Abstract
The human breast cancer resistance protein (BCRP/ABCG2) mediates efflux of drugs and xenobiotics out of cells. In this study, we investigated the role of five basic residues within or near transmembrane (TM) 2 of BCRP in transport activity. Lys452, Lys453, His457, Arg465, and Lys473 were replaced with Ala or Asp. K452A, K453D, H457A, R465A, and K473A were stably expressed in human embryonic kidney (HEK) cells, and their plasma membrane expression and transport activities were examined. All of the mutants were expressed predominantly on the plasma membrane of HEK cells. After normalization to BCRP levels, the activities of K452A and H457A in effluxing mitoxantrone, boron-dipyrromethene-prazosin, and Hoechst33342 were increased approximately 2- to 6-fold compared with those of wild-type BCRP, whereas the activities of K453D and R465A were decreased by 40 to 60%. Likewise, K452A and H457A conferred increased resistance to mitoxantrone and 7-ethyl-10-hydroxy-camptothecin (SN-38), and K453D and R465A exhibited lower resistance. The transport activities and drug-resistance profiles of K473A were not changed. These mutations also differentially affected BCRP ATPase activities with a 2- to 4-fold increase in Vmax/Km for K452A and H457A and a 40 to 70% decrease for K453D and R465A. These mutations may induce conformational changes as manifested by the altered binding of the 5D3 antibody to BCRP in the presence of prazosin and altered trypsin digestion. Molecular modeling and docking calculations indicated that His457 and Arg465 might be directly involved in substrate binding. In conclusion, we have identified several basic residues within or near TM2 that may be important for interaction of substrates with BCRP.
A major obstacle of successful cancer chemotherapy is the multidrug resistance that is frequently associated with overexpression of multidrug transporters by pumping many drugs out of cancer cells. One such efflux transporter is the breast cancer resistance protein (BCRP) (Doyle et al., 1998; Miyake et al., 1999). Structurally unrelated chemotherapeutic agents that are BCRP substrates include, among others, mitoxantrone (MX), irinotecan, and the active metabolite of irinotecan, 7-ethyl-10-hydroxy-camptothecin (SN-38) (Robey et al., 2009). BCRP also transports nonchemotherapy substances such as prazosin, Hoechst33342, glyburide, and rosuvastatin (Mao and Unadkat, 2005; Robey et al., 2009).
BCRP is the second member of the G subfamily of the large ATP-binding cassette (ABC) transporter superfamily and hence is designated as ABCG2. BCRP is highly expressed in the apical membrane of placental syncytiotrophoblasts and the epithelium of the small intestine and colon, in the liver canalicular membrane, and in the apical membrane of the brain microvessel endothelium (Maliepaard et al., 2001). BCRP also plays an important role in drug disposition and xenobiotic exposure because of its broad substrate specificity and the pattern of tissue localization (Mao and Unadkat, 2005; Robey et al., 2009).
BCRP is a half-ABC transporter containing one nucleotide binding domain followed by one membrane-spanning domain (MSD). Thus, BCRP likely functions as a homodimer or homooligomer (Xu et al., 2004; McDevitt et al., 2006). The MSD of BCRP consists of six transmembrane (TM) α-helices (Fig. 1A), which has been confirmed in a recent study that experimentally determined the topology structure of BCRP (Wang et al., 2008). Because BCRP is a medically important ABC efflux transporter, understanding its structure–function relationship and transport mechanism will aid in the rational design of novel modulators that could be used to overcome multidrug resistance in cancers and help predict BCRP-mediated drug disposition and xenobiotic exposure. Structure–function studies have only recently begun to identify domains and individual amino acid residues that are important for substrate specificity and transport activity of BCRP. For example, amino acid substitutions of Arg482 have been shown to significantly alter substrate specificity and transport activity. Rhodamine123 [Rho123; 2′-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-5′-bi-1h-benzimidazole] and anthracyclines such as doxorubicin (Dox) and daunorubicin are not substrates of wild-type BCRP, but can be efficiently transported by the mutants with Gly or Thr substitution of Arg482 (Honjo et al., 2001; Miwa et al., 2003; Ozvegy-Laczka et al., 2005). In contrast, methotrexate is a substrate only for wild-type BCRP, but not for the mutants of Arg482 (Mitomo et al., 2003). Mutation analysis has also revealed that Gly553 might be involved in the dimerization of BCRP (Polgar et al., 2006), and the domain comprising residues 528 to 655 may be involved in oligomerization and thus important for BCRP function (Xu et al., 2007).
