table 1), and so, we infer that the two enzymes are subject to positive allosteric regulation and exhibit half of sites activity with AA (Yuan et al., 2009). It should also be noted that as determined by radio thin-layer chromatography with [1-14C]AA, the products formed by S530A/S530A huPGHS-2 (Supplemental Fig. S1) and native huPGHS-2 (vide infra) contain the same relative amounts of monohydroxy fatty acids (∼10%) and PGH2 (∼90%).
TABLE 1.
Properties of native huPGHS-2 and S530A huPGHS-2
Specific activities were determined using a standard O2 electrode assay with 100 μM AA as substrate. Km values were determined using AA concentrations from 1 to 100 μM. The percentage of inhibition by diclofenac was determined in a standard O2 electrode assay with 100 μM AA after a 10-min preincubation of the enzymes at 24°C with 10 μM diclofenac versus without diclofenac; 10 μM diclofenac was also added to the assay chamber in the case of enzyme pretreated with diclofenac. Activation of activity by 20:1ω9 was measured using 5 μM AA plus 25 μM 20:1ω9 in the assay mixtures. Activation of activity by palmitic acid (16:0) was measured using 5 μM AA plus 25 μM 16:0 in the assay mixtures. The rates reported are maximum rates occurring after a lag phase. Igor Pro version 6.0 was used for graphing rates versus AA concentrations in obtaining Km and Vmax values. Errors are reported as standard deviations from at least three kinetic measurements for each substrate or inhibitor concentration.
| Property | huPGHS-2 |
|
|---|---|---|
| Native | S530A/S530A | |
| Specific activity with AA (U/mg protein = μmol O2/min/mg protein) | 40 ± 1 | 14 ± 1 |
| Km with AA (μM) | 5 ± 1 | 9 ± 1 |
| Inhibition by diclofenac (%) | 100 | 15 ± 1 |
| Stimulation by 20:1ω9 (%) | 132 ± 5 | 135 ± 7 |
| Stimulation by 16:0 (%) | 176 ± 5 | 155 ± 11 |
Effect of Diclofenac on huPGHS-2.
Ser530 but not Arg120 is important for inhibition of the COX activity of murine PGHS-2 by diclofenac (Rowlinson et al., 2003). Consistent with this, we found that diclofenac was an ineffective inhibitor of S530A/S530A huPGHS-2 but completely inhibited native huPGHS-2 (Table 1). Diclofenac caused a time-dependent inhibition of native huPGHS-2 (Table 1 and Fig. 1). As shown in Fig. 1A, the activity of enzyme that had been pretreated with diclofenac was not immediately regained—as it would be for a freely reversible inhibitor—when the enzyme was assayed in the presence of a noninhibitory concentration of diclofenac. This behavior is similar to that observed with indomethacin (Fig. 1A), which has been well documented as a time-dependent COX inhibitor. When titrated with diclofenac, maximal inhibition of native huPGHS-2 occurred at a ratio of one diclofenac molecule per molecule of huPGHS-2 dimer (Fig. 1B). Thus, binding of diclofenac to a single monomer of huPGHS-2 inhibits the activity of both monomers. Comparison of the results obtained with native huPGHS-2 versus mutant huPGHS-2 imply that Ser530 needs to be present in at least a single monomer for time-dependent inhibition by diclofenac to occur (Table 1). Ser530 is not required for inhibition by indomethacin, flurbiprofen, or meclofenamate, which are also maximally effective at one inhibitor molecule per dimer (Kulmacz and Lands, 1985; DeWitt et al., 1990; Yuan et al., 2006, 2009; Prusakiewicz et al., 2009) and mediate an inhibition requiring interaction with Arg120 (Bhattacharyya et al., 1996; Rieke et al., 1999; Rowlinson et al., 2003; Yuan et al., 2006) but not Ser530 (Rowlinson et al., 2003). Inhibition by some nsNSAIDs is associated with decreased formation of the Tyr385 radical involved in catalysis (Rogge et al., 2006, 2009). It is not known whether this is the case with diclofenac, but Ser530 does lie across from Tyr385 in the COX active site channel, perhaps with an intervening water molecule (Selinsky et al., 2001).
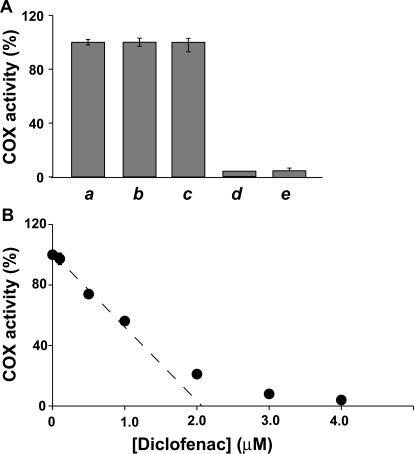
Fig. 1.
