Abstract
N-Methyl-d-aspartate (NMDA) receptors are ligand-gated ion channels that mediate a slow, Ca2+-permeable component of excitatory synaptic transmission in the central nervous system and play a pivotal role in synaptic plasticity, neuronal development, and several neurological diseases. We describe a fluorescence-based assay that measures NMDA receptor-mediated changes in intracellular calcium in a BHK-21 cell line stably expressing NMDA receptor NR2D with NR1 under the control of a tetracycline-inducible promoter (Tet-On). The assay selectively identifies allosteric modulators by using supramaximal concentrations of glutamate and glycine to minimize detection of competitive antagonists. The assay is validated by successfully identifying known noncompetitive, but not competitive NMDA receptor antagonists among 1800 screened compounds from two small focused libraries, including the commercially available library of pharmacologically active compounds. Hits from the primary screen are validated through a secondary screen that used two-electrode voltage-clamp recordings on recombinant NMDA receptors expressed in Xenopus laevis oocytes. This strategy identified several novel modulators of NMDA receptor function, including the histamine H3 receptor antagonists clobenpropit and iodophenpropit, as well as the vanilloid receptor transient receptor potential cation channel, subfamily V, member 1 (TRPV1) antagonist capsazepine. These compounds are noncompetitive antagonists and the histamine H3 receptor ligand showed submicromolar potency at NR1/NR2B NMDA receptors, which raises the possibility that compounds can be developed that act with high potency on both glutamate and histamine receptor systems simultaneously. Furthermore, it is possible that some actions attributed to histamine H3 receptor inhibition in vivo may also involve NMDA receptor antagonism.
N-methyl-d-aspartate (NMDA) receptors, a class of the ionotropic glutamate receptors, are ligand-gated cation-selective channels that mediate excitatory synaptic transmission in the central nervous system (Traynelis et al., 2010). These receptors are involved in many normal brain functions, such as neuronal development (Nacher and McEwen, 2006), learning, and memory (Lisman, 2003). NMDA receptors have also been implicated in a wide range of neuropathological conditions, including stroke, epilepsy, schizophrenia, depression, Huntington's disease, Alzheimer's disease, Parkinson's disease, and multiple sclerosis (Bräuner-Osborne et al., 2000; Waxman and Lynch, 2005; Traynelis et al., 2010).
The majority of native NMDA receptors are tetramers made up of two glycine-binding NR1 subunits and two glutamate-binding NR2 subunits, of which there are four family members (NR2A, NR2B, NR2C, and NR2D). The four NR2 subunits each endow the receptor with different pharmacological and kinetic properties (Vicini et al., 1998; Yuan et al., 2009). The NR2 subunits show distinct anatomical localization, which provides an opportunity to modify specific neuronal circuit function for therapeutic benefit using subunit-selective antagonists and potentiators.
Identification of the first potent and subunit-selective inhibitor of NMDA receptor function, namely, the NR2B-selective antagonist ifenprodil, has allowed not only evaluation of the role this subunit plays in many brain functions and diseases but also has led to clinical trials of NR2B-selective antagonists for use in treatment-resistant depression (Preskorn et al., 2008). However, despite the importance of NMDA receptors in many neurological disorders, antagonists that are more than 10-fold selective for NR2A, NR2C, or NR2D have not yet been identified. This largely reflects research efforts that have focused on the design of subunit-selective competitive antagonists. However, the agonist binding site is highly conserved across all glutamate-binding NR2 subunits, which has confounded attempts to engineer subunit selectivity among competitive antagonists. Similarly, channel blockers are similarly poorly selective for the various subunits (Dravid et al., 2007), presumably due to structural conservation of the permeation pore. The lack of subunit-selective pharmacological tools for this receptor class has precluded experiments that might otherwise provide greater understanding of the functional roles of the NR2A, NR2C, and NR2D subunits. To address the need for new pharmacological tools, we developed an assay that can identify noncompetitive antagonists and modulators of NMDA receptor function. Here, we studied the NR2D subunit, which gives rise to a nondesensitizing response and is suited for assays based on fluorescence measurements of intracellular calcium. Validation screens with small focused libraries confirmed the utility of this assay and identified two classes of compounds (TRPV1 and histamine H3 receptor antagonists) as novel NMDA receptor antagonists. TRPV1 receptors, also known as vanilloid-1 receptors or capsaicin receptors, are nonselective cation channels that are activated by a range of physical and chemical stimuli, including heat, extracellular protons, and vanilloids (Romanovsky et al., 2009). Histamine H3 receptors are expressed at high levels in the central nervous system, where they are presynaptic G protein-coupled receptors that regulate the release of a variety of neurotransmitters, including biogenic amines, acetylcholine, GABA, and glutamate (Haas et al., 2008). The actions of histamine H3 receptor antagonists on NR2B-containing NMDA receptors could present potentially interesting opportunities in terms of drug development.
Materials and Methods
Generation and Selection of a Stable Cell Line Expressing NR1/NR2D Receptors.
Baby hamster kidney (BHK)-21 cell lines stably expressing NR2D as well as NR1-1a (hereafter called NR1) under the control of a tetracycline-inducible promoter (Tet-On; Clontech, Mountain View, CA) were generated as described in Hansen et al. (2008). The DNA constructs used were the rat NR1-1a in pTRE2 vector and rat NR2D in the pCI-IRES-bla vector (GenBank accession numbers U11418 and D13214, respectively) (Hansen et al., 2008).
BHK-21 cells were maintained at 37°C, 5% CO2, and 95% relative humidity in Dulbecco's modified eagle medium containing GlutaMAX-I, 4500 mg/l glucose, and 110 mg/l sodium pyruvate (Invitrogen, Carlsbad, CA) supplemented with penicillin (100 units/ml), streptomycin (100 μg/ml) (Invitrogen), 10% dialyzed fetal bovine serum (Invitrogen), 1 mg/ml G418 (Invitrogen), and 10 μg/ml blasticidin S (Invivogen, San Diego, CA), as well as 200 μM dl-2-amino-5-phosphonopentanoate (AP5) (Ascent Scientific, Princeton, NJ) and 200 μM 7-chloro-kyneurenate (7-CKA) (Ascent Scientific) to prevent NMDA receptor-mediated cell death. Fura-2 imaging was used to confirm that the clonal cell line expressed functional NR1/NR2D receptors. Individual clones were grown for 48 h to approximately 90% confluence on 12-mm glass coverslips in the presence or absence of 2 μg/ml doxycycline. Before imaging, the medium was removed and the cells were washed thoroughly with imaging buffer containing 150 mM NaCl, 3 mM KCl, 22 mM sucrose, 10 mM glucose, 2 mM CaCl2, and 10 mM HEPES at pH 7.4. The cells were loaded for 30 min at 37°C with 5 μM Fura-2/acetoxymethyl ester and 1 μM Pluronic F-127 solution made in imaging buffer. The glass coverslips were placed in a perfusion chamber and continually perfused with imaging buffer. NMDA receptors are Ca2+-permeable; thus, their activation gives rise to Ca2+ influx and an increase in the intracellular Ca2+ concentration. NMDA receptor responses were activated using 50 μM glutamate and 50 μM glycine in imaging buffer. As a positive control, 10 μM ATP was used to activate endogenously expressed P2Y receptors, which are known to increase intracellular calcium. Intracellular Fura-2 was excited at 340 nm and 380 nm, and the ratio of the emissions at 510 nm was monitored using a BX51WI microscope (Olympus America Inc., Center Valley, PA) and a PTI IC200 intensified camera (PTI, Birmingham, NJ). The cell line that displayed a robust response to glutamate and glycine was expanded and frozen at −130°C.
NR1 protein expression was visualized by immunostaining. Cells were grown on glass coverslips as described above. The cells were washed with Tris-buffered saline (TBS) composed of 135 mM NaCl, 2.5 mM KCl, 50 mM Tris, and 0.1% Tween 20 at pH 7.4, and then they were fixed by incubating on ice for 10 min with 4% paraformaldehyde in phosphate-buffered saline, pH 7.4. Nonspecific binding of the antibody was blocked by incubating the cells in TBS containing 3% bovine serum albumin (BSA) for 1 h at room temperature. Cells were then incubated overnight at 4°C with anti-NMDAR1 mAb54.1 (BD Biosciences, San Jose, CA) diluted 1:1000 (0.5 μg/ml) in 3% BSA/TBS. After washing with TBS to remove unbound primary antibody, the cells were incubated for 2 h at room temperature with fluorescein isothiocyanate-conjugated anti-mouse IgG (Invitrogen) diluted 1:1000 (2 μg/ml) in 3% BSA/TBS. The cells were washed with TBS and NR1 immunofluorescence was visualized using an IX51 inverted microscope (Olympus America Inc.) and a PTI IC200 intensified camera.
Development and Optimization of the Screening Assay.
