Abstract
RNA editing is a post-transcriptional modification in which adenosine residues are converted to inosine (adenosine-to-inosine editing). Commonly used methodologies to quantify RNA editing levels involve either direct sequencing or pyrosequencing of individual cDNA clones. The limitations of these methods lead to a small number of clones characterized in comparison to the number of mRNA molecules in the original sample, thereby producing significant sampling errors and potentially erroneous conclusions. We have developed an improved method for quantifying RNA editing patterns that increases sequence analysis to an average of more than 800,000 individual cDNAs per sample, substantially increasing accuracy and sensitivity. Our method is based on the serotonin 2C receptor (5-hydroxytryptamine2C; 5HT2C) transcript, an RNA editing substrate in which up to five adenosines are modified. Using a high-throughput multiplexed transcript analysis, we were able to quantify accurately the expression of twenty 5HT2C isoforms, each representing at least 0.25% of the total 5HT2C transcripts. Furthermore, this approach allowed the detection of previously unobserved changes in 5HT2C editing in RNA samples isolated from different inbred mouse strains and dissected brain regions, as well as editing differences in alternatively spliced 5HT2C variants. This approach provides a novel and efficient strategy for large-scale analyses of RNA editing and may prove to be a valuable tool for uncovering new information regarding editing patterns in specific disease states and in response to pharmacological and physiological perturbation, further elucidating the impact of 5HT2C RNA editing on central nervous system function.
The 2C-subtype of serotonin receptor (5HT2C) is a member of the G-protein coupled receptor superfamily and has been implicated in the central regulation of mood, appetite, metabolism, and sleep-wake cycle regulation (Tecott et al., 1995; Nonogaki et al., 1998; Backstrom et al., 1999; Sodhi et al., 2001; Schmauss, 2003; Monti and Jantos, 2006). Transcripts encoding the 5HT2C receptor are subject to an RNA processing event in which specific adenosine residues are modified by hydrolytic deamination and converted to inosine moieties (adenosine-to-inosine editing) (Burns et al., 1997). Inosine is recognized as guanosine by the cellular translation machinery (Higuchi et al., 1993), leading to alterations in the coding potential of the transcript and subsequent alterations in receptor function. 5HT2C pre-mRNAs can be edited at up to five sites (sites A, B, E, C, and D) that can alter codons for three critical amino acids in the putative second intracellular loop of the receptor. Combinatorial editing at these five positions can generate up to 32 mRNA isoforms encoding 24 different receptor proteins (Burns et al., 1997; Niswender et al., 1998). RNA editing profiles have been shown to vary significantly between brain regions (Burns et al., 1997; Wang et al., 2000), among inbred mouse strains (Englander et al., 2005; Du et al., 2006; Hackler et al., 2006), in response to pharmacological treatment (Gurevich et al., 2002; Englander et al., 2005; Sodhi et al., 2005), and in affective disorders (Backstrom et al., 1999; Sodhi et al., 2001; Schmauss, 2003), implicating the modulation of RNA editing levels in the regulation of 5HT2C receptor function. The fully edited (VGV) isoform of the human 5HT2C receptor (encoding valine, glycine, and valine at amino acid positions 156, 158, and 160, respectively) exhibits reduced constitutive activity and decreased G protein-coupling efficacy compared with the genomically encoded (INI) isoform in heterologous expression systems (Burns et al., 1997; Fitzgerald et al., 1999; Niswender et al., 1999; Wang et al., 2000; Berg et al., 2001). Because the increased editing of 5HT2C RNAs generates receptors that are less likely to assume a G-protein coupled high-affinity conformation (R*) (Niswender et al., 1999), the observed potency of agonists with increased affinity for this receptor state is disproportionately reduced (Werry et al., 2008), and the specificity of agonist-directed trafficking of receptor stimulus is lost (Berg et al., 2001).
The most common methodology to quantify site-specific editing has involved reverse-transcription polymerase chain reaction (RT-PCR) amplification of specific RNAs followed by a modified primer-extension analysis (Rueter et al., 1995; Burns et al., 1997); however, this analytical paradigm is less useful for analyses of editing patterns in transcripts with multiple editing sites, such as 5HT2C mRNAs. The two most commonly used methods to quantify the editing of 5HT2C RNA isoforms involve either direct sequencing or pyrosequencing analysis of individual cDNA clones (Iwamoto et al., 2005; Sodhi et al., 2005; Werry et al., 2008). Although these methods yield unambiguous results, the accuracy of such methods is directly related to the number of clones sequenced, which can vary widely between studies and has been reported to be as few as 10 clones per sample (Sodhi et al., 2001). The limited number of clones analyzed can produce significant sampling errors that may either obscure or overestimate differences between experimental groups. Recent studies have analyzed 5HT2C editing using 454 deep-sequencing technology, increasing the number of 5HT2C cDNAs sequenced to nearly 800 while simultaneously avoiding the laborious subcloning, bacterial transformation, and preparation of individual cDNA clones (Wahlstedt et al., 2009). Despite the increased throughput, from 7 to 16 predicted mRNA isoforms remained undetectable in mouse brain samples between embryonic day 15 and postnatal day 21, possibly missing rare isoforms that could represent important species in minor neuronal subpopulations.
We have developed a high-throughput multiplexed transcript analysis (HTMTA) based on the massively parallel, short-read sequencing technology available on the Illumina/Solexa platform (Bentley et al., 2008). This approach allows an improved method for quantifying 5HT2C receptor RNA editing patterns with substantially increased accuracy and sensitivity. Using eight barcodes to maintain sample identity, this multiplexed strategy has been used to analyze RNA editing profiles for 5HT2C transcripts from up to 56 RNA samples simultaneously using a single Illumina Genome Analyzer flow cell. This adaptation of new sequencing technology provides a substantial improvement in the quantification of isoform-specific 5HT2C mRNA expression compared with methods used previously and should provide a powerful strategy for identifying previously undetected variations in RNA editing patterns in specific physiological or disease states and in response to pharmacological manipulation.