Fig. 1.
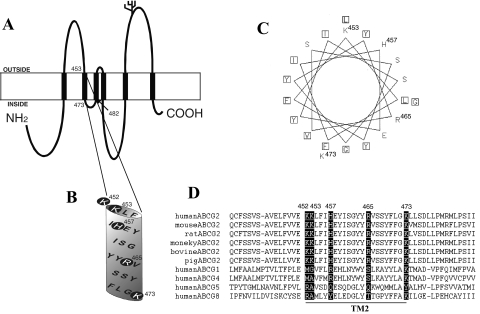
Topology model of human BCRP, location of Lys452, Lys453, His457, Arg465, and Lys473 in BCRP, helical wheel projection of TM2, and sequence alignments. A, the experimentally determined topology model of human BCRP with six TM α-helices. B, an expanded view of the putative TM2. The basic residues Lys452, Lys453, His457, Arg465, and Lys473 are indicated by shaded circles. C, helical wheel projection of the amino acid residues in the putative TM2. Squares indicate the relatively hydrophobic residues. The basic residues are labeled with their amino acid position numbers. D, the human BCRP sequence containing the putative TM2 (underlined) has been aligned with the corresponding sequences in its ABCG2 homologs and orthologs by using Clustal W. Residues in BCRP homologs and orthologs that are aligned with Lys452, Lys453, His457, Arg465, and Lys473 in human BCRP are highlighted.
Arg482 located within TM3 of BCRP at a position near the cytosolic membrane interface (Fig. 1A) appears to be absolutely crucial for substrate specificity and transport activity. Likewise, basic residues within or near TMs have been reported to play an important role in determining substrate specificity and/or overall transport activity of other ABC transporters as well, including MRP1 (Conseil et al., 2006), MRP2 (Ryu et al., 2000), and P-gp (Taguchi et al., 1997). Authors of those studies have suggested that substitutions of such basic residues may cause alterations in transporter–substrate interactions and/or subtle structural changes in the transporters at a distance. According to the recent topology structure of BCRP we determined (Wang et al., 2008), it is worth noting that, compared with other TMs of BCRP, TM2 is rich in basic residues. Five basic residues, Lys452, Lys453, His457, Arg465, and Lys473, cluster within or near the TM2 of BCRP (Fig. 1B). These residues are fully conserved among the BCRP/ABCG2 orthologs of various species (Fig. 1D). In this study, we investigated the functional importance of the five basic residues within or near TM2 of BCRP that were replaced with either the neutral amino acid Ala or the amino acid Asp of opposite charge. The mutants were stably expressed in HEK cells, and their plasma membrane expression and function were characterized. We showed that mutations of these basic residues differentially affected the transport activity of BCRP. We further demonstrated that these mutations affected BCRP ATPase activities and altered the binding of the 5D3 antibody to BCRP. Homology modeling and docking calculations indicated that His457 and Arg465 might be directly involved in BCRP–substrate interactions.
Materials and Methods
Materials.