For the screening assay, the BHK-21 cells were seeded into black-walled clear-bottomed 96-well plates at a density of 20,000 cells/well. At the time of plating, NR1 subunit expression was induced by supplementing the medium with 2 μg/ml doxycycline. The induction time for optimal expression was experimentally determined to be 36 to 48 h for the cell line used here. Before initiating the screen, the medium was removed and the cells were washed twice with assay buffer containing 2 mM CaCl2, 138 mM NaCl, 5.33 mM KCl, 0.34 mM Na2HPO4, 0.44 mM KH2PO4, 4.17 mM NaHCO3, 5.56 mM d-glucose, and 20 mM HEPES at pH 7.4. The cells were then incubated in assay buffer supplemented with 100 μl of Fluo-4 no-wash calcium assay dye solution (Invitrogen) for 30 min at 37°C according to the manufacturer's protocol. Unless otherwise stated, the dye solution also contained 30 μM 7-CKA, a competitive glycine site antagonist, to prevent NMDA receptor-mediated cytotoxicity during dye loading. After incubation at 37°C, the cells were allowed to equilibrate to room temperature for 10 min before initiation of the assay. Changes in Fluo-4 fluorescence in response to addition of agonists, and test compounds were measured at 538 nm using a FlexStation II multimode plate reader (Molecular Devices, Sunnyvale, CA) with excitation at 485 nm and emission cut-off at 530 nm to minimize background fluorescence. Baseline fluorescence was measured for 20 s before the addition of test compound to the assay plate, and the peak fluorescence was measured after addition of the agonists and the test compounds. Peak fluorescence was represented as percent of the preagonist baseline fluorescence (100% × F/Fbaseline). Agonists and the test compound in a volume of 25 μl were added to 100 μl of assay buffer in the assay plate. Compound plates for the assay were prepared with assay buffer and five times the final compound concentration to account for the 1:5 dilution from the compound plate to the assay plate. The final concentrations of the agonist were kept at 100 μM glutamate and 1 mM glycine. Test compounds were added to each well with agonists to yield a final well concentration of 10 μM test compound in 0.9% DMSO.
Compounds, pKa Prediction, and Structural Overlay.
All compounds from the library of pharmacologically active compounds (LOPAC) were obtained from Sigma-Aldrich (St. Louis, MO). All NMDA receptor-like antagonist (NRA) focused library compounds shown in Tables 1 to 3 were from Tocris Bioscience (Ellisville, MO), except for direct red and N-((((4-chloroanalino)carbothioyl) amino)methyl) benzamide (Sigma-Aldrich), 4-[(2-pyridylthio)methyl]-2-(3-thienyl)-1,3-thiazole (Ryan Scientific, Mt. Pleasant, SC), and N-{2-[(4-chlrophenyl) sulfanyl]ethyl}-2-(3-methylphenyl)acetamide and 3-(1,3-benzoxazol-2-ylthio)-1-(4-chlorophenyl)prop-2-en-1-one (Key Organics, Cornwall, UK), and levallorphan and phenylcyclohexamine derivatives (gifts from Dr. Stephen Holtzman, Emory University, Atlanta, GA). pKa values for capsazepine, clobenpropit, and iodophenpropit were calculated using ACD/pKa DB, version 9.03 (Advanced Chemistry Development, Inc., Toronto, ON, Canada). The Maestro suite, version 8.5 (Schrödinger, New York, NY) was used to do conformational search for ifenprodil using the OPLS-2005 force field (Jorgensen and Tirado-Rives, 2005). The conformationally best energy-minimized structure of ifenprodil was chosen as reference compound, and clobenpropit, iodophenpropit, and capsazepine were subjected to conformational search by using OMEGA, version 2.0 (OpenEye Scientific Software, Santa Fe, NM). The resulting confirmations were superimposed with ifenprodil using rapid overlay of chemical structures (ROCS) (Hawkins et al., 2007).
TABLE 1.
Detection of non- and uncompetitive NMDA receptor antagonists
Fluorescence response from a single well in screens of the BHK-21 cell line expressing NR1/NR2D shown as percentage of control (100 μM glutamate plus 1 mM glycine alone) in the presence of test ligand (10 μM). TEVC responses from the secondary screen are shown as a percentage of control in the presence of test ligand (10 μM) determined using TEVC recordings from X. laevis oocytes expressing recombinant NR1/NR2D. TEVC responses are mean ± S.E.M., and numbers in parentheses are the number of oocytes.
| Antagonist | Library | Fluorescence Response | TEVC Response |
|---|---|---|---|
| % control | |||
| (±)-N-Allylnormetazocine | LOPAC | 9* | 16 ± 2 (6)* |
| (+)-N-Allylnormetazocine | LOPAC | 1* | 19 ± 3 (6)* |
| Arcaine | LOPAC | 87 | 114 ± 3 (6) |
| CNS-1102 | LOPAC | 2* | 13 ± 8 (7)* |
| Dextromethorphan | LOPAC | 53* | 26 ± 8 (4)* |
| Dextrorphan | LOPAC | 4* | 11 ± 2 (3)* |
| Ifenprodil | LOPAC | 45* | 85 ± 2 (4) |
| Levallorphan | LOPAC, NRA | 0, 11* | 22 ± 2 (4)* |
| Memantine | LOPAC | 0* | 6 ± 2 (4)* |
| Metaphit | LOPAC, NRA | 86, 40* | 44 ± 5 (4)* |
| 3-Methoxy-morphanin | LOPAC | 6* | 19 ± 2 (6)* |
| (−)-MK-801 | LOPAC, NRA | 16, 5* | 9 ± 2 (4)* |
| (+)-MK-801 | LOPAC, NRA | 0, 4* | 0 ± 0 (3)* |
| N-Methyl-1-phenylcyclohexylamine | NRA | 4* | 19 ± 4 (5)* |
| N-Dimethyl-1-phenylcyclohexylamine | NRA | 11* | 32 ± 4 (6)* |
| N-Ethyl-1-phenylcyclohexylamine | NRA | 1* | 2 ± 1 (6)* |
| N-(n-Propyl)-1-phenylcyclohexylamine | NRA | 0* | 2 ± 1 (6)* |
| N-(s-Butyl)-1-phenylcyclohexylamine | NRA | 8* | 1 ± 1 (6)* |
| Pentamidine | LOPAC, NRA | 66, 59* | 50 ± 0 (3)* |
Asterisk indicates that the compound met the criteria for hit in the primary screen (≤60% of control) or in the secondary TEVC screen (≤75% control; see Materials and Methods).
TABLE 3.
LOPAC and NRA compounds not previously known to inhibit NMDA receptors
See legend to Table 1. Only compounds not previously known to inhibit NMDA receptors that met criteria for hit in both primary and secondary TEVC screen of NR1/NR2D are shown.
| Antagonist | Library | Fluorescence Response | TEVC Response |
|---|---|---|---|
| % control | |||
| Alaproclate | LOPAC | 3 | 46 ± 3 (6) |
| N-(4-Aminobutyl)-5-chloro-2-naphthalenesulfonamide | LOPAC | 36 | 50 ± 3 (6) |
| R-(+)-Butylindazone | LOPAC | 36 | 70 ± 5 (4) |
| Calmidazolium | LOPAC | 32 | 36 ± 4 (4) |
| (±)-Chlorpheniramine | LOPAC | 39 | 72 ± 3 (4) |
| (+)-Cyclazocine | LOPAC | 5 | 13 ± 2 (6) |
| Dequalinium | LOPAC | 5 | 24 ± 4 (5) |
| Isoliquiritigenin | LOPAC | 33 | 73 ± 5 (14) |
| 5-(N-Methyl-N-isobutyl)amiloride | LOPAC | 41 | 52 ± 3 (8) |
| MG 624 | LOPAC | 20 | 48 ± 6 (5) |
| Nortriptyline | LOPAC | 44 | 64 ± 2 (5) |
| Reactive blue 2 | LOPAC | 37 | 67 ± 2 (4) |
| Ruthenium red | LOPAC | 0 | 13 ± 3 (8) |
| TMB-8 | LOPAC | 39 | 74 ± 2 (4) |
| W-7 | LOPAC | 16 | 23 ± 4 (10) |
| Capsazepine | NRA | 63 | 75 ± 3 (12) |
| Clobenpropit | NRA | 43 | 37 ± 5 (5) |
| Iodophenpropit | NRA | 11 | 7 ± 2 (4) |
| Tiotidine | NRA | 47 | 49 ± 5 (4) |
| Direct red 20 | NRA | 25 | 61 ± 9 (8) |
| BW723C86 | NRA | 54 | 70 ± 7 (6) |
| Antazoline | NRA | 59 | 70 ± 4 (5) |
| N-{2-[(4-Chlrophenyl)sulfanyl]ethyl}-2-(3-methylphenyl) acetamide | NRA | 60 | 63 ± 5 (17) |
| N-((((4-Chloroanalino)carbothioyl) amino)methyl) benzamide | NRA | 21 | 49 ± 4 (15) |
| 4-[(2-Pyridylthio)methyl]-2-(3-thienyl)-1,3-thiazole | NRA | 53 | 75 ± 3 (4) |
| 1-Benzo[b]thien-2-yl-N-cyclopropylmethylcyclohexanamine | NRA | 40 | 75 ± 6 (3) |
| (±)-1-(1,2-Diphenylethyl)piperidine | NRA | 7 | 35 ± 3 (4) |
| Nisoxetine | NRA | 46 | 55 ± 2 (6) |
| 3-(1,3-Benzoxazol-2-ylthio)-1-(4-chlorophenyl)prop-2-en-1-one | NRA | 44 | 70 ± 4 (3) |
Two-Electrode Voltage-Clamp Electrophysiology.
Two-electrode voltage-clamp (TEVC) recordings were performed on Xenopus laevis oocytes expressing NMDA (NR1/NR2A-D), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (GluR1), and kainate (GluR6) receptors. cDNAs for rat NR1-1a (GenBank accession numbers U11418 and U08261), NR2A (GenBank accession number D13211), NR2B (GenBank accession number U11419), NR2C (GenBank accession number M91563), NR2D (GenBank accession number L31611), GluR1 (GenBank accession number X17184), and GluR6 (GenBank accession number Z11548) were provided by Drs. S. Heinemann (Salk Institute for Biological Studies, San Diego, CA), S. Nakanishi (Kyoto University, Kyoto, Japan), and P. Seeburg (University of Heidelberg, Heidelberg, Germany). The NR1(N616R) mutation and the NR2B mutant subunits were generated using the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) according to the manufacturer's protocol and verified by DNA sequencing. The DNA construct encoding the amino-terminal domain deletion of the NR2B subunit (NR2B-ΔATD) has been described previously (Yuan et al., 2009). Oocyte isolation and RNA injection were completed as described in detail previously (Traynelis et al., 1998); all protocols involving X. laevis were approved by the Emory University Institutional Animal Care and Use Committee. During TEVC recordings, oocytes were placed into a perfusion chamber and continually washed with recording solution containing 90 mM NaCl, 1 mM KCl, 0.5 mM BaCl2, 0.005 mM EDTA, and 10 mM HEPES at pH 7.4 (23°C). Glass electrodes had a tip resistance of 0.5 to 2.5 MΩ and were pulled from thin-walled glass capillary tubes using a PP-83 puller (Narashige, East Meadow, NY). Voltage and current electrodes were filled with 0.3 and 3 M KCl, respectively. The current and voltage electrodes were connected to an OC-725C amplifier (Warner Instruments, Hamden, CT), which held the membrane potential of the oocytes at −40 mV during recording (unless otherwise stated). In the secondary screen, the inhibitors identified in the calcium imaging screen were purchased as powder, made into 20 mM stocks in DMSO, diluted to reach a final concentration of 10 μM in recording solution containing 100 μM glutamate and 30 μM glycine. The final DMSO concentration was 0.05% (v/v).