Materials and Methods
Mice.
Inbred male mice of different strains were purchased from The Jackson Laboratory (Bar Harbor, ME; C57BL/6, DBA/2J, and BALB/cJ) or Taconic Farms (Germantown, NY; 129S6). Mice were maintained on a 12-h light/dark cycle in a humidity- and temperature-controlled environment with ad libitum access to water and standard laboratory chow. All animal studies were approved by the Institutional Animal Care and Use Committee of Vanderbilt University.
RNA Isolation.
Animals were sacrificed at 8 weeks of age, and brain tissue was either flash-frozen (whole-brain samples) or dissected into specific brain regions (Glowinski and Iversen, 1966) and homogenized immediately. All tissue was homogenized in Tri-Reagent (Ambion, Austin, TX); total RNA was isolated according to the manufacturer's instructions and quantified by spectrophotometry at A260 (Nanodrop; Thermo Fisher Scientific, Wilmington, DE).
HTMTA Primer Design.
Each antisense oligonucleotide primer was designed to contain a 24-nucleotide (nt) region complementary to exon 6 (72–95 nt downstream from the D site) of the 5HT2C gene preceded by a distinct 5-nt barcode sequence, a 37-nt region for “read 2” sequencing primer hybridization, and a 24-nt adapter sequence that can anneal to the complementary oligonucleotide bound to the Illumina flow cell surface (Bentley et al., 2008). Specifically, the eight barcodes consist of palindromic 5-nt sequences that allow the identity of a sample to be determined unambiguously with up to two errors in the barcode sequence. The sense oligonucleotide primer contains 30 nt complementary to exon 5 (13–42 nt upstream from the A site) and a 29-nt adapter sequence that can anneal to a second complementary oligonucleotide bound to the flow-cell surface (Bentley et al., 2008) (Fig. 1A). The “read 1” sequencing primer corresponds to the 30-nt region of 5HT2C complementarity in the sense primer. The adapter sequences and “read 2” sequencing primer sequences were modeled for use in paired-end sequencing with the Illumina Genome Analyzer (Illumina Inc., San Diego, CA) as described previously (Bentley et al., 2008). Primer sequences are detailed in Supplementary Table S1.
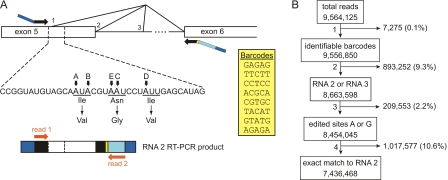
Fig. 1.
Summary of the HTMTA sequencing strategy. A, the structure and alternative splicing of 5HT2C pre-mRNA in the exon 5–6 region is presented (top). The locations of RT-PCR primers are indicated (black arrows). Adapter sequences for paired-end sequencing on the Illumina Genome Analyzer (dark blue) were incorporated into each primer. An annealing region for a sequencing primer (light blue) and one of eight distinct barcode sequences (yellow) were incorporated into the antisense primer. The sequence of the nonedited RNA, beginning immediately after the primer sequence, is specified below the pre-mRNA RT-PCR diagram. The positions of the five edited adenosines are indicated in boldface type (sites A, B, E, C, and D), and the amino acids encoded by the nonedited (top) and fully edited transcripts (bottom) are indicated below their respective codons. The position of the 36-nt sequence obtained from read 1 is indicated between the dashed lines on both the RT-PCR and pre-mRNA diagrams. Two sequencing primers (read 1 and read 2, orange arrows) were used for paired-end sequence analysis of RT-PCR amplicons generated from 5HT2C mRNA (bottom). Each of eight unique barcode sequences incorporated into the RT-PCR amplicon for sample identification is provided in the yellow inset. B, flow chart of data-sorting criteria. Total sequence reads obtained per RNA sample from HTMTA were culled to an appropriate data set by meeting a series of inclusion criteria, including the following: 1) barcode sequences are identifiable; 2) RNA 2 or RNA 3 alternatively spliced isoforms are identifiable; 3) nucleotides at the editing site are either A or G; and 4) the sequence of read 1 precisely matches the mouse 5HT2C RNA 2 reference sequence (Supplemental Table S2). The representative example is derived from the analysis of eight distinct barcoded samples. The number of reads that fail to meet each inclusion criterion is indicated to the right, followed by the percentage of the total reads that these excluded sequences represent. The number of sequence reads meeting each criterion is indicated below the inclusion description in each rectangle.
Sequence Analysis.