MX, prazosin hydrochloride, Dox, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide were purchased from Sigma (St. Louis, MO). Rho123 and diphenylcarbamyl chloride-treated trypsin were obtained from MP Biomedicals (Solon, OH). SN-38 was from Tocris Bioscience (Ellisville, MO). Fumitremorgin C (FTC) was obtained from the National Institutes of Health (Bethesda, MD). High-performance liquid chromatography-grade dimethyl sulfoxide was from Thermo Fisher Scientific (Waltham, MA). Fetal bovine serum (FBS) was purchased from HyClone Laboratories (Logan, UT). Eagle's minimal essential medium (MEM), 4′,6-diamidino-2-phenylindole (DAPI), phosphate-buffered saline (PBS), BODIPY FL-prazosin, Hoechst33342, trypsin-EDTA solution, and geneticin (G418) were purchased from Invitrogen (Carlsbad, CA). FuGENE HD transfection reagent was from Roche Applied Science (Indianapolis, IN). All restriction enzymes were from New England Biolabs (Ipswich, MA). Oligonucleotides used for site-directed mutagenesis were ordered from Integrated DNA Technologies, Inc. (Coralville, IA). The mouse anti-BCRP monoclonal antibody (mAb) BXP-21 was from Kamiya Biomedical (Seattle, WA). The mouse anti-human Na+/K+-ATPase mAb was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The goat anti-mouse horseradish peroxidase-conjugated mAb was from Bio-Rad Laboratories (Hercules, CA). Alexa-Fluor 488-conjugated goat anti-mouse IgG (H + L) (Fab′)2 fragment was obtained from Invitrogen. The phycoerythrin-conjugated anti-BCRP surface mAb 5D3 and the phycoerythrin-conjugated IgG2b antibody were from eBioscience (San Diego, CA). The pcDNA3.1 vector containing full-length wild-type human BCRP cDNA was kindly provided by Dr. Susan E. Bates (National Cancer Institute, Bethesda, MD).
Site-Directed Mutagenesis.
All mutations were generated by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The template for mutagenesis was the pcDNA3.1 vector containing full-length wild-type human BCRP cDNA. The polymerase chain reaction-based mutagenesis was performed according to the manufacturer's instructions with the following forward primers: K452A (5′-gaa ctc ttt gtg gta gag GCg aag ctc ttc ata cat gaa-3′), K453D (5′-ctc ttt gtg gta gag aag GaC ctc ttc ata cat gaa tac-3′), H457A (5′-gag aag aag ctc ttc ata GCt gaa tac atc agc gga tac-3′), R465A (5′-tac atc agc gga tac tac GCa gtg tca tct tat ttc ctt-3′), and K473A (5′-tca tct tat ttc ctt gga GCa ctg tta tct gat tta tta-3′). The mutated nucleotides are capitalized, and the reverse primers are complementary to the respective forward primers. All mutations were confirmed by DNA sequencing, and the entire BCRP cDNA in each mutant construct was also sequenced to make sure that no additional mutations were introduced during polymerase chain reaction mutagenesis.
Cell Culture and Stable Transfection of Mutant BCRP Constructs into HEK Cells.
Parent HEK293 cells obtained from American Type Culture Collection (Manassas, VA) were cultured in MEM containing 10% FBS at 37°C in a humidified incubator with 5% CO2. For stable transfection, approximately 2 μg of plasmid DNA in 1 ml of serum-free medium was used to transfect 4 × 105 cells with 5 μl of FuGENE HD transfection reagent in each well of 12-well plates, according to the manufacturer's instructions. After 12 h, cells were incubated in fresh medium containing 10% FBS without G418 for an additional 48 h. Cells were then cultured in 1 mg/ml G418 for 3 days. Subsequently, cells were pooled and subcultured in limited dilutions in 1 mg/ml G418 by frequently removing dead cells and changing medium until G418-resistant colonies appeared. Approximately 3 weeks after transfection, individual cell colonies were isolated and grown with 500 μg/ml G418. The BCRP levels in the G418-resistant cell clones were examined by immunoblotting. All of the G418-resistant cell lines were maintained in MEM supplemented with 10% FBS in the presence of 500 μg/ml G418 at 37°C in a 5% CO2 incubator. Cells grown to 80 to 90% confluence were harvested for subculturing, immunoblotting, confocal microscopy analysis, flow cytometric assays, or other functional studies. Only cells within 10 passages were used in these experiments.
Preparation of Plasma Membrane Samples.