Radioligand Binding.
Human embryonic kidney 293 cells were transfected with human histamine H3 receptor cDNA [full-length isoform (445 amino acids) in pCI-neo; GenBank accession number NM_007232] using calcium phosphate precipitation. The plasmid RSV.TAg that encodes the simian virus 40 T antigen was used in transfections to increase receptor expression. Cells were harvested and homogenized in ice-cold TE buffer (50 mM Tris-HCl and 5 mM EDTA, pH 7.4) approximately 48 h after transfection, followed by 30-min centrifugation at 20,000g. The membrane pellets were homogenized and centrifuged one more time as described above. Final membrane pellets were frozen at −80°C until needed. For competition binding, membrane preparations were incubated with ∼1.0 nM [3H]N-α-methylhistamine (PerkinElmer Life and Analytical Sciences, Boston, MA) in the presence or absence of increasing concentrations of ifenprodil. The binding reactions were incubated for 30 min at 23°C in a final volume of 0.5 ml of TE buffer. Nonspecific binding was defined with 30 μM thioperamide. Binding reactions were terminated by filtration under vacuum onto Whatman GF/B filters (Brandel Inc., Gaithersburg, MD) followed by two washes with ∼4 ml of ice-cold TE buffer, and bound radioligand was determined by liquid scintillation counting; for each of three independent experiments, measurements were made in duplicate. IC50 values were determined by fitting to the Hill equation (one-site binding), and Ki values were calculated using the Cheng-Prusoff equation. The previously published Kd value of 0.5 nM for [3H]N-α-methylhistamine binding to the human histamine H3 receptor was used to derive the Ki value for ifenprodil (Witte et al., 2006).
Data Analysis.
Data generated from measurements using the FlexStation II were analyzed using BioAssay software (CambridgeSoft Corporation, Cambridge, MA) to identify NR1/NR2D modulators. Systematic plate errors (a progressive increase in signal intensity across the plate moving from columns 1 to 12) were subtracted from raw data, and the average and standard deviation of the responses across each 96-well plate were calculated.
Concentration-response data for individual oocytes normalized to the response in the absence of inhibitor (100%) were fitted to the Hill equation:
where nH is the Hill slope, [X] is the compound concentration, and IC50 is the half-maximally effective concentration of inhibitor. Fitted IC50 values and Hill slopes from individual oocytes were used to calculate the mean and S.E.M. For graphical presentation, data points from individual oocytes were averaged and then fitted to the Hill equation and plotted together with the resulting curve. Unless otherwise stated, data are represented as mean ± S.E.M.
Results
Assay Design and Optimization.
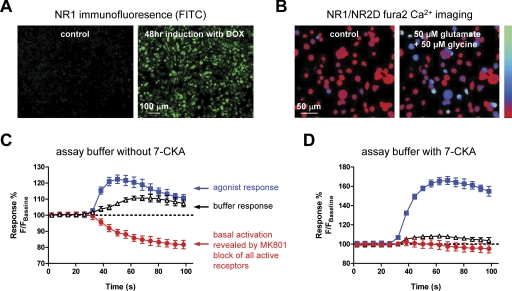
We developed a screening assay specifically designed to identify novel ligands that modulate the function of NR2D-containing NMDA receptors by acting at sites other than the agonist binding sites. NR2D-containing recombinant NMDA receptors show little desensitization and are Ca2+-permeable—two properties that should render them amenable to fluorescence-based optical assays that measure agonist-induced Ca2+ accumulation in mammalian cells using multiwell formats. We previously made a BHK-21 cell line that expresses the NR1 subunit under the control of the Tet-On inducible expression system and constitutively expresses of the NR2D subunit (Hansen et al., 2008). Previous studies indicate that stable expression of NMDA receptor subunits in heterologous expression systems can be highly cytotoxic (Cik et al., 1993). To avoid this, cell culture medium was supplemented with competitive NMDA receptor antagonists (200 μM AP5 and 200 μM 7-CKA). NR1 expression, a requirement for functional NMDA receptors, was induced by doxycycline approximately 48 h before the assay (Fig. 1A). Fura-2 calcium imaging in the doxycycline-induced NR1/NR2D cell line produced a glutamate EC50 value (0.34 μM) that was similar to that measured from TEVC recordings in oocytes (0.46 μM), suggesting this cell line reproduces the pharmacological properties of NR1/NR2D (Fig. 1B).
Fig. 1.
A, induction of NR1 visualized by immunofluorescence using anti-NR1 antibody after 48 h of doxycycline (DOX) treatment. Secondary antibody is fluorescein isothiocyanate-conjugated. Control refers to cells that were not treated with doxycycline and therefore do not express NR1. B, Fura-2 imaging of a BHK-21 cell line expressing NR1/NR2D during challenge with glutamate and glycine (50 μM each). Ratiometric images at 340/380-nm excitation and 510-nm emission for Fura-2 before (control) and after challenge with glutamate and glycine are shown. The bar (left) shows Ca2+ signal intensity ranging from 0 to 4 mM Ca2+ (bottom is 0 mM Ca2+). C and D, time course of fluorescence responses in the NR1/NR2D cell line in the absence and presence of 30 μM of the competitive glycine antagonist 7-CKA in the assay buffer. Agonist (blue squares), buffer (black triangles), or (+)-MK801 (red circles) were added to the well at time 20 s. The data points are displayed as percentage of baseline fluorescence 100% × F/Fbaseline, where F is the fluorescence measured after addition to the well, and Fbaseline is the average fluorescence measured before addition to the well. Data points are mean ± S.D. of four to five wells. Final agonist concentrations were 100 μM glutamate and 1 mM glycine. The uncompetitive open channel blocker (+)-MK801 (10 μM) was used to inhibit any receptors that were active before addition of the agonist and thereby reveal the basal activity of the receptors. The basal activity was virtually absent in the assay when 30 μM 7-CKA was added to the assay buffer.
The development of a multiwell assay using Fluo-4 calcium imaging to assess NR1/NR2D receptor function is associated with problems due to activation of NR1/NR2D receptors by glutamate and glycine at submicromolar concentrations (Hansen et al., 2008). Even low nanomolar concentrations of glutamate (with trace amounts of glycine) present in the assay buffer from the time of washing through the dye loading period are sufficient to activate NR1/NR2D receptors. This tonic activation can also elevate the baseline Ca2+ concentration, which reduces the signal-to-noise ratio (Fig. 1C). To circumvent this problem, we added the competitive glycine site antagonist 7-CKA (30 μM) during dye loading. This low-affinity antagonist prevents cytotoxicity during dye loading and subsequent experiments by preventing prolonged NR1/NR2D receptor activation. In the assay, the antagonist 7-CKA virtually eliminated basal receptor activation, the amount of which can be quantified as the difference between the preagonist baseline fluorescent response and the fluorescent responses after addition of an uncompetitive open channel blocker (+)-MK-801 (10 μM). In addition, 30 μM 7-CKA is easily displaced in the assay by addition of a high concentration of glycine (1 mM) coapplied with glutamate (100 μM), which together lead to full activation of the NR1/NR2D receptors (Fig. 1D). Concentration-response data were generated within the assay format for the coagonists glutamate and glycine (data not shown). This showed that 1 mM glycine was sufficient to completely overcome the inhibition produced by the competitive glycine site antagonist 7-CKA (30 μM) that was present during dye loading. The glutamate EC50 value (0.23 μM) is similar to that measured from Fura-2 calcium imaging using the same NR1/NR2D cell line (0.34 μM) and TEVC recordings in oocytes expressing NR1/NR2D (0.46 μM). The Z′ value is a standard measure of variability for multiwell assays, and values above 0.5 are considered a good indication that an assay is suitable for single-well screening of test compounds (Zhang et al., 1999). The Z′ value is calculated using the following equation:
Our assays yielded favorable Z′ values of 0.59 ± 0.12 (mean of 16 assay plates ± S.D.). Consistent with the acceptable Z′ values, the secondary screen using a different assay (see below) confirmed the majority of the hits that were identified.
Assay Validation and Screening for Known NMDA Receptor Antagonists.
The reliability of the assay was validated by screening the commercially available LOPAC-1280 library (1280 compounds; Sigma-Aldrich) and our own NMDA NRA-focused library of 520 compounds, many of which contained biaryl ring systems separated by nitrogen-containing linkers of varied length and rigidity. We anticipated that these compounds should have a higher probability of inhibiting NMDA receptors because they were selected based on similarity with the NR2B-selective antagonist ifenprodil. Compounds that altered the response of any given well compared with control wells on the same plate by more than 2.5-fold of the S.D. (calculated from all wells on the plate for LOPAC) or by more than 40% of the control response on the same plate (NRA-focused library) were selected for secondary screening using TEVC recordings of X. laevis oocytes expressing recombinant NR1/NR2D receptors. These selection criteria were empirically determined to reduce false positives, while maintaining a throughput that could reasonably be evaluated in the secondary screen.