Pyrosequencing analyses of 5HT2C receptor RNA editing profiles were performed as described previously (Sodhi et al., 2005). For HTMTA, first-strand cDNA was synthesized in a 10-μl reaction from 1 μg of RNA using avian myoblastosis virus reverse transcriptase (Promega, Madison, WI) according to the manufacturer's instructions in a reaction containing 0.75 μg of a specific bar-coded antisense primer. Parallel control reactions lacking reverse transcriptase were performed for all samples. The total reverse-transcription volume was included in a 25-μl PCR amplification reaction with Phusion DNA polymerase (Finnzymes, Woburn, MA) according to the manufacturer's recommendations using 0.75 μg of the HTMTA sense primer. All reactions were amplified for 28 cycles unless otherwise specified. Amplified products were separated on a 2% agarose gel and purified from excised gel slices using the Wizard SV Gel and PCR Purification Kit (Promega). PCR amplicons corresponding to 5HT2C RNA 2 (249 base pairs) were isolated for all experiments except for comparisons of editing in alternatively spliced 5HT2C mRNAs. For these studies, PCR amplicons corresponding to both 5HT2C RNA 2 and RNA 3 (334 base pairs) were isolated in a single excised gel slice. The concentration of gel-purified fragments was measured by spectrophotometry (A260), and ∼20 ng of each individual sample was pooled with seven other products containing unique barcodes and subjected to paired-end sequencing with the Illumina Genome Analyzer as described previously (Bentley et al., 2008) using sequencing primers detailed in Supplementary Table S1. Sequencing data were filtered as indicated in Fig. 1B to eliminate sequences derived from either RNA 3 or human transcripts, where appropriate. Read 2 sequences were used to distinguish both barcode and 5HT2C splice form identities, whereas sequences obtained from read 1 were used to determine the editing profile of individual cDNA clones.
Statistical Analysis.
Statistical differences between the editing profiles of individual edited isoforms were determined using a Poisson linear model. This model included the number of reads for each isoform as the outcome variable, with the main independent variable being group (e.g., brain region 1 versus brain region 2). To allow for sample comparisons, the logarithm of total isoform count was calculated for each sample and was included in the model as an offset variable so that statistical significance of the variable group would indicate whether the average proportion (observed count/total count) of the isoform is significantly different for the two samples. In addition, an overdispersion parameter was included in the model to account for greater variability in the data set than those accounted by assuming the Poisson model. The editing level at each individual site (A–E) was analyzed similarly, except for replacing the individual isoform count with the count at each site as the outcome variable.
To estimate the number of samples needed to detect an isoform with abundance level p with probability t, we assumed a binomial model and used the formula Pr (an editing pattern occurs for at least 1 sample among n samples) = t = 1 − Pr (an editing pattern does not occur for all n samples) = 1 − (1 − p) n .
Results
Validation of HTMTA Analyses for 5HT2C mRNA Editing Profiles.
To develop a high-throughput method for quantifying 5HT2C receptor RNA editing patterns, we adapted the Illumina Genome Analyzer whole-transcriptome sequencing technology (Bentley et al., 2008) to report sequence data from an average of 828,354 (±93,282) individual RT-PCR amplicons derived solely from 5HT2C receptor transcripts (Fig. 1A). To decrease the total cost of analysis, we included five nucleotide barcode sequences in the antisense primers, allowing multiplexing of sequencing reactions derived from eight separate RNA samples while maintaining sample identity. HTMTA data from a representative flow cell lane yielded 9,564,125 raw sequence reads, and we used a bidirectional sequencing approach that allowed us to distinguish and eliminate any RNA 1- or RNA 3-derived 5HT2C sequences from read 2 (Supplemental Table S2). The data were further filtered to isolate 7,438,012 reads with identifiable barcodes precisely matching the mouse 5HT2C RNA 2 reference sequence (77.8% recovery rate; Fig. 1B).
Because extensive RT-PCR amplification can selectively bias template-to-product ratios in multitemplate PCR (Kanagawa, 2003), we determined whether the number of amplification cycles could affect the observed editing profile by performing control studies in which identical PCR reactions were amplified for 19, 22, 25, 28, 31, or 34 cycles before cluster generation. Sequence analysis revealed that editing profiles did not change significantly between 19 and 34 cycles (data not shown). All subsequent PCR amplification was performed for 28 cycles, because this was the lowest cycle number for which a robust PCR product could routinely be observed for gel purification (data not shown).
To determine whether the increased sensitivity provided by HTMTA results in the false discovery of contaminating DNA molecules or RNA misidentification because of polymerase errors, a nonedited human 5HT2C cDNA clone (INI) was used as a control for detection of other potential edited species within the sample. Results from this analysis revealed the identification of non-INI-encoding species representing 0.25% of total 5HT2C expression, indicating that HTMTA is accurate within 0.1% when analyzing the level of editing at each individual site (A–E) and within 0.25% when determining the relative expression of individual isoforms (Supplemental Figure S1). Furthermore, repeated analysis of a single RNA sample isolated from mouse whole brain (C57BL/6J) revealed a low level of intrasample variability for editing at each site (n = 3; mean percentage of editing at each site ± S.E.M.: A = 79.48 ± 0.34; B = 68.16 ± 0.35; E = 4.01 ± 0.09; C = 23.91 ± 0.34; and D = 62.98 ± 0.15%).
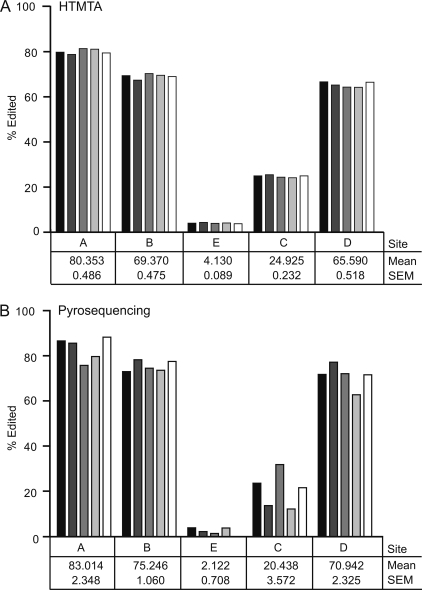
We compared 5HT2C RNA editing profiles determined by HTMTA with pyrosequencing analysis (Iwamoto et al., 2005; Sodhi et al., 2005) of 426 total cDNA clones generated from five separate C57BL/6 whole-brain mouse RNA samples. Although each technique detected similar levels of site-specific editing upon analyses of the same RNA samples (Fig. 2; p > 0.05), HTMTA revealed substantially decreased interanimal variability compared with pyrosequencing, suggesting that the variance detected by pyrosequencing among these inbred mice was largely due to insufficient sampling (Fig. 2 and Table 1). Of the 32 possible mRNA isoforms, HTMTA detected 20 isoforms at frequencies >0.25% of total 5HT2C transcripts (the error threshold for these experiments are shown in Supplemental Figure S1). RNAs encoding the ISI, VSI, and VGI receptor isoforms were readily observed, although they were absent from pyrosequencing analyses, and RNAs encoding the VDI and VDV isoforms were present at levels 3-fold greater than determined by pyrosequencing.