Time-dependent inhibition of native huPGHS-2 by diclofenac and indomethacin. Purified huPGHS-2 (2 μM) was pretreated with no inhibitor (a, b, and c) for 10 min at 37°C or 12 μM diclofenac (d) for 10 min at 37°C or 100 μM indomethacin (e) for 5 min at 37°C. No inhibitor was added to the O2 electrode assay chamber when the sample (a) was assayed. Diclofenac (0.16 μM) was included in the assay chamber when the sample (b) was assayed; sample b served as the negative control for sample d. Indomethacin (1.3 μM) was included in the assay chamber when sample c was assayed; sample c served as the negative control for sample e. Dilution of the aliquot of sample d yielded a final concentration of 0.16 μM diclofenac in the assay chamber, and no additional diclofenac was added to this assay chamber. Dilution of the aliquot of sample e yielded a final concentration of 1.3 μM indomethacin in the assay chamber, and no additional indomethacin was added to this assay chamber. Assays were performed at 37°C in 3 ml of 0.1 M Tris-HCl, pH 8.0, containing 100 μM arachidonic acid, 1 mM phenol, and 1 μM hematin. B, purified huPGHS-2 (2.1 μM) was pretreated with the indicated concentrations of diclofenac at 37°C for 10 min and then assayed for COX activity with 100 μM AA essentially as described above. Error bars show standard deviations from multiple kinetic trials.
Aspirin Acetylation of huPGHS-2.
Figure 2A shows a time course for the inhibition of native huPGHS-2 by 0.5 mM aspirin. Aspirin treatment had no detectable effect on the activity of S530A/S530A huPGHS-2. After 150 min at 24°C, native huPGHS-2 retained approximately 40% of its original oxygenase activity as measured by O2 consumption with an O2 electrode assay. Native huPGHS-2 was acetylated by aspirin on only one of its monomers; specifically, 0.92 ± 0.29 mole of acetate was incorporated per mole of native huPGHS-2 dimer (n = 5) during a 150-min incubation with [1-14C]acetylsalicylate. No additional 14C-acetyl groups were incorporated into native huPGHS-2 during an overnight incubation at 4°C, and there was no additional loss of enzyme activity. As anticipated, no 14C-label was incorporated above enzyme control levels into the S530A/S530A huPGHS-2 homodimer upon incubation with [1-14C]acetylsalicylate.
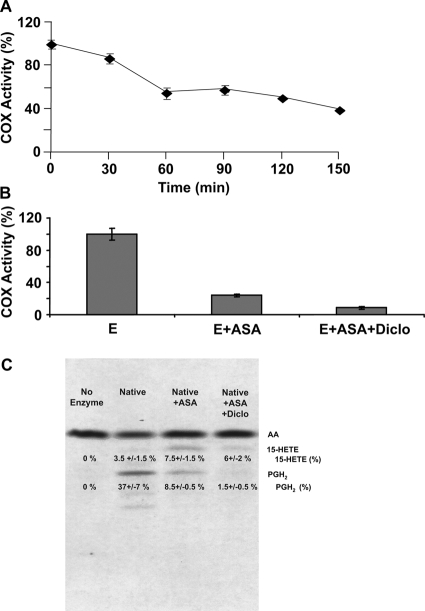
Fig. 2.
Inhibition of huPGHS-2 by aspirin. A, purified native huPGHS-2 homodimer was incubated at 24°C with or without freshly prepared aspirin (500 μM) for the indicated times and then assayed for COX activity using an O2 electrode. Values are derived from the average of triplicate determinations ± S.E. Similar experiments were performed at least three times with different preparations of enzyme and yielded quantitatively similar results. B, purified native huPGHS-2 was pretreated with 500 μM aspirin for 1 h at 37°C, which causes maximal inhibition. The sample was then incubated without (E + ASA) or with (E + ASA + Diclo) 12.5 μM diclofenac (Diclo) for 10 min and assayed for COX activity using an O2 electrode. For the samples pretreated with diclofenac for 10 min, the assay mixture also contained 12.5 μM diclofenac. A control with huPGHS-2 without inhibitor (E) was treated for 1 h at 37°C with vehicle (in place of aspirin) and then incubated for an additional 10 min without any inhibitor. C, purified native huPGHS-2 was incubated with or without ASA (500 μM) at 37°C for 1 h, and the oxygenation of [1-14C]AA was assayed by radio thin-layer chromatography in the presence and absence of 12.5 μM Diclo as described under Materials and Methods. Thin-layer chromatography was used to separate the endoperoxide (PGH2) and monohydroxy acid products, mainly 15-HETE. To perform the assays, [1-14C]AA (100 μM) was mixed with ∼9 μg of native huPGHS-2 in an assay volume of 0.10 ml, and the reactions were allowed to proceed for 40 s. The reactions were stopped, and the products were extracted, separated, and visualized by autoradiography as shown. The thin-layer plates were subsequently scraped, and the amount of radioactivity cochromatographing with AA, PGH2, and 15-HETE standards was determined by scintillation counting. Numbers obtained from scintillation counting were used to calculate the percentage of total radioactivity found in each product. The experiment was performed three times with consistent results.