Plasma membrane samples were prepared essentially the same as described previously (Vethanayagam et al., 2005) with minor modifications. In brief, HEK cells stably expressing wild-type and mutant BCRP and the vector control cells were harvested and resuspended in homogenization buffer [250 mM sucrose, 50 mM Tris/HCl (pH 7.4), 0.25 mM CaCl2, and the protease inhibitor cocktail and benzamidine]. Cells were disrupted by N2 cavitation and centrifuged at 1,900g for 10 min at 4°C. The supernatant was layered over 35% (w/w) sucrose and centrifuged at 100,000g for at least 1 h at 4°C. The interface layer was collected and washed twice with the buffer containing 50 mM Tris/HCl (pH 7.5) and 250 mM sucrose by centrifugation. The membrane pellet was resuspended in the same buffer and passed 20 times through a 27-gauge needle. Protein concentrations of plasma membranes were determined by the Pierce BCA protein assay kit (Thermo Fisher Scientific). Relative levels of BCRP expression in plasma membrane preparations were determined by immunoblotting using the mAb BXP-21 and densitometric analysis after normalization to Na+/K+-ATPase as described below.
SDS-Polyacrylamide Gel Electrophoresis and Immunoblotting.
Whole-cell lysates or plasma membrane samples were prepared from HEK cell lines stably expressing wild-type and mutant BCRP as described previously (Vethanayagam et al., 2005). Protein concentrations were measured by the Pierce BCA protein assay kit using bovine serum albumin as standard. Whole-cell lysates (20 μg each lane) or plasma membranes (2.5 μg each lane) were subjected to SDS-polyacrylamide gel electrophoresis and immunoblotting using the BCRP-specific mAb BXP-21 as described previously (Wang et al., 2008). Human β-actin in whole-cell lysates was detected as an internal control. Human Na+/K+-ATPase in plasma membrane samples was detected as an internal standard in the plasma membranes by using a mouse anti-Na+/K+-ATPase mAb at a 1:2000 dilution. Relative BCRP levels were determined by densitometric analysis of the immunoblots using National Institutes of Health Scion Image software (Scion Corporation, Frederick, MD).
Confocal Microscopy.
HEK cells expressing wild-type and mutant BCRP and the vector control cells were seeded in a four-chamber glass slide (Falcon; BD Biosciences Discovery Labware, Bedford, MA) at approximately 5 × 105 cells/well. Cells were grown to 80% confluence and washed twice with PBS at room temperature. Cells were then fixed with 4% paraformaldehyde in PBS at room temperature for 30 min, washed twice with PBS, and incubated in PBS containing 0.1% Triton X-100 and 2% horse serum at room temperature for 60 min. Subsequently, cells were incubated with BXP-21 (1:500 dilution in PBS containing 0.1% Triton X-100 and 2% horse serum) for 60 min and washed three times with PBS containing 0.02% Tween 20. Cells were then incubated in the dark with Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) (Fab′)2 fragment (1:1000 dilution in PBS containing 0.1% Triton X-100 and 2% horse serum) for 1 h. In the same preparations, cell nuclei were stained with 300 nM DAPI. Finally, cells were washed twice with PBS, mounted in Fluoromount G (Southern Biotechnology Associates, Birmingham, AL), and observed at 488-nm excitation and 519-nm emission wavelengths by using a Zeiss TCS SP1 MP multiphoton confocal microscope (Zeiss Microsystems, Exton, PA).
Detection of Cell Surface Expression of Wild-Type and Mutant BCRP Using Flow Cytometry.
The flow cytometric assay used to determine cell surface expression of BCRP was essentially the same as described previously (Vethanayagam et al., 2005). Cells were grown to 80 to 90% confluence, collected, and washed once with PBS. Approximately 5 × 105 cells were incubated with the phycoerythrin-conjugated anti-BCRP mAb 5D3 (20 μl) or the phycoerythrin-conjugated mouse IgG2b negative control (20 μl) in 0.75 ml of PBS containing 2% bovine serum albumin for 30 min at room temperature. Cells were then washed once with PBS and placed on ice in the dark until analysis. Within 1 h, cells were analyzed on a BD FACScan flow cytometer (BD Biosciences, San Jose, CA) equipped with a 488-nm argon laser and a 585/42-nm bandpass filter. A total of 10,000 (104) events were collected. Cell debris was eliminated by gating on forward versus side scatter. The differences in phycoerythrin fluorescence between cells incubated with 5D3 and the IgG2b control were used to express the relative cell surface expression of wild-type BCRP and its mutants.