The NRA-focused library included 13 known NMDA receptor antagonists, three competitive antagonists and 10 uncompetitive use-dependent channel blockers. The screen identified all 10 uncompetitive inhibitors but none of the competitive antagonists (Tables 1 and 2). The LOPAC library contains 14 known noncompetitive and uncompetitive NMDA receptor antagonists. The screen of the LOPAC library using NR1/NR2D expressing BHK-21 cells successfully identified the known noncompetitive NMDA receptor antagonist ifenprodil, which shows low potency at the NR2D subunit (Table 1). In addition, this screen identified the known uncompetitive NMDA receptor channel blockers (+)-MK-801, (−)-MK-801, CNS-1102, memantine, dextramethorphan, dextrorphan, levallorphan, 3-methoxy-morphanin, (±)-allylnormetazoline, and (+)-allylnormetazoline (Table 1). Thus, the primary screen of the LOPAC library identified 11 of 14 of the known NMDA receptor non- and uncompetitive antagonists present in the library, all of which were subsequently validated by showing at least 25% inhibition in the TEVC secondary screen. In addition, two more NMDA receptor antagonists (metaphit and pentamidine) that missed the threshold of detection in the screen of the LOPAC library were identified in the screen of the NRA focused library (Table 1). The LOPAC library also contains 17 well known competitive NMDA receptor antagonists that act at either the glycine or glutamate binding site. Only one of these known competitive antagonists (5-fluoroindole-2-carboxylic acid) surpassed the 2.5 S.D. from the mean (Table 2).
TABLE 2.
Detection of competitive NMDA receptor antagonists
Fluorescence response from a single well in screens of the BHK-21 cell line expressing NR1/NR2D shown as percentage of control (100 μM glutamate plus 1 mM glycine alone) in the presence of test ligand (10 μM).
| Antagonist | Library | Fluorescence Response |
|---|---|---|
| % control | ||
| (dl-)-2-AP3 | LOPAC | 106 |
| (dl)-2-AP4 | LOPAC | 102 |
| (dl)-2-AP5 | LOPAC | 92 |
| (d)-2-AP5 | NRA | 103 |
| (dl)-2-AP7 | LOPAC | 95 |
| (d)-2-AP7 | LOPAC, NRA | 119, 89 |
| 7-Chlorokynurenic acid | LOPAC | 79 |
| CNQX | LOPAC | 123 |
| (R,S)-CPP | LOPAC | 119 |
| (R)-CPP | NRA | 101 |
| 5,7-Dichlorokynurenic acid | LOPAC | 117 |
| 6,7-Dichloroquinoxaline-2,3-dione | LOPAC | 112 |
| 5-Fluoroindole-2-carboxylic acid | LOPAC | 56* |
| Flupirtine | LOPAC | 111 |
| (R,S)-HA-966 | LOPAC | 108 |
| Kynurenic acid | LOPAC | 117 |
| L-701,324 | LOPAC | 80 |
| LY-235,959 | NRA | 93 |
| MDL 105,519 | LOPAC | 114 |
| O-Phospho-l-serine | LOPAC | 137 |
Asterisk indicates that the compound met the criteria for hit in the primary screen (≤60% of control).
Together, the combined data obtained with these two small libraries suggest that the use of high concentrations of both glutamate and glycine identifies only 5% (1 of 19) of the competitive antagonists screened, whereas 95% (18 of 19) of the noncompetitive and uncompetitive antagonists tested were identified. The known uncompetitive antagonist (arcaine) that was not identified in the screen also did not inhibit NR1/NR2D receptor responses in TEVC recordings from oocytes. These results suggest the assay as described here is robust and should identify noncompetitive and uncompetitive NMDA receptor inhibitors with a high level of confidence.
Compound Screening for Novel NR2D Modulators.
The primary screen of the NRA focused library of 520 compounds identified 70 inhibitors using a criteria of 40% inhibition (hit rate, 13.5%), including the 10 uncompetitive NMDA receptor antagonists. In total, 24 inhibitors were active in the TEVC secondary screen as defined by showing at least 25% inhibition at 10 μM (confirmed hit rate 4.6%), 14 of which were not previously known to inhibit NMDA receptors. In total, 44 inhibitors (hit rate 3.4%) were identified in the primary screen of the LOPAC library, of which 27 compounds (confirmed hit rate, 2.1%) tested were found to be active in the secondary TEVC screen. This included 15 compounds that were not previously known to inhibit NMDA receptors. Table 3 shows all compounds in both libraries not previously reported to inhibit NMDA receptors, and Table 4 summarizes the screening hit rates for both test libraries.
TABLE 4.
Summary of screening results on NR2D-containing NMDA receptors
Numbers in parenthesis are percentage of the number of compounds in the individual libraries or the total number of compounds screened. Only hits from the primary screening assay that was commercially available as powder were tested in the secondary screen (TEVC recordings).
| Library | Compounds Screened | Hits from Screening Assay | Hits Tested in TEVC Recordings | Hits Verified in TEVC Recordings |
|---|---|---|---|---|
| NRA | 520 | 70 (13.5) | 62 (11.9) | 24 (4.6) |
| LOPAC | 1280 | 44 (3.4) | 41 (3.2) | 27 (2.1) |
| Total | 1800 | 114 (6.3) | 103 (5.7) | 51 (2.8) |
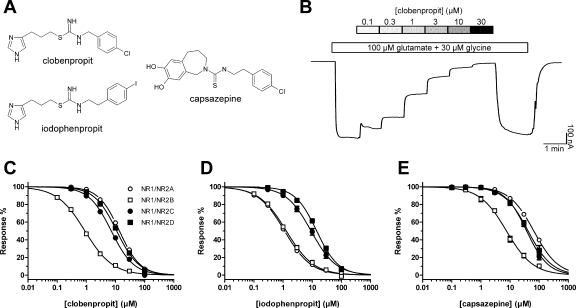
Several of the compounds identified in these validation screens that were not previously known to inhibit NMDA receptors represent particularly intriguing classes of compounds, including the prototypical vanilloid receptor TRPV1 antagonist capsazepine and the histamine H3 receptor antagonists clobenpropit and iodophenpropit (Fig. 2A). We subsequently generated concentration-response data for these ligands at recombinant NR1/NR2A, NR1/NR2B, NR1/NR2C, NR1/NR2D, GluR1, and GluR6 receptors expressed in X. laevis oocytes (Fig. 2). Table 5 summarizes the IC50 values of these compounds for inhibiting recombinant NMDA receptors.
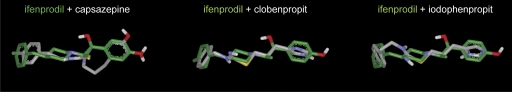
Fig. 2.
A, chemical structures of histamine H3 receptor antagonists clobenpropit and iodophenpropit, as well as the vanilloid receptor TRPV1 antagonist capsazepine. B, representative TEVC recordings showing inhibition by increasing concentrations of clobenpropit of responses to 100 μM glutamate plus 30 μM glycine on NR1/NR2B receptors expressed in X. laevis oocytes. The bars above the trace indicate the duration of ligand applications. C to E, concentration-response data for clobenpropit, iodophenpropit, and capsazepine at NR1/NR2A (open circles), NR1/NR2B (open squares), NR1/NR2C (closed circles), and NR1/NR2D (closed squares) determined by TEVC recordings. Data are mean ± S.E.M. from 5 to 15 oocytes. IC50 values are listed in Table 5.
TABLE 5.
Concentration-response data for novel NMDA receptors antagonists
IC50 ± S.E.M. and Hill slopes (nH) determined by TEVC recordings from X. laevis oocytes expressing the indicated receptors. n is the number of oocytes. At NR1/NR2A–D and mutant NR1/NR2B receptors, the compounds were coapplied with 100 μM glutamate and 30 μM glycine. At GluR1 and GluR6, the compounds were coapplied with 100 μM glutamate. Oocytes expressing GluR6 were incubated for 5 min in 10 μM concanavalin A. The oocytes were voltage-clamped at −40 mV. IC50 > 100 μM indicates that the compound showed some inhibition but less than 50% inhibition at 100 μM.
| Clobenpropit |
Iodophenpropit |
Capsazepine |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| IC50 | nH | n | IC50 | nH | n | IC50 | nH | n | |
| μM | μM | μM | |||||||
| Receptor | |||||||||
| NR1/NR2A | 14 ± 2 | 1.3 | 10 | 1.3 ± 0.2 | 1.0 | 13 | 66 ± 9 | 1.1 | 7 |
| NR1/NR2B | 1.0 ± 0.1 | 0.9 | 23 | 1.4 ± 0.2 | 0.9 | 15 | 8.1 ± 1.2 | 1.0 | 12 |
| NR1/NR2C | 7.3 ± 0.3 | 1.1 | 8 | 9.7 ± 1.7 | 1.0 | 5 | 44 ± 5 | 1.1 | 8 |
| NR1/NR2D | 11 ± 1 | 1.2 | 8 | 14 ± 1 | 1.2 | 7 | 42 ± 3 | 1.1 | 7 |
| GluR1 | >100 | 4 | >100 | 4 | >100 | 4 | |||
| GluR6 | 103 ± 1 | 0.7 | 12 | 53 ± 1 | 0.7 | 12 | N.E. | 4 | |
| Mutant | |||||||||
| NR1/NR2B-ΔATD | 7.4 ± 1.5 | 0.8 | 13 | 3.5 ± 1.1 | 0.6 | 7 | 74 ± 13 | 0.7 | 7 |
| NR1(N616R)/NR2B | 4.8 ± 0.6 | 0.7 | 6 | N.D. | N.D. | ||||
| NR1(N616R)/NR2B-ΔATD | 107 ± 29 | 0.5 | 6 | N.D. | N.D. | ||||
N.D., not determined; N.E., no significant effect at 100 μM.