As illustrated in Fig. 2B, diclofenac inhibited the rate of O2 consumption of aspirin-treated native huPGHS-2 by approximately two thirds. Interpretation of this result must take into account that after its acetylation by aspirin, huPGHS-2 forms 15R-HETE in addition to smaller amounts of PGH2-derived products (Holtzman et al., 1992; Lecomte et al., 1994; Xiao et al., 1997), and formation of 15-HETE consumes only one oxygen molecule per molecule of AA, whereas formation of PGH2 involves the incorporation of two oxygen molecules per AA molecule. The experiment depicted in Fig. 2C indicates that 15-HETE is formed by aspirin-treated native huPGHS-2 at significantly greater levels and in higher proportions relative to PGH2 than by untreated enzyme. It is important to note that diclofenac inhibits PGH2 formation by acetylated huPGHS-2 to a much greater extent than 15-HETE production. Based on studies with S530A PGHS-2, we assume that diclofenac binds to the nonacetylated monomer having an unaltered Ser530 to block PGH2 formation but not to the acetylated monomer to block 15-HETE formation. This implies that the acetylated monomer forms 15-HETE and the nonacetylated monomer forms primarily PGH2.
Oxygenation of EPA by huPGHS-2.
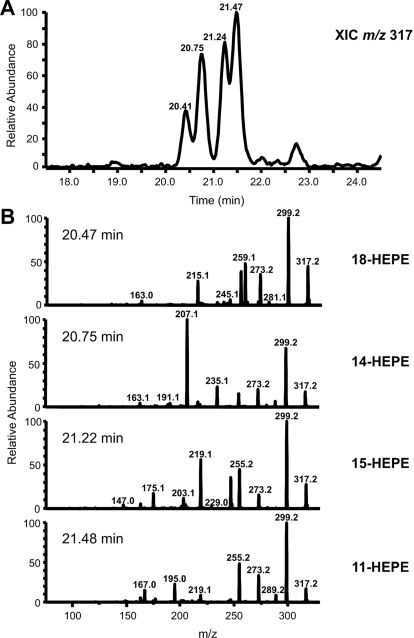
EPA and DHA are poorer substrates than AA for huPGHS-2 (Liu et al., 2006; Wada et al., 2007; Yuan et al., 2009). With saturating concentrations of fatty acid substrate (100 μM), the relative COX activities of huPGHS-2 with AA, EPA, and DHA were 100, 45, and 15%, respectively (Table 2). It should be noted that these values are normalized rates of formation of oxygenated fatty acids corrected for differences in rates of oxygen consumption that are intrinsically higher with fatty acids such as AA that are bis-oxygenated. Although there is some information in the literature (Serhan et al., 2002), the details concerning the nature and ratios of the products formed from EPA and DHA by highly purified huPGHS-2 have not been reported previously. We found that huPGHS-2 converts 100 μM EPA to an approximately equal mixture of PGH3 and monohydroxy fatty acids and that treatment of huPGHS-2 with aspirin inhibits the formation of both PGH3 and monohydroxy fatty acid by approximately 70%. LC-MS/MS analyses revealed that two mono-oxygenated products 11-HEPE (32%) and 14-HEPE (11%) are formed from EPA by native huPGHS-2 (Tables 2 and 3; Fig.3). The formation of 11-HEPE was confirmed by matching the retention time and MS/MS spectrum with an authentic 11-HEPE standard. No 14-HEPE standard is commercially available. The conclusion that the second monohydroxy acid is 14-HEPE is based on the information summarized in Table 3. The MS/MS spectrum is consistent with the fragments predicted for 14-HEPE, and reduction of the carbon-carbon double bonds of the unknown acid (predicted to be 14-HEPE) yields a product with a 10-Da increase in molecular mass and an MS/MS spectrum expected for 14-hydroxyeicosanoic acid. The only straightforward mechanism for the formation of 14-HEPE would seem to be abstraction of the omega-5 hydrogen from EPA; however, we are unaware of a precedent for this for native PGHS.
TABLE 2.
Comparison of maximal rates of oxygenation and product profiles for AA, EPA, and DHA incubated with huPGHS-2 and aspirin-acetylated huPGHS-2
Rates were determined by measuring O2 consumption using an O2 electrode as described under Materials and Methods. Values for rates of O2 consumption were converted to values for rates of FA consumption based on the percentages of mono-oxygenated and bis-oxygenated (endoperoxide) products. The latter percentages were determined by radioactive thin-layer chromatography and autoradiography with [1-14C]AA or [1-14C]EPA as substrates or by thin-layer chromatography and visualizing the products by staining in the case of DHA; only mono-oxygenated cyclooxygenase products of DHA were observed. Errors are standard deviations from at least three kinetic measurements for each substrate. Values for AA- and EPA-derived products were determined by measuring the percentage of radioactivity from [1-14C]AA and [1-14C]EPA, respectively, that cochromatographed with monohydroxy acid (15-HETE) and PGH2 upon radio thin-layer chromatography and using the ratio of hydroxy acids as determined by LC-MS/MS. Values for DHA were determined by LC-MS/MS directly because only mono-oxygenated cyclooxygenase products were observed by thin-layer chromatography.