Flow Cytometric Efflux Assay.
Flow cytometric efflux assays were performed essentially the same as described previously (Vethanayagam et al., 2005) with minor modifications. In brief, HEK cells expressing wild-type and mutant BCRP or the vector control cells were incubated with the following fluorescent compounds: 10 μM MX, 500 nM BODIPY-prazosin, or 0.05 μg/ml Hoechst33342 in the presence or absence of 10 μM FTC for 30 min at 37°C. Cells were then washed and resuspended in 1 ml of fluorescent compound-free incubation buffer with or without 10 μM FTC, and incubation was continued for 1 h at 37°C (efflux phase). Cells were immediately washed, resuspended in 1 ml of ice-cold PBS, and kept on ice in the dark. Intracellular fluorescence was measured within 1 h with a 488-nm argon laser and a 650-nm longpass filter for MX and a 488-nm argon laser and a 530-nm bandpass filter for BODIPY-prazosin in a BD FACScan flow cytometer. A FACSVantage flow cytometer equipped with a 360-nm UV laser was used to detect Hoechst33342 fluorescence. The difference (ΔF) in median fluorescence between the FTC/efflux and the corresponding efflux histograms was used as a measure of FTC-inhibitable efflux activity of BCRP and its mutants. The efflux activities were normalized to the BCRP levels to take into consideration the differences in BCRP expression. Statistical significance of the differences in efflux activities between wild-type and mutant BCRP was analyzed by Student's t test. A difference with a p value of <0.05 was considered statistically significant.
Cytotoxicity Assay.
In brief, cells were seeded in collagen-coated 96-well plates at a density of 4000 to 5000 cells per well with 200 μl of MEM supplemented with 10% FBS and 500 μg/ml G418. Resistance profiles of HEK cells expressing wild-type and mutant BCRP and the vector control cells to the drug MX, SN-38, Dox, or Rho123 were determined by using the standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide microtiter plate assay as described previously (Vethanayagam et al., 2005). The concentration ranges of MX, SN-38, Dox, and Rho123 used were 0.001 to 10, 0.0001 to 1, 0.001 to 10, and 0.1 to 200 μM, respectively. Time of exposure for all drugs was 72 h. Cells treated with the lowest drug concentrations were set as controls (100% cell survival). IC50 values were calculated by fitting the following model to the data points using nonlinear regression (Prism, version 3.03; GraphPad Software, Inc., San Diego, CA): I = Imax − [Imax − Io] × [Cγ /(Cγ + IC50γ)], where I is the cell survival as percentage of the optical density of the control cells, Imax is the maximal cell survival, Io is the lowest residual cell survival at the high drug concentrations, C is the drug concentration, γ is the slope factor, and IC50 is the drug concentration leading to 50% cell survival. Relative resistance factors were calculated as the ratios of the IC50 values of cells expressing wild-type and mutant BCRP to the IC50 values of the vector control cells. The relative resistance levels were finally normalized to the BCRP expression levels to take into account the differences in BCRP expression. Statistically significant differences in resistance factors between wild-type BCRP and its mutants were analyzed by Student's t test. A difference with a p value of <0.05 was considered significant.
Vanadate-Sensitive ATPase Activity.