Histamine H3 Receptor Antagonists Are NR2B-Selective NMDA Receptor Antagonists.
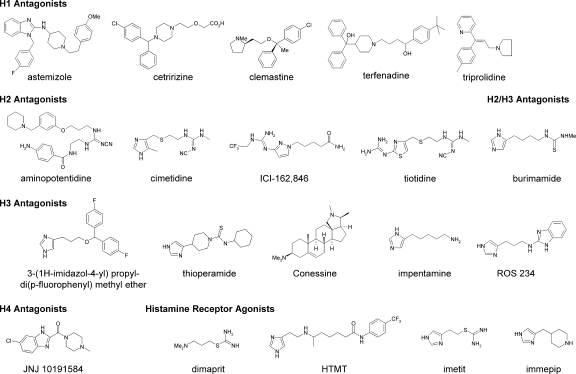
The histamine H3 receptor antagonist clobenpropit showed high potency (1.0 μM) at NMDA receptors that contained the NR2B subunit, even though the screen identified the compounds at cells expressing NR1/NR2D. The structurally related histamine H3 receptor antagonist iodophenpropit showed similar selectivity for NR1/NR2B. These two biaryl compounds show structural similarity to a series of thiosemicarbazide and amide derivatives recently described as NR2B antagonists (Mosley et al., 2009). To evaluate the extent to which the family of histamine receptor antagonists showed cross-reactivity with NMDA receptors, we assessed H1-, H2-, H3-, and H4-selective histamine receptor antagonists as well as some structurally related histamine receptor agonists against recombinant NR1/NR2A, NR1/NR2B, NR1/NR2C, and NR1/NR2D receptors expressed in X. laevis oocytes. Only aminopotentidine (Table 6) and the isothioureas, such as clobenpropit and iodophenpropit (Table 5), showed inhibition of recombinant NMDA receptors, with IC50 values less than 10 μM. The chemical structures of the histamine receptor ligands studied are shown in Figs. 2 and 3.
TABLE 6.
IC50 values of histamine receptor ligands on recombinant NMDA receptors
See legend to Table 5. IC50 values are in micromolar. IC50 > 100 μM indicates that the compound showed some inhibition, but less than 50% inhibition at 100 μM. Hill slopes were 0.7 to 1.5, and the number of oocytes were 3 to 12 for all compounds. Chemical structures are shown in Fig. 4.
| NR1/NR2A | NR1/NR2B | NR1/NR2C | NR1/NR2D | |
|---|---|---|---|---|
| H1 antagonists | ||||
| Astemizole | >100 | >100 | >100 | >100 |
| Cetririzine | >100 | >100 | >100 | 44 ± 3 |
| Clemastine | 64 ± 9 | >100 | >100 | >100 |
| Terfenadine | 60 ± 13 | 59 ± 6 | 61 ± 5 | >100 |
| Triprolidine | >100 | >100 | >100 | >100 |
| H2 antagonists | ||||
| Aminopotentidine | 5.1 ± 0.5 | 8.4 ± 1.6 | >100 | >100 |
| Cimetidine | N.E. | >100 | >100 | >100 |
| ICI-162,846 | N.E. | N.E. | >100 | >100 |
| Tiotidine | 43 ± 4 | 16 ± 1 | 51 ± 5 | 54 ± 9 |
| H2/H3 antagonist | ||||
| Burimamide | >100 | N.E. | N.E. | N.E. |
| H3 antagonists | ||||
| Conessine | 44 ± 9 | 54 ± 9 | >100 | >100 |
| 3-(1H-Imidazol-4-yl) propyl-di(p-fluorophenyl) methyl ether | >100 | >100 | >100 | >100 |
| Impentamine | N.E. | >100 | >100 | >100 |
| ROS 234 | 80 ± 6 | 37 ± 3 | >100 | >100 |
| Thioperamide | >100 | >100 | >100 | >100 |
| H4 antagonists | ||||
| JNJ-10191584 | N.E. | >100 | >100 | >100 |
| Histamine receptor agonists | ||||
| Dimaprit | >100 | >100 | >100 | >100 |
| Histamine trifluoromethyl toluidide | >100 | 37 ± 4 | >100 | >100 |
| Imetit | 52 ± 14 | >100 | >100 | >100 |
| Immepip | N.E. | >100 | >100 | >100 |
N.E., no significant effect at 100 μM.
Fig. 3.
Chemical structures of histamine receptor ligands characterized at recombinant NMDA receptors by TEVC recordings. Also see Table 6. HTMT, histamine trifluoromethyl toluidide.
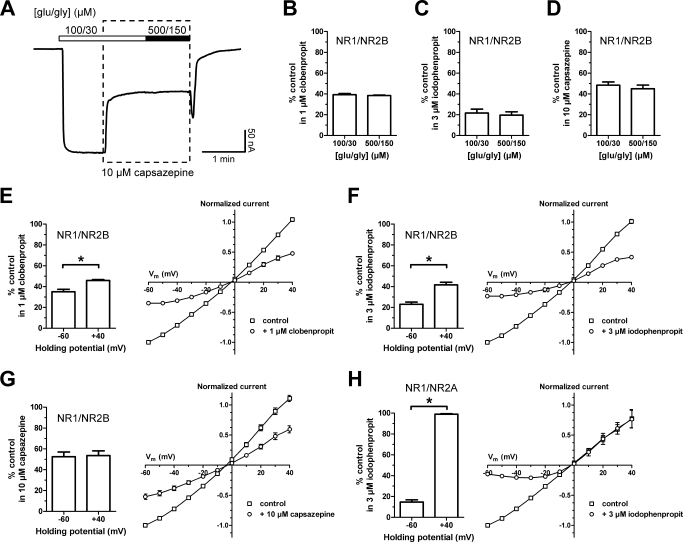
To evaluate the mechanism of action of clobenpropit and iodophenpropit as NMDA receptor antagonists, we first assessed whether inhibition could be surmounted by increasing concentration of the coagonists glutamate and glycine. Increasing the concentration of glutamate and glycine from 100 and 30 μM, respectively, to 500 and 150 μM did not reduce the extent of inhibition at NR1/NR2B receptors produced by a half-maximally effective concentration of clobenpropit or iodophenpropit, indicating block was insurmountable and thus noncompetitive (Fig. 4, A–C). Inhibition of NR1/NR2B by capsazepine was also noncompetitive (Fig. 4D). Evaluation of the current-voltage relationship suggested the majority of the inhibition of NR1/NR2B receptors by both clobenpropit and iodophenpropit was mediated by a voltage-independent mechanism (Fig. 4, E and F). However, there was a significantly higher degree of inhibition at −60 mV than at −40 mV for both compounds, suggesting that these compounds exhibit voltage-dependent block of NR1/NR2B receptors with low potency. This voltage-dependent component of the inhibition was undetectable for capsazepine on NR1/NR2B (Fig. 4G). Capsazepine is uncharged at physiological pH, whereas both clobenpropit and iodophenpropit show ionizable imidazole rings with similar predicted pKa values of 7.3. In addition, the isothiourea are predicted to show pKa values of 7.6 and 8.3 for clobenpropit and iodophenpropit, respectively (see Materials and Methods). Thus, more than 50% of the molecules will be positively charged at pH 7.4, consistent with the observation of voltage-dependent pore block. To determine whether voltage-dependent block of NR1/NR2A receptors, which lack the ifenprodil binding site, accounts for all inhibition observed for iodophenpropit, we evaluated the current-voltage relationship of iodophenpropit inhibition. In contrast to NR1/NR2B receptors, iodophenpropit showed inhibition of NR1/NR2A that was entirely voltage-dependent (Fig. 4H). These results suggest that clobenpropit and iodophenpropit act with high potency at the ifenprodil site on NR2B-containing receptors to inhibit function. They also inhibit both NR1/NR2A and NR1/NR2B receptors at lower potency by blocking the pore, and the potency of this block is enhanced at hyperpolarized potentials. Consistent with this idea, the potency of iodophenpropit at a membrane potential of −80 mV (IC50 = 0.4 ± 0.1 μM; n = 6) was 3.5-fold higher compared with the potency at a membrane potential of −40 mV (IC50 = 1.4 ± 0.2 μM; n = 15). For NR1/NR2A (and we predict NR1/NR2C and NR1/NR2D) that lack sensitivity to ifenprodil-like compounds, this voltage-dependent block fully accounts for the observed inhibition.
Fig. 4.
A, representative TEVC recordings of NR1/NR2B receptors expressed in X. laevis oocytes showing that increasing the concentration of glutamate and glycine from 100 and 30 μM (100/30), respectively, to 500 and 150 μM (500/150) did not reduce the extent of inhibition by 10 μM capsazepine. The bars above the trace indicate the duration of agonist applications. The dashed box indicates the duration of capsazepine application. B to D, bar graphs showing inhibition of responses to saturating agonist concentrations (100 μM glutamate and 30 μM glycine) and supersaturating agonist concentrations (500 μM glutamate and 150 μM glycine) of NR1/NR2B by clobenpropit, iodophenpropit, and capsazepine at the indicated concentrations. Responses are shown as percentage of the response to 100 μM glutamate plus 30 μM glycine alone (control) and are mean ± S.E.M. from six oocytes. E to G, bar graphs and current-voltage relationship curves showing inhibition of NR1/NR2B responses at membrane potentials of −60 and +40 mV by clobenpropit, iodophenpropit, and capsazepine at the indicated concentrations. Responses are shown as percentage of the response to 100 μM glutamate plus 30 μM glycine alone (control) and are mean ± S.E.M. from four oocytes. The current-voltage relationship curves are normalized to the response at −60 mV. H, bar graph showing inhibition of NR1/NR2A responses at membrane potentials of −60 and +40 mV by iodophenpropit (3 μM). Responses are shown as percentage of the response to 100 μM glutamate plus 30 μM glycine alone (control) and are mean ± S.E.M. from four oocytes. The current-voltage relationship curves are normalized to the response at −60 mV. Asterisk (*) indicates significantly different (t test, P < 0.05).