| Fatty Acid Substrate | huPGHS-2 |
Aspirin-Acetylated
huPGHS-2 |
||
|---|---|---|---|---|
| Rate of FA Oxygenation | Products | Rate of FA Oxygenation | Products | |
| μM FA/min/mg | % | μM FA/min/mg | % | |
| AA | 22 ± 0.2 | PGH2 (91) | 12 ± 0.5 | PGH2 (53) |
| HETEs (9) | 15-HETE (47) | |||
| EPA | 10.8 ± 0.5 | PGH3 (50) | 3.1 ± 0.2 | PGH3 (40) |
| 11-HEPE (32) | 11-HEPE (21) | |||
| 14-HEPE (18) | 14-HEPE (16) | |||
| 15-HEPE (16) | ||||
| 18-HEPE (7) | ||||
| DHA | 3.2 | 13-HDHA (100) | 0.9 | 13-HDHA (25) |
| 17-HDHA (75) | ||||
TABLE 3.
Summary of evidence that the peak with a retention time of 20.75 min in the HPLC separation of products derived from incubation of 5,8,11,14,17-eicosapentaenoic acid with huPGHS-2 is 14-HEPE
| Data | Result | Conclusion |
|---|---|---|
| UV, MS, and MS/MS data for peak at 20.75 min | ||
| [M-H]− = m/z 317 | Acidic functional group | Mr = 318 |
| Fragment ion at m/z 299 | Loss of H2O | Monohydroxy |
| Fragment ion at m/z 273 | Loss of CO2 | Carboxylic acid |
| UV absorbance at 230 nm | UV peak at 20.75 min | Conjugated diene |
| Intense fragment ion at m/z 207 | Probable α-hydroxy-β-ene rearrangement | 14-Hydroxyl group |
| MS/MS data after reduction of peak at 20.75 min with Adam's catalyst | ||
| [M-H]− = m/z 327 | Increase of 10 mass units | Five double bonds |
| Retention time increase of >10 min | More hydrophobic | Saturated chain |
| Fragment ion at m/z 309 | Loss of H2O | Monohydroxy acid |
| Fragment ion at m/z 283 | Loss of CO2 | Carboxylic acid |
| Fragment ion at m/z 325 | Loss of H2 | Formation of double bond |
| Fragment ion at m/z 211 | Probable α-hydroxy-β-ene rearrangement | 14-Hydroxy; C11–C12 double bond |
Fig. 3.
Products formed from EPA by aspirin-acetylated huPGHS-2. EPA (100 μM) was mixed with 9 μg of enzyme, and the reactions continued for 40 s. The products were extracted as described in Fig. 2C and reconstituted with 50% methanol for LC-MS/MS analysis.
Aspirin-acetylated huPGHS-2 converted 100 μM EPA to a mixture of four different hydroxy acids (60% of the products) and PGH3 (40% of the products) (Fig. 3). A minor product is 18-HEPE, which represents 7% of the total oxygenated products. Oxygenated products were not detected when [1-14C]EPA (100 μM) was incubated with aspirin-acetylated huPGHS-2 in the presence of diclofenac (12.5 μM) (data not shown). This is unlike what is seen with AA as a substrate (Fig. 2C).
In experiments similar to those reported in Table 2, we analyzed the efficiency of oxygenation of EPA by native huPGHS-2 and aspirin-treated huPGHS-2 at a concentration (5 μM) closer to that likely to be physiologically relevant (Table 4). We also tested the effect of 25 μM palmitic acid on EPA oxygenation because palmitic acid is the most abundant free fatty acid in cells and is a positive allosteric activator of AA oxygenation by PGHS-2 in vitro (Table 1) (Yuan et al., 2009). Palmitic acid had little or no effect on the rate of EPA oxygenation or on the pattern of products formed by native huPGHS-2; in the presence of palmitic acid, EPA is oxygenated at 25% of the rate of AA. Aspirin-treated huPGHS-2 has only 35% of native huPGHS-2 activity with 5 μM EPA. The product profile is shifted somewhat so that more monohydroxy fatty acids are formed, but the effect of aspirin to increase monohydroxy acid formation from EPA is much less pronounced than what is observed with AA. 18-HEPE is detectable as one of the products of EPA but comprises only 2% of the total products with 5 μM EPA (Table 4).
TABLE 4.
Comparison of rates of oxygenation and product profiles for 5 μM AA, EPA, and DHA with native huPGHS-2 and aspirin-acetylated huPGHS-2
Rates and products were determined as described in Table 2 using 5 μM fatty acid substrate and 25 μM palmitic acid (16:0).