Fig. 2.
Comparison of RNA editing patterns using HTMTA and pyrosequencing methodologies. A, quantitative analysis of site-specific 5HT2C editing in RNA isolated from five independent whole-brain mouse samples as determined by HTMTA. B, quantitative analysis of 5HT2C editing using the same RNA samples as above by pyrosequencing. The mean percentage of site-specific editing is listed below each site (±S.E.M.).
TABLE 1.
Comparison of 5HT2C mRNA editing profiles from C57BL/6J mouse whole brain samples using HTMTA and pyrosequencing methods
| Protein Isoform | RNA Isoform | HTMTA |
Pyrosequencing |
||||
|---|---|---|---|---|---|---|---|
| Total Reads | Percentage of Total | S.E.M. | Total Reads | Percentage of Total | S.E.M. | ||
| INI | Nonedited | 244,399 | 7.692 | 0.153 | 32 | 7.530 | 1.343 |
| VNI | A | 192,775 | 6.080 | 0.180 | 19 | 4.558 | 1.413 |
| AB | 346,950 | 10.975 | 0.358 | 48 | 11.356 | 1.245 | |
| MNI | B | 16,798 | 0.529 | 0.013 | 1 | 0.244 | 0.244 |
| IDI | E | 9593 | 0.303 | 0.009 | 2 | 0.494 | 0.494 |
| ISI | C | 35,986 | 1.133 | 0.036 | 0 | 0.000 | 0.000 |
| INV | D | 215,924 | 6.817 | 0.202 | 23 | 5.442 | 1.098 |
| VDI | AE | 6946 | 0.218 | 0.008 | 0 | 0.000 | 0.000 |
| ABE | 16,856 | 0.529 | 0.022 | 1 | 0.241 | 0.241 | |
| VSI | AC | 27,964 | 0.882 | 0.016 | 0 | 0.000 | 0.000 |
| ABC | 171,158 | 5.408 | 0.113 | 20 | 4.692 | 0.992 | |
| VNV | AD | 126,821 | 3.981 | 0.100 | 19 | 4.407 | 0.852 |
| ABD | 1,113,379 | 35.326 | 0.426 | 186 | 43.263 | 4.120 | |
| MDI | BE | 213 | 0.007 | 0.001 | 0 | 0.000 | 0.000 |
| MSI | BC | 1804 | 0.057 | 0.004 | 0 | 0.000 | 0.000 |
| MNV | BD | 28,591 | 0.901 | 0.021 | 4 | 0.940 | 0.238 |
| IGI | EC | 2795 | 0.088 | 0.003 | 0 | 0.000 | 0.000 |
| IDV | ED | 9221 | 0.290 | 0.008 | 1 | 0.208 | 0.208 |
| ISV | CD | 51,051 | 1.607 | 0.053 | 6 | 1.458 | 0.598 |
| VGI | AEC | 7525 | 0.237 | 0.004 | 0 | 0.000 | 0.000 |
| ABEC | 8637 | 0.273 | 0.010 | 0 | 0.000 | 0.000 | |
| VDV | AED | 5888 | 0.186 | 0.009 | 0 | 0.000 | 0.000 |
| ABED | 38,767 | 1.225 | 0.029 | 2 | 0.455 | 0.280 | |
| VSV | ACD | 28,514 | 0.896 | 0.025 | 3 | 0.717 | 0.477 |
| ABCD | 425,393 | 13.447 | 0.199 | 55 | 13.178 | 3.402 | |
| MGI | BEC | 56 | 0.002 | 0.001 | 0 | 0.000 | 0.000 |
| MDV | BED | 555 | 0.018 | 0.003 | 2 | 0.485 | 0.297 |
| MSV | BCD | 4439 | 0.141 | 0.007 | 1 | 0.247 | 0.247 |
| IGV | ECD | 1984 | 0.063 | 0.004 | 0 | 0.000 | 0.000 |
| VGV | AECD | 5116 | 0.160 | 0.004 | 0 | 0.000 | 0.000 |
| ABECD | 16,865 | 0.531 | 0.010 | 1 | 0.241 | 0.241 | |
| MGV | BECD | 72 | 0.002 | 0.000 | 0 | 0.000 | 0.000 |
Differential Editing of Alternatively Spliced 5HT2C Transcripts.