Histamine H3 Receptor Antagonists and Ifenprodil Share Structural Determinants for Selective Inhibition of NR2B-Containing NMDA Receptors.
To evaluate the possibility that capsazepine, clobenpropit, and iodophenpropit can inhibit NR2B-containing NMDA receptors by acting at the ifenprodil binding site, we compared the molecular shapes and electrostatic potentials of these compounds by performing a conformational search using the OMEGA conformation generator (see Materials and Methods). These conformer pools were then analyzed with the program ROCS that provides a measure of molecular shape complementarity and electrostatic potential distribution overlap through maximization of shape overlap between two structures. Shape complementarity is calculated by means of a simple Tanimoto comparison (Ts) resulting in a score between 0.0 (no overlap) and 1.0 (identical shape). A score of Ts >0.7 suggests significant shape complementarity (Nicholls et al., 2004). The structural alignments of ifenprodil with capsazepine, clobenpropit, and iodophenpropit are depicted in Fig. 5. Ts scores compared with ifenprodil were 0.72, 0.77, and 0.73 for capsazepine, clobenpropit, and iodophenpropit, respectively. The ROCS scores, which include components of both shape and functional similarity, were 1.09, 1.02, and 0.99 for capsazepine, clobenpropit, and iodophenpropit, respectively. The good structural similarities and complementarities of capsazepine, clobenpropit, and iodophenpropit with ifenprodil support the idea that the compounds and ifenprodil access a common binding site on NR2B. Similarly, competition binding experiments showed that ifenprodil is capable of inhibiting [3H]N-α-methylhistamine binding to recombinant histamine H3 receptors, with an IC50 value of 17 ± 3 μM (n = 3), corresponding to an estimated Ki value of 5.7 ± 0.9 μM. This result further suggests some similarity between the binding sites for clobenpropit, iodophenpropit, and ifenprodil on histamine H3 receptors and NR2B.
Fig. 5.
Structural alignment of ifenprodil (green) with the vanilloid receptor TRPV1 antagonist capsazepine, as well as the histamine H3 receptor antagonists clobenpropit and iodophenpropit. The conformationally best-minimized structure of ifenprodil was chosen as reference compound, and capsazepine, clobenpropit, and iodophenpropit were subjected to conformational search and superimposed with ifenprodil (see Materials and Methods). Shape complementarity is calculated by means of a simple Ts comparison, resulting in a score between 0.0 (no overlap) and 1.0 (identical shape). Ts scores compared with ifenprodil were 0.72, 0.77, and 0.73 for capsazepine, clobenpropit, and iodophenpropit, respectively. The ROCS scores, which include components of both shape and functional similarity, were 1.09, 1.02, and 0.99 for capsazepine, clobenpropit, and iodophenpropit, respectively.
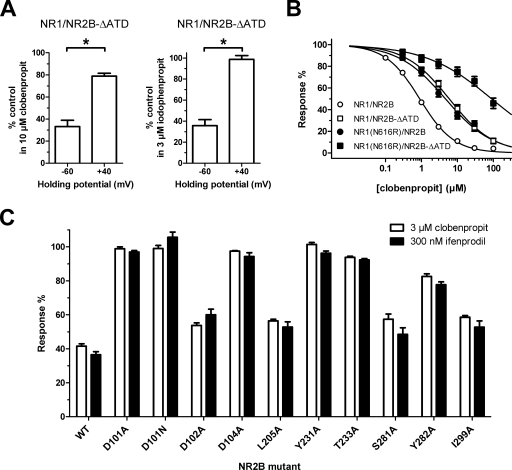
We further explored the structural basis of the NR1/NR2B-selective inhibition by the histamine H3 receptor antagonists. However, the dual action (voltage-dependent channel block and NR2B-selective noncompetitive antagonism) of these compounds at NMDA receptors complicates a straight-forward strategy using site-directed mutagenesis to identify the structural determinants for the NR2B-selective component of the inhibition. Initially, we compared the IC50 values for capsazepine, clobenpropit, and iodophenpropit at wild-type NR1/NR2B with those at the NR1/NR2B-ΔATD deletion mutant, which does not contain the amino-terminal domain of NR2B (Table 5). Consistent with ifenprodil binding to the amino-terminal domain of NR2B (Perin-Dureau et al., 2002), the IC50 values for all three compounds were increased (i.e., lower potency) at NR1/NR2B-ΔATD. However, the fold changes in IC50 values were only moderate for capsazepine (9.1-fold), clobenpropit (7.4-fold), and iodophenpropit (2.5-fold) and not as large as might be expected if they only inhibited NR1/NR2B through a mechanism involving the amino-terminal domain. Comparison of the inhibition at holding potentials of −60 and −40 mV for clobenpropit and iodophenpropit at NR1/NR2B-ΔATD revealed that the inhibition now display a pronounced voltage dependence (Fig. 6A; also see Fig. 4, E and F, for comparison with wild type NR1/NR2B), consistent with the hypothesis that the voltage-dependent channel block mechanism of action for the compounds is responsible for their activity at NR1/NR2B-ΔATD.
Fig. 6.
A, bar graphs showing inhibition by clobenpropit and iodophenpropit of NR1/NR2B-ΔATD, where the amino-terminal domain of NR2B has been deleted, at membrane potentials of −60 and +40 mV. Both compounds display pronounced voltage-dependent inhibition indicating that they are channel blockers of at NR1/NR2B-ΔATD. Responses are shown as percentage of the response to 100 μM glutamate plus 30 μM glycine alone (control) and are mean ± S.E.M. from six oocytes. Asterisk (*) indicates significantly different (t test, P < 0.05). B, concentration-response curves for clobenpropit on wild-type NR1/NR2B (open circles), NR1/NR2B-ΔATD (open squares), NR1(N616R)/NR2B (closed circles), and NR1(N616R)/NR2B-ΔATD (closed squares). The N616R mutation in NR1 relieves voltage-dependent inhibition by channel blockers and clobenpropit therefore inhibits NR1(N616R)/NR2B-ΔATD with low potency. IC50 values are listed in Table 5. C, bar graph showing mean inhibition of NR1/NR2B (mutant) by ifenprodil (300 nM) and of NR1(N616R)/NR2B (mutant) by clobenpropit (3 μM). These mutations have strikingly similar effects on ifenprodil and clobenpropit inhibition. Responses are shown as percentage of the response to 100 μM glutamate plus 30 μM glycine alone (control) and are mean ± S.E.M. from 4 to 10 oocytes.
To minimize the contribution of voltage-dependent channel block to the inhibition of NR1/NR2B by the histamine H3 receptor antagonists, we took advantage of the N616R mutation in NR1, which has previously been shown to drastically reduce the potency of channel blockers (Kashiwagi et al., 2002). The IC50 values for clobenpropit inhibition of NR1(N616R)/NR2B and NR1(N616R)/NR2B-ΔATD were increased 4.8- and 107-fold, respectively, which is consistent with the removal of channel block inhibition for NR1(N616R)/NR2B and removal of both channel block and NR2B-selective noncompetitive antagonism for NR1(N616R)/NR2B-ΔATD (Fig. 6B; Table 5). We further evaluated the clobenpropit sensitivity of NR1(N616R) coexpressed with NR2B subunits that harbor amino-terminal domain mutations that have either been shown to reduce ifenprodil sensitivity (Mony et al., 2009) or are adjacent to residues that influence ifenprodil sensitivity (Fig. 6C). All the evaluated mutations show a strikingly similar sensitivity to ifenprodil and clobenpropit inhibition, suggesting that the compounds share structural determinants for their activity. Together, these data are consistent with the idea that the histamine H3 receptor antagonists clobenpropit and iodophenpropit act by binding to the ifenprodil site on NR1/NR2B receptors.
Discussion
The most important finding of this study is that some ligands that bind the histamine H3 receptor can also bind the amino-terminal domain of the NR2B NMDA receptor subunit. The evidence for this is that highly potent histamine H3 receptor antagonists are NR2B-selective noncompetitive antagonists that are sensitive to the same mutations in the NR2B amino-terminal domain that alter the IC50 values of NR2B-selective ligands. This finding raises the possibility that compounds can be designed to potentially inhibit both receptors systems, which may have useful therapeutic effects on brain function.
Novel Assay for Noncompetitive NMDA Receptor Inhibitors.
Stable expression of NMDA receptor subunits has long been known to be cytotoxic, and this has complicated attempts to develop reliable and robust multiwell assays for NMDA receptor modulators. Our initial attempts to develop a robust NR1/NR2D assay were hampered by cytotoxic accumulation of intracellular divalent cations due to activation of NR1/NR2D by nanomolar levels of glutamate and glycine present in the external solution during dye loading. In addition, this accumulation of intracellular divalent ions increased the baseline signal of the Ca2+-sensitive dye, thereby decreasing the apparent response amplitude during addition of agonist (Fig. 1, C and D). We were able to circumvent this problem by adding the competitive glycine site antagonist 7-CKA during dye loading of the cells immediately preceding the fluorescence measurements. This reduced accumulation of intracellular Ca2+ and thus improved cell conditions and the signal-to-noise ratio, leading to acceptable measured Z′ values greater than 0.5. We further designed this assay to eliminate hits on the agonist binding sites, which increases the probability of selectively identifying noncompetitive antagonists. The competitive glycine site antagonist 7-CKA can be readily displaced by a high concentration of glycine (1 mM) to initiate the signal in the assay. This high concentration of glycine together with high concentration of glutamate (200-fold EC50) reduces the likelihood that test ligands at 10 μM will displace either coagonist. Implementation of the assay on two test libraries that contained known NMDA receptor antagonists revealed a high success rate in identifying noncompetitive inhibitors and successfully avoided hits from competitive antagonists, demonstrating that this strategy is effective.