| Fatty Acids | huPGHS-2 |
ASA-huPGHS-2 |
||
|---|---|---|---|---|
| Rate | Products | Rate | Products | |
| μM FA/min/mg | % | μM FA/min/mg | % | |
| AA | 9.3 ± 0.51 | 15-HETE (6.8) | 5.9 ± 0.43 | 15-HETE (52) |
| 11-HETE (9.2) | 11-HETE (9.0) | |||
| PGH2 (84) | PGH2 (39) | |||
| AA + 16:0 | 16 ± 1.0 | 15-HETE (7.3) | 6.4 ± 0.51 | 15-HETE (27) |
| 11-HETE (9.6) | 11-HETE (4.1) | |||
| PGH2 (83) | PGH2 (69) | |||
| EPA | 4.1 ± 0.03 | 14-HEPE (15) | 1.5 ± 0.07 | 18-HEPE (2.2) |
| 15-HEPE (1.9) | 14-HEPE (16) | |||
| 11-HEPE (22) | 15-HEPE (16) | |||
| PGH3 (61) | 11-HEPE (18) | |||
| PGH3 (47) | ||||
| EPA + 16:0 | 4.3 ± 0.14 | 14-HEPE (14) | 1.5 ± 0.18 | 18-HEPE (2.0) |
| 15-HEPE (3.3) | 14-HEPE (12) | |||
| 11- HEPE (18) | 15-HEPE (23) | |||
| PGH3 (65) | 11-HEPE (18) | |||
| PGH3 (45) | ||||
| DHA | 2.0 ± 0.08 | 13-HDHA (100) | 1.1 ± 0.23 | 17-HDHA (78) |
| 13-HDHA (22) | ||||
| DHA + 16:0 | 2.0 ± 0.11 | 13-HDHA (100) | 1.1 ± 0.04 | 17-HDHA (73) |
| 13-HDHA (27) | ||||
Oxygenation of DHA by huPGHS-2.
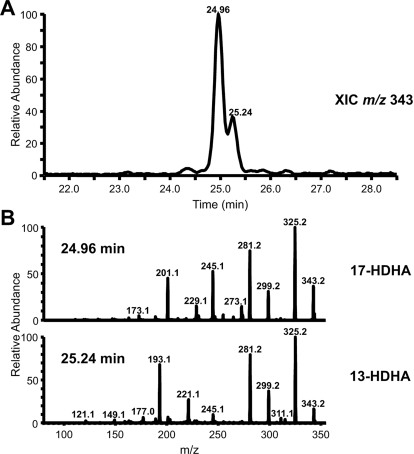
The specific activity of highly purified huPGHS-2 with 100 μM DHA is 15% of the specific activity observed with 100 μM AA (Table 2). After incubation of huPGHS-2 or aspirin-acetylated huPGHS-2 with DHA, the only products visualized after thin-layer chromatography cochromatographed with authentic 13-hydroxydocosahexaenoic acid (HDHA) and 17-HDHA standards, and the latter two hydroxydocosahexaenoic acid standards cochromatographed in the thin-layer chromatography system used in our experiments (data not shown). We subsequently analyzed the products by LC-MS/MS (Tables 2 and 4; Fig. 4). Consistent with what has been reported (Serhan et al., 2002), highly purified huPGHS-2 converts DHA exclusively to HDHA. The identity of 13-hydroxydocosahexaenoic acid was confirmed by matching the retention time and MS/MS spectrum with that of a 13-HDHA standard (Fig. 4). Aspirin-acetylated huPGHS-2 has less than one third of the activity of native huPGHS-2 with 100 μM DHA and forms a mixture of 13-HDHA (∼25%) and 17-HDHA (∼75%). As observed using EPA as the substrate, diclofenac (12.5 μM) completely inhibited the oxygenation of DHA (100 μM) by aspirin-acetylated huPGHS-2.
Fig. 4.
Products formed from DHA by aspirin-acetylated huPGHS-2. DHA (100 μM) was mixed with 9 μg of enzyme, and the reactions continued for 40 s. The products were extracted as described in Fig. 2C and reconstituted with 50% methanol for LC-MS/MS analysis.
Experiments were also performed with 5 μM DHA in the presence and absence of palmitic acid (Table 4). The results are qualitatively and quantitatively similar to those seen with 100 μM DHA (Table 2). The relative amounts of 13-HDHA versus 17-HDHA formed from 5 μM DHA by aspirin-treated huPGHS-2 was unaffected by the presence of 5 μM AA. There was also no appreciable effect of 5 μM DHA on the formation of AA-derived products (data not shown). The latter results are consistent with previous measurements of rates of O2 consumption when DHA and AA are coincubated with huPGHS-2 (Yuan et al., 2009).
Discussion
Earlier studies on the effect of aspirin on PGHS-2 showed that aspirin causes partial inhibition of the enzyme, that the aspirin-treated enzyme forms a new product 15R-HETE from AA, and that the acetyl group from aspirin is incorporated into the protein at Ser530 (Holtzman et al., 1992; Lecomte et al., 1994). Subsequent work has suggested that 15R-HETE is formed from an alternative conformation of AA by aspirin-treated PGHS-2 (Xiao et al., 1997). More recently, studies by Serhan and coworkers on trihydroxylated compounds dubbed resolvins have indicated that RvE1 and 17R-RvD1 can be derived from 18R-HEPE and 17R-HDHA that are formed from EPA and DHA, respectively, by aspirin-treated PGHS-2. In this report, we have expanded and refined the results obtained earlier with AA and have also examined the oxygenation of EPA and DHA by purified, aspirin-acetylated PGHS-2.