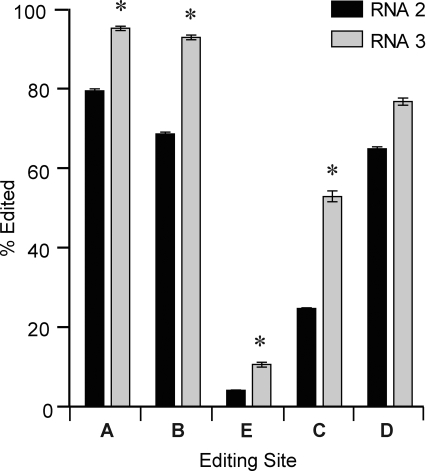
Alternative splicing of 5HT2C pre-mRNAs can generate three distinct mRNA species from the use of different 5′-splice sites located in exon 5 and intron 5. Use of the proximal and distal 5′-splice sites results in the generation of mRNAs encoding the truncated nonfunctional receptors 5HT2C-tr (Canton et al., 1996) and 5HT2C-COOHΔ (Wang et al., 2000), respectively, whereas use of the intermediate splice junction generates RNA 2, which encodes the full-length, functional 5HT2C receptor (Fig. 1A). 5HT2CR-tr transcripts (RNA 1) lack a 95-nt region encoding the second intracellular loop and the fourth transmembrane domain of the receptor, which encompasses the five editing sites. 5HT2C-COOHΔ mRNAs (RNA 3) include an additional 90-nt region of intron 5 containing a premature, in-frame stop codon (Wang et al., 2000). During the initial characterization of RNA 3, standard sequence analysis of 21 cDNA clones revealed that this mRNA was edited to completion at all five sites (Wang et al., 2000). Such increased editing was hypothesized to result from an increased time interval for adenosine-to-inosine conversion provided by the maintenance of an essential duplex structure in mature RNA 3 mRNAs (Wang et al., 2000). During the preparation of the C57BL/6 whole-brain samples for HTMTA analysis, RT-PCR amplicons derived from RNA 2 and RNA 3 were coexcised and purified to re-examine the RNA editing profiles between these two alternatively spliced 5HT2C mRNA species. Analyses of these transcripts revealed significant increases in editing at the A, B, E, and C sites in RNA 3 transcripts compared with RNA 2 (Fig. 3), corresponding to a significant increase in three highly edited isoforms [ABEC (+1.36%, p < 0.01), ABCD (+22.42%; p < 0.001), and ABECD (+4.15%; p < 0.000001)], confirming that editing of RNA 3 is increased significantly compared with RNA 2.
Fig. 3.
Quantitative analysis of 5HT2C RNA editing for alternatively spliced RNA 2 and RNA 3. HTMTA determination of site-specific editing is presented for RNA 2 and RNA 3 (mean ± S.E.M., n = 5; *, p < 0.05).
5HT2C RNA Editing in Commonly Used Inbred Mouse Strains.
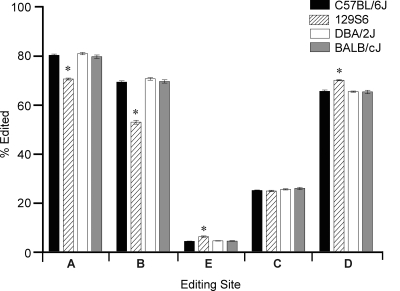
Previous studies have reported inconsistent variations in 5HT2C RNA editing profiles between inbred mouse strains (Englander et al., 2005; Du et al., 2006; Hackler et al., 2006). To reassess 5HT2C RNA editing patterns in these strains, HTMTA sequencing was performed using whole-brain samples isolated from 129S6, DBA/2J, and BALB/cJ mice and compared with results obtained from C57BL/6J samples (Fig. 4 and Supplemental Table S3). The C57BL/6J, DBA/2J, and BALB/cJ strains showed remarkable similarities in 5HT2C editing profiles with no significant differences in editing levels observed at any of the five sites (Fig. 4) and only a small decrease in expression of the AD isoform detected in BALB/cJ compared with C57BL/6J samples (Supplemental Table S3). Analysis of 5HT2C RNA editing patterns in 129S6 mice, however, provided a significantly different profile compared with C57BL/6 animals; editing at the A and B sites was significantly reduced (−9.75 and −16.4%, respectively), whereas editing at the E and D sites was significantly increased (+1.86% and + 4.52%, respectively) (Fig. 4). Similar differences were observed when 129S6 mice were compared with both the DBA/2J and BALB/cJ strains (data not shown). Quantitative analysis of specific 5HT2C RNA species revealed that the expression of seven isoforms was significantly decreased, and the expression of 18 isoforms was significantly increased in 129S6 mice compared with the C57BL/6J strain (Supplemental Table S3); similar changes were observed comparing 129S6 mice with both the DBA/2J and the BALB/cJ strains (Supplemental Table S3 and data not shown).
Fig. 4.
Analysis of 5HT2C mRNA profiles in inbred mouse strains. HTMTA of site-specific editing patterns from whole-brain RNA isolated from the C57BL/6J, 129S6, DBA/2J, and BALB/cJ inbred mouse strains (mean ± S.E.M., n = 5 per strain; *, p < 0.0001).
5HT2C RNA Editing in Dissected Brain Regions.