Vanadate-sensitive ATPase activities were determined as described previously (Mao et al., 2004). In brief, reactions were initiated by mixing plasma membranes (5 μg of protein per reaction) with 100 μl of the reaction buffer containing 50 mM HEPES (pH 7.0), 5 mM MgCl2, 10 mM Na3N, 2 mM EGTA, 2 mM ouabain, and varying concentrations of MgATP up to 5 mM in the presence or absence of 1 mM sodium orthovanadate and maintained at 37°C for 30 min. Reactions were terminated by adding 6% SDS, and the liberation of inorganic phosphate (Pi) was measured immediately by detecting the optical density at 650 nm. The differences between ATPase activities determined in the absence and presence of vanadate were used to express vanadate-sensitive ATPase activities. Vanadate-sensitive ATPase activities of wild-type and mutant BCRP were then calculated by subtracting the background activities of the vector control from those of wild-type and mutant BCRP samples. The Km and Vmax values for ATP hydrolysis were estimated by fitting the Michaelis-Menten equation to the data points by using nonlinear regression (GraphPad Prism, version 3.03).
Determining the Effects of Prazosin on the Binding of 5D3 to Wild-Type and Mutant BCRP.
The same procedure as for detection of BCRP cell surface expression was used in the absence and presence of increasing concentrations (0.1–100 μM) of prazosin. In preliminary experiments, we incubated cells with 5D3 and prazosin for up to 2 h, and the fluorescence did not change compared with incubation for 30 min. Therefore, the incubation time used in this assay was 30 min. The final concentration of dimethyl sulfoxide used to dissolve prazosin was less than 1% (v/v), and no effects of the vehicle on fluorescence detection were observed at this concentration. The normalized differences in fluorescence (ΔF/F0) between the cells incubated with 5D3 in the presence of prazosin and 5D3 alone (F0) were used to express the concentration-dependent effects of prazosin on the binding of 5D3 to wild-type and mutant BCRP. The apparent equilibrium dissociation constant (Ks) of the prazosin complex with wild-type or mutant BCRP was estimated according to the Michaelis-Menten analysis (ΔF/F0) = (ΔF/F0)max × [prazosin]/([prazosin] + Ks) by nonlinear regression (GraphPad Prism, version 3.03), where [prazosin] is the concentration of prazosin.
Limited Trypsin Digestion of Wild-Type and Mutant BCRP.
To provide additional evidence that point mutations may cause conformational changes in BCRP, we performed limited trypsin digestion analysis of plasma membranes as below. Plasma membrane preparations were diluted to 0.5 μg/μl in the buffer (50 mM Tris/HCl, pH 7.5, and 250 mM sucrose) and incubated at 37°C for 15 min with diphenylcarbamyl chloride-treated trypsin at trypsin/protein ratios ranging from 1:10,000 to 1:10 (w/w). Trypsin digestion was then stopped by adding the Laemmli sample buffer containing 10 mM phenylmethylsulfonyl fluoride and leupeptin (16 μg/ml), and the samples were immediately subjected to SDS- polyacrylamide gel electrophoresis on a 4 to 15% gradient gel (Bio-Rad Laboratories) with 2 μg of plasma membrane protein per lane. Immunoblotting was performed with the mAb BXP-21, and the intensity of BCRP bands was determined by densitometric analysis of the immunoblots using National Institutes of Health Scion Image software as described above.
Homology Modeling and Molecular Docking of BCRP.
The substrate-bound inward-facing form of mouse P-gp (Aller et al., 2009) (Protein Data Bank ID code 3G60) was used as a full-length template for homology modeling. Homology modeling was performed based on the membrane topology structure of BCRP we recently determined (Wang et al., 2008) as described previously (M. F. Rosenberg, Z. Bikadi, J. Chan, X. Liu, Z. Ni, X. Cai, R. C. Ford, and Q. Mao, submitted for publication).