In the case of PGHS-1, a single monomer of the homodimer is acetylated by aspirin. The result is complete inhibition of the cyclooxygenase activity of the enzyme (Rimon et al., 2010). This is an example of what is becoming recognized as a general mechanism of inhibition of PGHS by time-dependent inhibitors that include nsNSAIDs that can act on both PGHS-1 and PGHS-2 (Kulmacz and Lands, 1985; Yuan et al., 2006, 2009) and probably coxibs functioning on PGHS-2 (Walker et al., 2001). These inhibitors act as negative allosteric effectors by binding to one monomer of the enzyme to markedly reduce or even eliminate activity in the partner monomer. In a related manner, fatty acids such as palmitic acid function as positive allosteric effectors of PGHS-2 by binding one monomer of the dimer and promoting the binding and oxygenation of AA in the other monomer (Yuan et al., 2009). In contrast, time-independent, freely reversible inhibitors such as ibuprofen need to bind both monomers of PGHS simultaneously to cause enzyme inhibition, and the inhibition is complete (Prusakiewicz et al., 2009).
Our results demonstrate that only a single monomer of PGHS-2 is acetylated by aspirin but that the outcome is somewhat more complex than that seen with PGHS-1. The enzyme retains a significantly compromised ability to form PGH2 and produces an alternative product, 15-HETE. The fact that PGH2 formation is more than 80% inhibited by diclofenac whereas the formation of 15-HETE is reduced by only 20% suggests that diclofenac is binding to the nonacetylated monomer of aspirin-treated PGHS-2 and that this monomer is the one that forms PGH2, whereas the acetylated monomer forms primarily 15R-HETE. Thus, the effect of aspirin on PGHS-2 is an incomplete allosteric inhibitory effect compared with that seen with PGHS-1. Our ability to draw these conclusions depends on the use of diclofenac, which is an unusual anti-inflammatory drug that functions by interacting with Ser530 and not Arg120 (Rowlinson et al., 2003).
Diclofenac completely inhibited the oxygenation of EPA and DHA by aspirin-acetylated huPGHS-2. This suggests that all of the products derived from EPA and DHA are formed by the nonacetylated subunit; however, we cannot exclude the possibility that diclofenac also indirectly interferes with the binding of EPA and DHA to the aspirin-acetylated monomer and inhibits EPA and DHA oxygenation in both monomers. In the context of differential usage of AA, EPA, and DHA by the acetylated versus nonacetylated monomers, the differential effect of palmitic acid may be relevant. Palmitic acid had little or no effect on the pattern of products formed from EPA or DHA by aspirin-acetylated PGHS-2 but had a significant effect on the product profile with AA. With AA and palmitic acid and aspirin-treated huPGHS-2, there was a relative increase in PGH2 at the expense of 15-HETE formation. We speculate that palmitic acid preferentially binds the acetylated subunit and interferes with events occurring in that subunit.
RvE1 has potent biological activity in the resolution of inflammation (Serhan et al., 2008). It can be formed in complex tissue systems primed with aspirin pretreatment and exogenous EPA (Serhan et al., 2000; Arita et al., 2005). Moreover, RvE1 can be detected in plasma at a level of up to 1 nM in individuals given a bolus of EPA and treated with aspirin (Arita et al., 2005). A minimal concentration of 1 nM seems to be required for bioactivity (Arita et al., 2005).
The proposed pathway for RvE1 formation involves an initial production of 18R-HEPE via aspirin-acetylated PGHS-2 and subsequent further oxygenation. Although 18R-HEPE was reportedly formed by microsomes containing recombinant PGHS-2 (Serhan et al., 2000), in our hands, purified recombinant huPGHS-2 did not form this product at saturating concentrations of EPA (100 μM). In agreement with previous studies, 18R-HEPE was formed by purified huPGHS-2 after aspirin treatment. However, 18R-HEPE was the least abundant of five products formed from EPA, and aspirin-acetylated huPGHS-2 has 35% of the oxygenase activity of native huPGHS-2 with EPA. With concentrations of EPA (5 μM) approaching but likely to be even higher than those available under physiological conditions, 18R-HEPE represented only 2% of the products formed from EPA by aspirin-treated, purified huPGHS-2. 18-HEPE formation by aspirin-treated huPGHS-2 from EPA plus palmitic acid occurs at less than 0.25% of the rate of PGH2 formation from AA plus palmitic acid by native huPGHS-2. In short, although RvE1 is biologically active, we suspect that it could be formed via huPGHS-2 only in quite low abundance and only in highly unusual biological settings.
17R-RvD1 is reportedly formed from DHA, with the first step being the conversion of DHA to 17R-HDHA (Serhan et al., 2002, 2008). Again, using the extreme comparison of the relative rates of PGH2 formation by native huPGHS-2 in the presence of palmitic acid versus 17-HDHA formation from DHA in the presence of palmitic acid by aspirin-treated huPGHS-2 indicates that 17-HDHA synthesis by aspirin-treated huPGHS-2 occurs maximally at ∼5% of the rate of PGH2 synthesis by native huPGHS-2. Aspirin-acetylated huPGHS-2 showed no obvious preference for AA versus DHA when these fatty acids were tested together at 5 μM each.