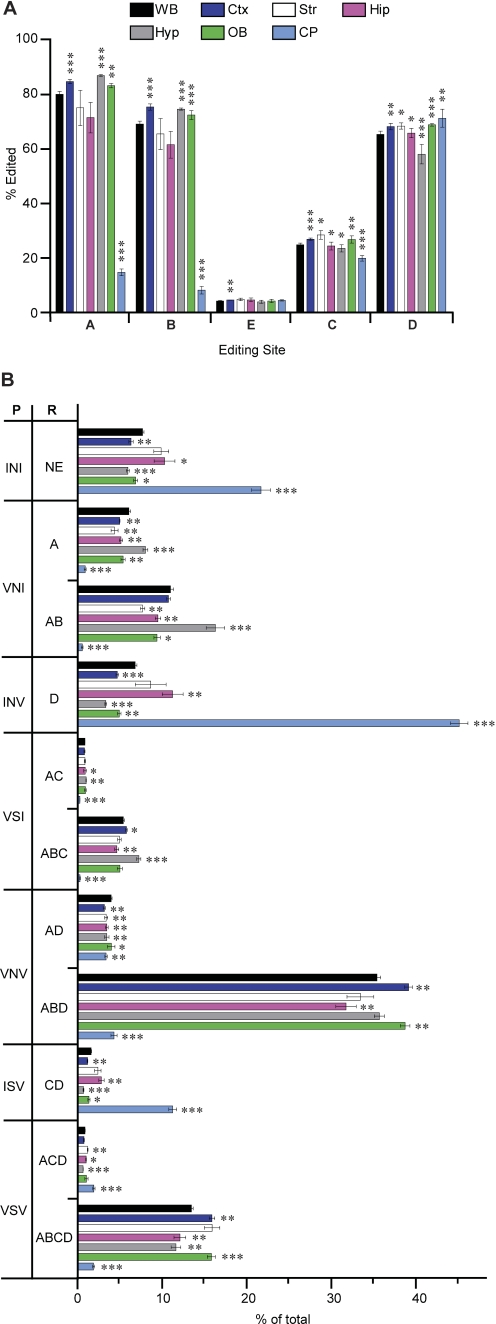
The expression pattern for 5HT2C receptors in the central nervous system is suggestive of specific roles in normal physiology and, when dysregulated, in the development of certain disease states such as obesity, anxiety, epilepsy, sleep disorders, and motor dysfunction (Wright et al., 1995; Werry et al., 2008). Previous studies using conventional sequence analyses of cDNAs isolated from dissected rat, mouse, and human brains identified the region-specific expression of as many as 12 major 5HT2C receptor isoforms (Burns et al., 1997; Niswender et al., 1999; Werry et al., 2008) that may allow the differential modulation of region-specific responses to ambient or stimulated levels of neurotransmitter. Decreased editing of 5HT2C mRNAs in the choroid plexus represents the most consistent difference observed previously (Burns et al., 1997; Wang et al., 2000). Because a failure to identify significant changes in isoform distribution between other dissected brain regions may have been limited by a reduced statistical power, resulting from the relatively small number of sequences used for these analyses, we sought to evaluate 5HT2C mRNA editing in six distinct brain regions isolated from C57BL/6J animals. Significant increases in site-specific RNA editing were identified in the olfactory bulb, frontal cortex, and striatum compared with data obtained from whole-brain RNA (Fig. 5A), and further analysis of 5HT2C mRNA profiles from these dissected regions revealed numerous changes (both increases and decreases) in the relative expression of specific mRNA isoforms (Fig. 5B). Although fewer differences were observed in the striatum compared with whole-brain RNA editing profiles, the expression of five isoforms was significantly decreased, and the expression of four isoforms was significantly increased in the striatum (Fig. 5B and Supplemental Table S4). On the other hand, less 5HT2C RNA editing was observed in the hippocampus, with significant reductions in editing at the A, B, and C sites (Fig. 5A) that were reflected as subsequent changes in the encoded 5HT2C receptor profile (Fig. 5B).
Fig. 5.
Analysis of brain region-specific 5HT2C RNA editing profiles in C57BL/6J mice. 5HT2C editing profiles were determined by HTMTA from RNA isolated from whole brain (WB) and dissected brain regions including cortex (Ctx), striatum (Str), hippocampus (Hip), hypothalamus (Hyp), olfactory bulb (OB), and choroid plexus (CP) (mean ± S.E.M.; n ≥ 4; *, p < 0.05, **, p < 0.01, ***, p < 0.001). A, site-specific editing patterns from whole brain and dissected brain regions; the percentage of editing at each site is indicated. B, quantitative analysis of the major 5HT2C RNA (R) and encoded protein (P) isoforms resulting from RNA editing are represented as percentage of total sequence reads; NE, nonedited.
RNA samples isolated from hypothalamus revealed significantly increased levels of editing at the A and B site sites compared with whole-brain RNA and a reduction in editing at the E, C, and D sites (Fig. 5A). Consistent with previous results (Burns et al., 1997; Werry et al., 2008; Pandey et al., 2006; Wang et al., 2000), characterization of choroid plexus RNA revealed the largest differences in 5HT2C RNA editing. Although editing was significantly decreased at the A, B, and C sites compared with whole-brain RNA, there was a small but significant increase in the level of editing at the D site (Fig. 5). These alterations in site-specific editing were reflected as a change in the overall profile of 5HT2C mRNA expression in the choroid plexus in which the relative level of all but two (BEC and BECD) of the 32 mRNA isoforms were affected (Fig. 5B and Supplemental Table S4).
Discussion
Here we show that quantitative analysis of 5HT2C RNA editing profiles using a high-throughput strategy based on the Illumina sequencing platform (Bentley et al., 2008) represents a significant improvement over low-throughput sequencing technologies developed previously. This approach is rapid, efficient, and cost-effective compared with currently existing paradigms involving the sequencing or pyrosequencing of isolated cDNA clones and is easily adaptable to any RNA editing substrate, providing a reduction in both inter- and intrasample variability (Fig. 2 and data not shown). By generating an average of more than 800,000 sequence reads per individual RNA sample, this approach provides both increased sensitivity for the detection of rare 5HT2C isoforms and increased statistical power compared with conventional sequencing or pyrosequencing methodologies (Burns et al., 1997; Wang et al., 2000; Iwamoto et al., 2005; Sodhi et al., 2005; Werry et al., 2008). To estimate the minimum number of individual cDNA sequences necessary to detect a specific 5HT2C isoform, we used a binomial model to determine that a minimum of 2994 clones must be sequenced to simply detect a rare isoform (0.1% abundance) with 95% probability (Table 2). This suggests that the ability of previous sequencing technologies to accurately quantify rare isoforms or demonstrate small changes in editing profiles between different RNA samples may have been limited.