Novel Noncompetitive NMDA Receptor Antagonists.
Application of this assay to two small test libraries identified two histamine H3 receptor antagonists with structural similarity to known NR2B-selective antagonists with submicromolar IC50 values. NR1/NR2B inhibition involved an intriguing mixed mechanism; components of inhibition involved both interactions with the ifenprodil binding site in the NR2B subunit and voltage-dependent channel block.
Histamine H3 receptors are primarily expressed in nervous tissue and clobenpropit binds to and competitively inhibits human histamine H3 receptors, with a pKi value of 8.5 (i.e., Ki = 3 nM); iodophenpropit acts as an antagonist at histamine H3 receptors, with a pKi value of 8.1 (Ki = 8 nM) (Lim et al., 2005). Clobenpropit and iodophenpropit are well known pharmacological tools that are often used as histamine H3 receptor antagonists, although they are also known to act at other targets. In addition to their ability to interact with high potency and activate the closely related histamine H4 receptors, these ligands also have relatively high affinity for the serotonin 5-hydroxytryptamine3, α2-adrenergic, and σ receptors (Leurs et al., 1995; Liu et al., 2001; Lim et al., 2005). Furthermore, imidazole-containing compounds, which include clobenpropit and iodophenpropit, are often associated with an inherent risk of cytochrome P450 inhibition (Lin and Lu, 1998). These and other off-target effects led to reduced interest in the development of this structural class of histamine H3 receptor antagonists for therapeutical purposes.
In the present study, we evaluated the extent to which the histamine receptor ligands other than clobenpropit and iodophenpropit showed cross-reactivity with NMDA receptors (Table 6; Fig. 3). We evaluated histamine H1, H2, H3, and H4 receptor antagonists as well as various agonists. Among these ligands, only clobenpropit and iodophenpropit showed the most potent inhibition of recombinant NMDA receptors (IC50 < 2 μM). These results suggest that histamine receptor ligands in general are not antagonists at NR2B-containing NMDA receptors. Whereas NR2B-selective effect is not a general feature of histamine receptor ligands, it is intriguing that histamine itself has been proposed to potentiate NMDA receptors in an NR2B-selective manner (Bekkers, 1993; Vorobjev et al., 1993; Williams, 1994; Burban et al., 2010). The site of action of histamine on NMDA receptors is not fully resolved.
Our finding that clobenpropit and iodophenpropit can inhibit NR2B-containing NMDA receptors at submicromolar concentrations raises a number of interesting ideas. For example, an NR2B-selective antagonist (CP-101,606) has recently been shown to significantly improve symptoms in patients with treatment-resistant depression (Preskorn et al., 2008). The onset of antidepressive effects was rapid, and the effect was sustained in patients for days after a single drug infusion. Compounds with histamine H3 receptor antagonist activity combined with inhibition of serotonin re-uptake have been considered as a useful antidepressant strategy (Barbier et al., 2007). Combination of NR2B antagonist activity with histamine H3 receptor block and serotonin reuptake inhibition may improve the antidepressant properties of these classes of compounds.
Inhibition of histamine H3 receptors increases release of glutamate in corticostriatal pathway and hippocampus (for review, see Haas et al., 2008), which would be detrimental under neuropathological conditions that involve excitotoxicity in these regions. However, inhibition of histamine H3 receptors also increases release of the inhibitory transmitter GABA that could be considered beneficial under the same conditions (for review, see Haas et al., 2008). Consequently, it could be hypothesized that synergistic effects may be obtained by combining NMDA receptor inhibition with histamine H3 receptor antagonism. Histamine H3 receptor antagonists are known to be anticonvulsant in several models of seizures (Fischer and van der Goot, 1998). Similarly, NR2B-selective NMDA receptor antagonists also show anticonvulsant properties (Wlaź et al., 1999; Mares and Mikulecka, 2009). These results support the idea that a compound that can block both of these two receptor classes may possess synergistic anticonvulsant activity. Furthermore, both clobenpropit (Dai et al., 2007) and NR2B antagonists show neuroprotective properties (Tamura et al., 1993), further creating the opportunity for synergistic actions of dual antagonists of both histamine H3 receptors and NR2B-containing NMDA receptors.
Pharmacological Selectivity of Histamine H3 Receptor Antagonists.
Our findings reinforce the idea that care is warranted in the interpretation of data with clobenpropit and iodophenpropit (Leurs et al., 1995; Liu et al., 2001; Lim et al., 2005). Although clobenpropit has a Ki value at histamine H3 receptors that suggests at least 100-fold higher potency than the measured IC50 value at NR1/NR2B, the extent of histamine H3 receptor inhibition by the competitive antagonist clobenpropit will depend on the extracellular concentration of histamine. This means that the IC50 value for inhibition of neuronal responses to histamine may lie closer to the IC50 value for actions at the NR1/NR2B receptor. Thus, studies that use clobenpropit or iodophenpropit at unknown brain concentrations may inadvertently be causing some degree of inhibition of NR2B-containing NMDA receptors. It is possible that analogs of histamine H3 receptor antagonists currently under development could have higher potency at NR1/NR2B receptors, which could have important implications. Finally, the vanilloid receptor TRPV1 antagonist capsazepine is often used at a concentration of 10 μM, which is half-maximally effective at inhibiting NR2B-containing NMDA receptors. This cross-reactivity may contribute to results in reports that capsazepine shows interactions with the glutamatergic system or is neuroprotective (Ray et al., 2003; Zanchet and Cury, 2003; Peng et al., 2008).
In summary, we have identified intriguing cross-reactivity between certain histamine H3 receptor antagonists and NR2B-containing NMDA receptors. The activity of histamine H3 receptor ligands at NMDA receptors presents several potentially interesting opportunities in terms of drug development.
Acknowledgments
We thank Drs. Ray Dingledine, Jan Egebjerg, and Timothy A. Esbenshade for helpful discussion and commentary on the project. We are grateful to OpenEye Scientific Software for the generous provision of a no-cost license for the ROCS software.
This work was supported in part by the Michael J. Fox Foundation (to S.F.T.); the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS036654] (to S.F.T.); the National Institutes of Health [Grant 5U54-HG003918] (to I.L., Y.D., S.K.); the Carlsberg Foundation (to H.B.-O.); the GluTarget Center of Excellence (to T.K. and H.B.-O.); the Alfred Benzon Foundation (to K.B.H.); the Villum Kann Rasmussen Foundation (to K.B.H.); and the Lundbeck Foundation (to K.B.H.).
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.110.166256.
- NMDA
- N-methyl-d-aspartate
- TRPV1
- vanilloid receptor transient receptor potential cation channel, subfamily V, member 1
- BHK
- baby hamster kidney
- AP5
- dl-2-amino-5-phosphonopentanoate
- 7-CKA
- 7-chloro-kyneurenate
- TBS
- Tris-buffered saline
- BSA
- bovine serum albumin
- CNQX
- 7-nitro-2,3-dioxo-1,4-dihydroquinoxaline-6-carbonitrile
- CPP
- 3-((R)-2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid
- HA-966
- 3-amino-1-hydroxypyrrolidin-2-one
- DMSO
- dimethyl sulfoxide
- LOPAC
- library of pharmacologically active compounds
- NRA
- N-methyl-d-aspartate receptor antagonist-like
- ROCS
- rapid overlay of chemical structures
- TEVC
- two-electrode voltage-clamp
- MK-801
- (−)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine
- CNS-1102
- aptiganel
- Ts
- Tanimoto comparison
- CP-101,606
- traxoprodil mesylate
- ATD
- amino-terminal domain
- LY-235,959
- decahydro-6-(phosphonomethyl)-3-isoquinolinecarboxylic acid
- L-701,324
- 7-chloro-4-hydroxy-3-(3-phenoxy)phenyl-2(1H)-quinolone
- MDL 105,519
- 3-(2-phenyl-2-carboxyethenyl)-4,6-dichloro-1H-indole-2-carboxylic acid
- MG 624
- triethyl-(β-4-stilbenoxyethyl)ammonium
- TMB-8
- 8-(N,N-diethylamino)octyl-3,4,5-trimethoxybenzoate
- W-7
- N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide
- BW723C86
- 1-(5-(2-thenyloxy)-1H-indol-3-yl)propan-2-amine.