Supplementary Material
Acknowledgments
We thank Dr. Yu H. Hong for technical assistance.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
These studies were supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM068848]; and the National Institutes of Health National Heart, Lung, and Blood Institute [Grant HL085149].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.063115.
- PGHS
- prostaglandin endoperoxide H synthase
- COX
- cyclooxygenase
- AA
- arachidonic acid
- hu
- human
- nsNSAID
- nonspecific nonsteroidal anti-inflammatory drug
- EPA
- eicosapentaenoic acid
- DHA
- docosahexaenoic acid
- TCA
- trichloroacetic acid
- 11-HETE
- 11-hydroxyeicosatetraenoic acid
- 15-HETE
- 15-hydroxyeicosatetraenoic acid
- 11-HEPE
- 11-hydroxyeicosapentaenoic acid
- 15-HEPE
- 15-hydroxyeicosapentaenoic acid
- 18-HEPE
- 18-hydroxyeicosapentaenoic acid
- 13-HDHA
- 13-hydroxydocosahexaenoic acid
- 17-HDHA
- 17-hydroxydocosahexaenoic acid
- RvE1
- Resolvin E1, 5S,12R,18R-trihydroxy-6Z,8E,10E,14Z,16E-eicosapentaenoic acid
- 17R-RvD1
- 17R-resolvin D1, 7S,8R,17R-trihydroxy-4Z,9E,13Z,15E,19Z-docosahexaenoic acid
- PGH2
- prostaglandin H2
- ASA
- acetylsalicylic acid
- HPLC
- high-performance liquid chromatography
- MS/MS
- tandem mass spectrometry
- LC-MS/MS
- liquid chromatography/tandem mass spectrometry
- 15R-HETE
- 15R-hydroxy-eicosatetraenoic acid.
References
- Arita M, Bianchini F, Aliberti J, Sher A, Chiang N, Hong S, Yang R, Petasis NA, Serhan CN. (2005) Stereochemical assignment, antiinflammatory properties, and receptor for the omega-3 lipid mediator resolvin E1. J Exp Med 201:713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala M, Chin CN, Logan AT, Amin T, Marnett LJ, Boutaud O, Oates JA. (2008) Acetylation of prostaglandin H2 synthases by aspirin is inhibited by redox cycling of the peroxidase. Biochem Pharmacol 75:1472–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya DK, Lecomte M, Rieke CJ, Garavito M, Smith WL. (1996) Involvement of arginine 120, glutamate 524, and tyrosine 355 in the binding of arachidonate and 2-phenylpropionic acid inhibitors to the cyclooxygenase active site of ovine prostaglandin endoperoxide H synthase-1. J Biol Chem 271:2179–2184 [DOI] [PubMed] [Google Scholar]
- Blobaum AL, Marnett LJ. (2007) Structural and functional basis of cyclooxygenase inhibition. J Med Chem 50:1425–1441 [DOI] [PubMed] [Google Scholar]
- DeWitt DL. (1999) Cox-2-selective inhibitors: the new super aspirins. Mol Pharmacol 55:625–631 [PubMed] [Google Scholar]
- DeWitt DL, el-Harith EA, Kraemer SA, Andrews MJ, Yao EF, Armstrong RL, Smith WL. (1990) The aspirin and heme-binding sites of ovine and murine prostaglandin endoperoxide synthases. J Biol Chem 265:5192–5198 [PubMed] [Google Scholar]
- Grosser T, Fries S, FitzGerald GA. (2006) Biological basis for the cardiovascular consequences of COX-2 inhibition: therapeutic challenges and opportunities. J Clin Invest 116:4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman MJ, Turk J, Shornick LP. (1992) Identification of a pharmacologically distinct prostaglandin H synthase in cultured epithelial cells. J Biol Chem 267:21438–21445 [PubMed] [Google Scholar]
- Kulmacz RJ, Lands WE. (1985) Stoichiometry and kinetics of the interaction of prostaglandin H synthase with anti-inflammatory agents. J Biol Chem 260:12572–12578 [PubMed] [Google Scholar]
- Lecomte M, Laneuville O, Ji C, DeWitt DL, Smith WL. (1994) Acetylation of human prostaglandin endoperoxide synthase-2 (cyclooxygenase-2) by aspirin. J Biol Chem 269:13207–13215 [PubMed] [Google Scholar]
- Liu J, Seibold SA, Rieke CJ, Song I, Cukier RI, Smith WL. (2007) Prostaglandin endoperoxide H synthases: peroxidase hydroperoxide specificity and cyclooxygenase activation. J Biol Chem 282:18233–18244 [DOI] [PubMed] [Google Scholar]
- Liu W, Cao D, Oh SF, Serhan CN, Kulmacz RJ. (2006) Divergent cyclooxygenase responses to fatty acid structure and peroxide level in fish and mammalian prostaglandin H synthases. FASEB J 20:1097–1108 [DOI] [PubMed] [Google Scholar]
- Prusakiewicz JJ, Duggan KC, Rouzer CA, Marnett LJ. (2009) Differential sensitivity and mechanism of inhibition of COX-2 oxygenation of arachidonic acid and 2-arachidonoylglycerol by ibuprofen and mefenamic acid. Biochemistry 48:7353–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieke CJ, Mulichak AM, Garavito RM, Smith WL. (1999) The role of arginine 120 of human prostaglandin endoperoxide H synthase-2 in the interaction with fatty acid substrates and inhibitors. J Biol Chem 274:17109–17114 [DOI] [PubMed] [Google Scholar]