TABLE 2.
Estimated sample sizes to detect a low abundant isoform with high probability (confidence) using the HTMTA method
| Isoform Abundance | Probability (Confidence) of
Detecting Isoform |
|||||
|---|---|---|---|---|---|---|
| 99% | 95% | 90% | 80% | 50% | 10% | |
| 10% | 44 | 28 | 22 | 15 | 7 | 1 |
| 1% | 458 | 298 | 229 | 160 | 69 | 10 |
| 0.10% | 4603 | 2994 | 2301 | 1609 | 693 | 105 |
| 0.01% | 46,049 | 29,956 | 23,025 | 16,094 | 6931 | 1054 |
| 0.001% | 460,515 | 299,572 | 230,257 | 160,943 | 69,314 | 10,536 |
HTMTA primers were developed initially to amplify all three possible spliced forms of 5HT2C mRNA simultaneously (Canton et al., 1996; Wang et al., 2000) (Fig. 1A), thereby allowing quantification of both RNA editing and alternative splicing patterns. However, the relative expression levels for RNA 1 and RNA 2 varied significantly between identical RT-PCR reactions and with amplification cycle number, preventing a reliable measurement of splicing ratios from RT-PCR amplicons (data not shown). Because the five editing sites are not contained within RNA 1 because of the use of an upstream 5′-splice junction (Canton et al., 1996), only the editing profiles of RNA 2 and RNA 3 can be compared (Fig. 3). Comparisons of editing patterns between these alternatively spliced 5HT2C mRNAs revealed significant increases in the editing of RNA 3, most likely resulting from the retention of a 90-nt region within intron 5 that is essential for adenosine-to-inosine conversion to allow continued editing of RNA 3 after splicing has been completed. Despite these editing differences, however, the potential consequences of increased editing for RNA 3 mRNAs have yet to be elucidated (Wang et al., 2000).
Previous comparisons of 5HT2C RNA editing profiles in different mouse strains have yielded conflicting results, although these studies did not feature overlapping experiments that could be compared directly. Such studies reported either no changes in whole-brain RNA editing between the C57BL/6, DBA/2J, and 129S1 strains (Du et al., 2006), decreased editing in the amygdala of both DBA/2J and BALB/cJ mice compared with C57BL/6 mice (Hackler et al., 2006) or substantial decreases in editing in the forebrain neocortex for BALB/cJ (but not 129S6) compared with C57BL/6 animals (Englander et al., 2005). In the current study, we found significant alterations in whole-brain RNA editing in 129S6 mice compared with the C57BL/6, DBA/2J, or BALB/cJ strains (Fig. 4 and Supplementary Table S3). Observed changes in 129S6 whole-brain 5HT2C RNA editing seemed comparable with results published previously, although the present method revealed numerous statistically significant differences that were not identified previously (Du et al., 2006). Binomial probability analysis (Table 2) suggests that the strain-specific differences observed previously in dissected brain regions (Englander et al., 2005; Hackler et al., 2006) may require further analysis because of the small number of cDNA clones analyzed and because of a lack of adequate statistical power.
HTMTA analysis of 5HT2C RNA editing profiles in dissected mouse brain regions supported previous results (Burns et al., 1997; Niswender et al., 1999; Wang et al., 2000; Werry et al., 2008) yet also identified additional differences in region-specific RNA editing patterns. Compared with whole-brain samples, editing levels were increased in the olfactory bulb, frontal cortex, and striatum, decreased in the hippocampus, or showed mixed changes in the hypothalamus and choroid plexus (Fig. 5 and Supplementary Table S4). Subtle variations in editing profiles between brain regions could reflect small neuronal subpopulations or specific pathways whose editing patterns deviate substantially from the surrounding tissue. Although it has not yet been determined whether small changes in editing profiles produce detectable changes in the function of cells expressing 5HT2C receptors, recent studies have described genetically modified mice expressing only the fully edited (VGV) isoform of the receptor that exhibit phenotypic alterations, including severe reductions in fat mass, hyperphagia. and increased energy expenditure (Kawahara et al., 2008), demonstrating the importance of normal patterns of 5HT2C RNA editing in vivo.
HTMTA represents a new adaptation of the massively parallel, short-read sequencing technology available on the Illumina/Solexa Genome Analyzer to accurately determine the RNA editing profile of multiple biological samples simultaneously. This assay is easily adaptable to any RNA editing substrate, providing a facile strategy to quantify editing of numerous ADAR substrates. The improved sensitivity and statistical power provided by this methodology increases the ability to accurately quantify variations in 5HT2C RNA editing that may be important in affective disorders or in response to pharmacological manipulation.
Supplementary Material
Acknowledgments
We thank the Vanderbilt University Genome Technology Core for technical assistance. We are particularly grateful to Dr. Shawn E. Levy and Christian Shaffer for aid in designing the barcoded RT-PCR primers and computational sorting of sequence data, respectively.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Institute of Mental Health [Grants R21-MH77942, P50-MH078028], the latter grant through the Vanderbilt University Silvio O. Conte Center for Neuroscience Research.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.061903.
- 5HT2C
- 2C subtype of 5-hydroxytryptamine receptor
- HTMTA
- high-throughput multiplexed transcript analysis
- nt
- nucleotide
- PCR
- polymerase chain reaction
- RT-PCR
- reverse-transcription polymerase chain reaction.