References
- Barbier et al., 2007.Barbier AJ, Aluisio L, Lord B, Qu Y, Wilson SJ, Boggs JD, Bonaventure P, Miller K, Fraser I, Dvorak L, et al. (2007) Pharmacological characterization of JNJ-28583867, a histamine H(3) receptor antagonist and serotonin reuptake inhibitor. Eur J Pharmacol 576:43–54 [DOI] [PubMed] [Google Scholar]
- Bekkers, 1993.Bekkers JM. (1993) Enhancement by histamine of NMDA-mediated synaptic transmission in the hippocampus. Science 261:104–106 [DOI] [PubMed] [Google Scholar]
- Bräuner-Osborne et al., 2000.Bräuner-Osborne H, Egebjerg J, Nielsen EO, Madsen U, Krogsgaard-Larsen P. (2000) Ligands for glutamate receptors: design and therapeutic prospects. J Med Chem 43:2609–2645 [DOI] [PubMed] [Google Scholar]
- Burban et al., 2010.Burban A, Faucard R, Armand V, Bayard C, Vorobjev V, Arrang JM. (2010) Histamine potentiates N-methyl-D-aspartate receptors by interacting with an allosteric site distinct from the polyamine binding site. J Pharmacol Exp Ther 332:912–921 [DOI] [PubMed] [Google Scholar]
- Cik et al., 1993.Cik M, Chazot PL, Stephenson FA. (1993) Optimal expression of cloned NMDAR1/NMDAR2A heteromeric glutamate receptors: a biochemical characterization. Biochem J 296:877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai et al., 2007.Dai H, Fu Q, Shen Y, Hu W, Zhang Z, Timmerman H, Leurs R, Chen Z. (2007) The histamine H3 receptor antagonist clobenpropit enhances GABA release to protect against NMDA-induced excitotoxicity through the cAMP/protein kinase A pathway in cultured cortical neurons. Eur J Pharmacol 563:117–123 [DOI] [PubMed] [Google Scholar]
- Dravid et al., 2007.Dravid SM, Erreger K, Yuan H, Nicholson K, Le P, Lyuboslavsky P, Almonte A, Murray E, Mosely C, Barber J, et al. (2007) Subunit-specific mechanisms and proton sensitivity of NMDA receptor channel block. J Physiol 581:107–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer and van der Goot, 1998.Fischer W, van der Goot H. (1998) Effect of clobenpropit, a centrally acting histamine H3-receptor antagonist, on electroshock- and pentylenetetrazol-induced seizures in mice. J Neural Transm 105:587–599 [DOI] [PubMed] [Google Scholar]
- Haas et al., 2008.Haas HL, Sergeeva OA, Selbach O. (2008) Histamine in the nervous system. Physiol Rev 88:1183–1241 [DOI] [PubMed] [Google Scholar]
- Hansen et al., 2008.Hansen KB, Bräuner-Osborne H, Egebjerg J. (2008) Pharmacological characterization of ligands at recombinant NMDA receptor subtypes by electrophysiological recordings and intracellular calcium measurements. Comb Chem High Throughput Screen 11:304–315 [DOI] [PubMed] [Google Scholar]
- Hawkins et al., 2007.Hawkins PC, Skillman AG, Nicholls A. (2007) Comparison of shape-matching and docking as virtual screening tools. J Med Chem 50:74–82 [DOI] [PubMed] [Google Scholar]
- Jorgensen and Tirado-Rives, 2005.Jorgensen WL, Tirado-Rives J. (2005) Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc Natl Acad Sci USA 102:6665–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi et al., 2002.Kashiwagi K, Masuko T, Nguyen CD, Kuno T, Tanaka I, Igarashi K, Williams K. (2002) Channel blockers acting at N-methyl-d-aspartate receptors: differential effects of mutations in the vestibule and ion channel pore. Mol Pharmacol 61:533–545 [DOI] [PubMed] [Google Scholar]
- Leurs et al., 1995.Leurs R, Tulp MT, Menge WM, Adolfs MJ, Zuiderveld OP, Timmerman H. (1995) Evaluation of the receptor selectivity of the H3 receptor antagonists, iodophenpropit and thioperamide: an interaction with the 5-HT3 receptor revealed. Br J Pharmacol 116:2315–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim et al., 2005.Lim HD, van Rijn RM, Ling P, Bakker RA, Thurmond RL, Leurs R. (2005) Evaluation of histamine H1-, H2-, and H3-receptor ligands at the human histamine H4 receptor: identification of 4-methylhistamine as the first potent and selective H4 receptor agonist. J Pharmacol Exp Ther 314:1310–1321 [DOI] [PubMed] [Google Scholar]
- Lin and Lu, 1998.Lin JH, Lu AY. (1998) Inhibition and induction of cytochrome P450 and the clinical implications. Clin Pharmacokinet 35:361–390 [DOI] [PubMed] [Google Scholar]
- Lisman, 2003.Lisman J. (2003) Long-term potentiation: outstanding questions and attempted synthesis. Philos Trans R Soc Lond B Biol Sci 358:829–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al., 2001.Liu C, Ma X, Jiang X, Wilson SJ, Hofstra CL, Blevitt J, Pyati J, Li X, Chai W, Carruthers N, et al. (2001) Cloning and pharmacological characterization of a fourth histamine receptor (H(4)) expressed in bone marrow. Mol Pharmacol 59:420–426 [DOI] [PubMed] [Google Scholar]
- Mares and Mikulecka, 2009.Mares P, Mikulecka A. (2009) Different effects of two N-methyl-d-aspartate receptor antagonists on seizures, spontaneous behavior, and motor performance in immature rats. Epilepsy Behav 14:32–39 [DOI] [PubMed] [Google Scholar]
- Mony et al., 2009.Mony L, Krzaczkowski L, Leonetti M, Le Goff A, Alarcon K, Neyton J, Bertrand HO, Acher F, Paoletti P. (2009) Structural basis of NR2B-selective antagonist recognition by N-methyl-d-aspartate receptors. Mol Pharmacol 75:60–74 [DOI] [PubMed] [Google Scholar]
- Mosley et al., 2009.Mosley CA, Myers SJ, Murray EE, Santangelo R, Tahirovic YA, Kurtkaya N, Mullasseril P, Yuan H, Lyuboslavsky P, Le P, et al. (2009) Synthesis, structural activity-relationships, and biological evaluation of novel amide-based allosteric binding site antagonists in NR1A/NR2B N-methyl-d-aspartate receptors. Bioorg Med Chem 17:6463–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacher and McEwen, 2006.Nacher J, McEwen BS. (2006) The role of N-methyl-d-aspartate receptors in neurogenesis. Hippocampus 16:267–270 [DOI] [PubMed] [Google Scholar]
- Nicholls et al., 2004.Nicholls A, MacCuish NE, MacCuish JD. (2004) Variable selection and model validation of 2D and 3D molecular descriptors. J Comput Aided Mol Des 18:451–474 [DOI] [PubMed] [Google Scholar]
- Peng et al., 2008.Peng HY, Huang PC, Liao JM, Tung KC, Lee SD, Cheng CL, Shyu JC, Lai CY, Chen GD, Lin TB. (2008) Estrous cycle variation of TRPV1-mediated cross-organ sensitization between uterus and NMDA-dependent pelvic-urethra reflex activity. Am J Physiol Endocrinol Metab 295:E559–E568 [DOI] [PubMed] [Google Scholar]
- Perin-Dureau et al., 2002.Perin-Dureau F, Rachline J, Neyton J, Paoletti P. (2002) Mapping the binding site of the neuroprotectant ifenprodil on NMDA receptors. J Neurosci 22:5955–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preskorn et al., 2008.Preskorn SH, Baker B, Kolluri S, Menniti FS, Krams M, Landen JW. (2008) An innovative design to establish proof of concept of the antidepressant effects of the NR2B subunit selective N-methyl-D-aspartate antagonist, CP-101,606, in patients with treatment-refractory major depressive disorder. J Clin Psychopharmacol 28:631–637 [DOI] [PubMed] [Google Scholar]
- Ray et al., 2003.Ray AM, Benham CD, Roberts JC, Gill CH, Lanneau C, Gitterman DP, Harries M, Davis JB, Davies CH. (2003) Capsazepine protects against neuronal injury caused by oxygen glucose deprivation by inhibiting I(h). J Neurosci 23:10146–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanovsky et al., 2009.Romanovsky AA, Almeida MC, Garami A, Steiner AA, Norman MH, Morrison SF, Nakamura K, Burmeister JJ, Nucci TB. (2009) The transient receptor potential vanilloid-1 channel in thermoregulation: a thermosensor it is not. Pharmacol Rev 61:228–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura et al., 1993.Tamura Y, Sato Y, Yokota T, Akaike A, Sasa M, Takaori S. (1993) Ifenprodil prevents glutamate cytotoxicity via polyamine modulatory sites of N-methyl-d-aspartate receptors in cultured cortical neurons. J Pharmacol Exp Ther 265:1017–1025 [PubMed] [Google Scholar]
- Traynelis et al., 1998.Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. (1998) Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci 18:6163–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis et al., 2010.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 61:doi:10.1124/pr.109.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicini et al., 1998.Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. (1998) Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J Neurophysiol 79:555–566 [DOI] [PubMed] [Google Scholar]
- Vorobjev et al., 1993.Vorobjev VS, Sharonova IN, Walsh IB, Haas HL. (1993) Histamine potentiates N-methyl-d-aspartate responses in acutely isolated hippocampal neurons. Neuron 11:837–844 [DOI] [PubMed] [Google Scholar]
- Waxman and Lynch, 2005.Waxman EA, Lynch DR. (2005) N-methyl-D-aspartate receptor subtypes: multiple roles in excitotoxicity and neurological disease. Neuroscientist 11:37–49 [DOI] [PubMed] [Google Scholar]
- Williams, 1994.Williams K. (1994) Subunit-specific potentiation of recombinant N-methyl-d-aspartate receptors by histamine. Mol Pharmacol 46:531–541 [PubMed] [Google Scholar]
- Witte et al., 2006.Witte DG, Yao BB, Miller TR, Carr TL, Cassar S, Sharma R, Faghih R, Surber BW, Esbenshade TA, Hancock AA, et al. (2006) Detection of multiple H3 receptor affinity states utilizing [3H]A-349821, a novel, selective, non-imidazole histamine H3 receptor inverse agonist radioligand. Br J Pharmacol 148:657–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlaź et al., 1999.Wlaź P, Ebert U, Löscher W. (1999) Anticonvulsant effects of eliprodil alone or combined with the glycineB receptor antagonist L-701,324 or the competitive NMDA antagonist CGP 40116 in the amygdala kindling model in rats. Neuropharmacology 38:243–251 [DOI] [PubMed] [Google Scholar]
- Yuan et al., 2009.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. (2009) Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci 29:12045–12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchet and Cury, 2003.Zanchet EM, Cury Y. (2003) Peripheral tachykinin and excitatory amino acid receptors mediate hyperalgesia induced by Phoneutria nigriventer venom. Eur J Pharmacol 467:111–118 [DOI] [PubMed] [Google Scholar]
- Zhang et al., 1999.Zhang JH, Chung TD, Oldenburg KR. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73 [DOI] [PubMed] [Google Scholar]