- Rimon G, Sidhu RS, Lauver DA, Lee JY, Sharma NP, Yuan C, Frieler RA, Trievel RC, Lucchesi BR, Smith WL. (2010) Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1. Proc Natl Acad Sci USA 107:28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge CE, Ho B, Liu W, Kulmacz RJ, Tsai AL. (2006) Role of Tyr348 in Tyr385 radical dynamics and cyclooxygenase inhibitor interactions in prostaglandin H synthase-2. Biochemistry 45:523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge CE, Liu W, Kulmacz RJ, Tsai AL. (2009) Peroxide-induced radical formation at TYR385 and TYR504 in human PGHS-1. J Inorg Biochem 103:912–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzer CA, Marnett LJ. (2003) Mechanism of free radical oxygenation of polyunsaturated fatty acids by cyclooxygenases. Chem Rev 103:2239–2304 [DOI] [PubMed] [Google Scholar]
- Rouzer CA, Marnett LJ. (2008) Non-redundant functions of cyclooxygenases: oxygenation of endocannabinoids. J Biol Chem 283:8065–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlinson SW, Kiefer JR, Prusakiewicz JJ, Pawlitz JL, Kozak KR, Kalgutkar AS, Stallings WC, Kurumbail RG, Marnett LJ. (2003) A novel mechanism of cyclooxygenase-2 inhibition involving interactions with Ser-530 and Tyr-385. J Biol Chem 278:45763–45769 [DOI] [PubMed] [Google Scholar]
- Schneider C, Pratt DA, Porter NA, Brash AR. (2007) Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol 14:473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinsky BS, Gupta K, Sharkey CT, Loll PJ. (2001) Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations. Biochemistry 40:5172–5180 [DOI] [PubMed] [Google Scholar]
- Serhan CN, Chiang N, Van Dyke TE. (2008) Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol 8:349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Clish CB, Brannon J, Colgan SP, Chiang N, Gronert K. (2000) Novel functional sets of lipid-derived mediators with antiinflammatory actions generated from omega-3 fatty acids via cyclooxygenase 2-nonsteroidal antiinflammatory drugs and transcellular processing. J Exp Med 192:1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Hong S, Gronert K, Colgan SP, Devchand PR, Mirick G, Moussignac RL. (2002) Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J Exp Med 196:1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WL. (2008) Nutritionally essential fatty acids and biologically indispensable cyclooxygenases. Trends Biochem Sci 33:27–37 [DOI] [PubMed] [Google Scholar]
- Smith WL, DeWitt DL. (1999) Cyclooxygenase inhibitors, in Therapeutic Immunology ( Austen KF, Burakoff SJ, Rosen FS, Strom TB. eds), Blackwell Science, Cambridge, MA [Google Scholar]
- Smith WL, DeWitt DL, Garavito RM. (2000) Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem 69:145–182 [DOI] [PubMed] [Google Scholar]
- van der Donk WA, Tsai AL, Kulmacz RJ. (2002) The cyclooxygenase reaction mechanism. Biochemistry 41:15451–15458 [DOI] [PubMed] [Google Scholar]
- Wada M, DeLong CJ, Hong YH, Rieke CJ, Song I, Sidhu RS, Yuan C, Warnock M, Schmaier AH, Yokoyama C, et al. (2007) Enzymes and receptors of prostaglandin pathways with arachidonic acid-derived versus eicosapentaenoic acid-derived substrates and products. J Biol Chem 282:22254–22266 [DOI] [PubMed] [Google Scholar]
- Walker MC, Kurumbail RG, Kiefer JR, Moreland KT, Koboldt CM, Isakson PC, Seibert K, Gierse JK. (2001) A three-step kinetic mechanism for selective inhibition of cyclo-oxygenase-2 by diarylheterocyclic inhibitors. Biochem J 357:709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Tsai AL, Palmer G, Boyar WC, Marshall PJ, Kulmacz RJ. (1997) Analysis of hydroperoxide-induced tyrosyl radicals and lipoxygenase activity in aspirin-treated human prostaglandin H synthase-2. Biochemistry 36:1836–1845 [DOI] [PubMed] [Google Scholar]
- Yuan C, Rieke CJ, Rimon G, Wingerd BA, Smith WL. (2006) Partnering between monomers of cyclooxygenase-2 homodimers. Proc Natl Acad Sci USA 103:6142–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Sidhu RS, Kuklev DV, Kado Y, Wada M, Song I, Smith WL. (2009) Cyclooxygenase Allosterism, Fatty Acid-mediated Cross-talk between Monomers of Cyclooxygenase Homodimers. J Biol Chem 284:10046–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.