References
- Backstrom JR, Chang MS, Chu H, Niswender CM, Sanders-Bush E. (1999) Agonist-directed signaling of serotonin 5-HT2C receptors: differences between serotonin and lysergic acid diethylamide (LSD). Neuropsychopharmacology 21(2 Suppl):77S–81S. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg KA, Cropper JD, Niswender CM, Sanders-Bush E, Emeson RB, Clarke WP. (2001) RNA-editing of the 5-HT(2C) receptor alters agonist-receptor-effector coupling specificity. Br J Pharmacol 134:386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB . (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387:303–308 [DOI] [PubMed] [Google Scholar]
- Canton H, Emeson RB, Barker EL, Backstrom JR, Lu JT, Chang MS, Sanders-Bush E. (1996) Identification, molecular cloning, and distribution of a short variant of the 5-hydroxytryptamine2C receptor produced by alternative splicing. Mol Pharmacol 50:799–807 [PubMed] [Google Scholar]
- Du Y, Davisson MT, Kafadar K, Gardiner K. (2006) A-to-I pre-mRNA editing of the serotonin 2C receptor: comparisons among inbred mouse strains. Gene 382:39–46 [DOI] [PubMed] [Google Scholar]
- Englander MT, Dulawa SC, Bhansali P, Schmauss C. (2005) How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J Neurosci 25:648–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald LW, Iyer G, Conklin DS, Krause CM, Marshall A, Patterson JP, Tran DP, Jonak GJ, Hartig PR. (1999) Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacology 21(2 Suppl):82S–90S [DOI] [PubMed] [Google Scholar]
- Glowinski J, Iversen LL. (1966) Regional studies of catecholamines in the rat brain. I. The disposition of [3H]norepinephrine, [3H]dopamine and [3H]dopa in various regions of the brain. J Neurochem 13:655–669 [DOI] [PubMed] [Google Scholar]
- Gurevich I, Englander MT, Adlersberg M, Siegal NB, Schmauss C. (2002) Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J Neurosci 22:10529–10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackler EA, Airey DC, Shannon CC, Sodhi MS, Sanders-Bush E. (2006) 5-HT(2C) receptor RNA editing in the amygdala of C57BL/6J, DBA/2J, and BALB/cJ mice. Neurosci Res 55:96–104 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. (1993) RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75:1361–1370 [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Kato T. (2005) Estimating RNA editing efficiency of five editing sites in the serotonin 2C receptor by pyrosequencing. RNA 11:1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagawa T. (2003) Bias and artifacts in multitemplate polymerase chain reactions (PCR). J Biosci Bioeng 96:317–323 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, Nishikura K. (2008) Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J Neurosci 28:12834–12844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti JM, Jantos H. (2006) Effects of activation and blockade of 5-HT2A/2C receptors in the dorsal raphe nucleus on sleep and waking in the rat. Prog Neuropsychopharmacol Biol Psychiatry 30:1189–1195 [DOI] [PubMed] [Google Scholar]
- Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E. (1999) RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem 274:9472–9478 [DOI] [PubMed] [Google Scholar]
- Niswender CM, Sanders-Bush E, Emeson RB. (1998) Identification and characterization of RNA editing events within the 5-HT2C receptor. Ann N Y Acad Sci 861:38–48 [DOI] [PubMed] [Google Scholar]
- Nonogaki K, Strack AM, Dallman MF, Tecott LH. (1998) Leptin-independent hyperphagia and type 2 diabetes in mice with a mutated serotonin 5-HT2C receptor gene. Nat Med 4:1152–1156 [DOI] [PubMed] [Google Scholar]
- Pandey GN, Dwivedi Y, Ren X, Rizavi HS, Faludi G, Sarosi A, Palkovits M. (2006) Regional distribution and relative abundance of serotonin(2c) receptors in human brain: effect of suicide. Neurochem Res 31:167–176 [DOI] [PubMed] [Google Scholar]
- Rueter SM, Burns CM, Coode SA, Mookherjee P, Emeson RB. (1995) Glutamate receptor RNA editing in vitro by enzymatic conversion of adenosine to inosine. Science 267:1491–1494 [DOI] [PubMed] [Google Scholar]
- Schmauss C. (2003) Serotonin 2C receptors: suicide, serotonin, and runaway RNA editing. Neuroscientist 9:237–242 [DOI] [PubMed] [Google Scholar]
- Sodhi MS, Airey DC, Lambert W, Burnet PW, Harrison PJ, Sanders-Bush E. (2005) A rapid new assay to detect RNA editing reveals antipsychotic-induced changes in serotonin-2C transcripts. Mol Pharmacol 68:711–719 [DOI] [PubMed] [Google Scholar]
- Sodhi MS, Burnet PW, Makoff AJ, Kerwin RW, Harrison PJ. (2001) RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol Psychiatry 6:373–379 [DOI] [PubMed] [Google Scholar]
- Tecott LH, Sun LM, Akana SF, Strack AM, Lowenstein DH, Dallman MF, Julius D. (1995) Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature 374:542–546 [DOI] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Ensterö M, Ohman M. (2009) Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res 19:978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, O'Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K. (2000) Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem 74:1290–1300 [DOI] [PubMed] [Google Scholar]
- Werry TD, Loiacono R, Sexton PM, Christopoulos A. (2008) RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol Ther 119:7–23 [DOI] [PubMed] [Google Scholar]
- Wright DE, Seroogy KB, Lundgren KH, Davis BM, Jennes L. (1995) Comparative localization of serotonin1A, 1C, and 2 receptor subtype mRNAs in rat brain. J Comp Neurol 351:357–373 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.