To identify the potential interaction mode of substrates with BCRP and determine whether the basic residues analyzed in this study might be directly involved in substrate binding, molecular docking of MX, prazosin, SN-38, and Hoechst33342 to the substrate-bound and nucleotide-free inward-facing model of BCRP was performed by using DockingServer (www.dockingserver.com). We have shown that 80% of the first-rank docking results are most similar to the experimental data using the same DockingServer methodology (Bikadi and Hazai, 2009). All of the docking calculations described here involved flexible docking of the substrates to the rigid receptor, which is the BCRP model in this study. In brief, input structures of both the substrates and the BCRP model were prepared by using the PM6 charges calculated with MOPAC2009 software (Stewart, 2009). Semiempirical assignments were performed by using the PM6 method by the Mozyme function of the MOPAC2009 program integrated in DockingServer. Docking calculations were then performed by using AutoDock 4 (Huey et al., 2007) integrated in DockingServer. Grid parameter files were built, and atom-specific affinity maps were constructed by using the built-in AutoGrid 4 program (Huey et al., 2007). These map files were generated by using 80 × 80 × 80 grid points and 0.375-Å spacing, with the maps centered on the TM region of BCRP. Docking simulations were carried out by using the Lamarckian Genetic Algorithm. The initial position, orientation, and torsions of the substrate molecules were set randomly, and all rotatable torsions were released during docking. Each docking experiment was derived from 100 different runs that were set to terminate after a maximum of 10,000,000 energy evaluations and had a population size of 500. The conformer with the lowest docking energy calculated with AutoDock 4 scoring function was selected as the final binding conformation of BCRP for these substrates, and the higher ranked results were not considered in this study.
Results
Expression of Wild-Type and Mutant BCRP in Stably Transfected HEK Cells.
Based on the recent topology of BCRP (Wang et al., 2008), TM2 comprises amino acid residues approximately 453 to 475 in which there are four basic residues Lys453, His457, Arg465, and Lys473 (Fig. 1, A and B). Lys452 is at the extracellular membrane interface of TM2 (Fig. 1B). These basic residues are strictly conserved in the human, mouse, rat, monkey, bovine, and porcine orthologs of BCRP (Fig. 1D). Although they are not fully conserved in the human ABCG subfamily members, most of them seem to share certain common characters, either the same charge or a high polarity (Fig. 1D). Helical wheel projection indicates that TM2 is highly amphiphatic with Lys453, His457, Arg465, and other hydrophilic residues concentrating on one face of the TM2 α-helix and relatively hydrophobic residues on the opposite face (Fig. 1C), suggesting that Lys453, His457, and Arg465 might be facing toward the substrate-binding cavity if TM2 is part of the translocation pathway. To evaluate the role of these basic residues in BCRP activity, we generated mutants in which Lys452, His457, Arg465, and Lys473 were replaced with Ala. Because substitution of Lys453 with Asp showed substantially higher levels of expression than substitution with Ala in all the clones obtained in this study, we examined substitution of Lys453 with Asp that may affect BCRP activity if the positive charge of Lys453 is functionally essential.
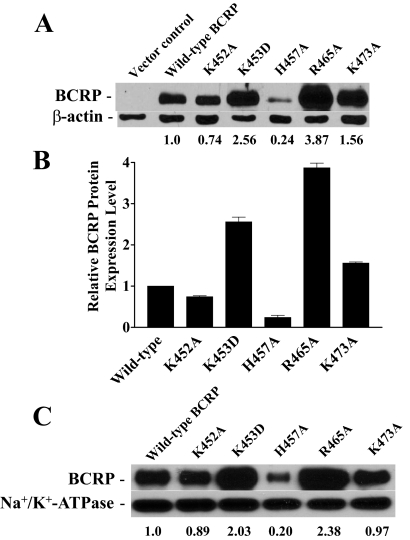
The pcDNA3.1 vectors containing cDNAs of wild-type and mutant BCRP were used to stably transfect HEK cells, and stable cell lines were generated by limited dilution and selection in G418. Expression levels of BCRP mutants relative to wild-type protein in the stable cell clones were determined by immunoblotting. The resultant single stable cell clones (10–15 for each mutant) expressed substantially variable levels of BCRP, and only the clones expressing the highest levels of BCRP were maintained for subsequent studies. Endogenous BCRP in HEK cells transfected with the empty vector was not detectable under the conditions used (Fig. 2A). The expression levels of the mutants K452A, K453D, H457A, R465A, and K473A, determined by immunoblotting of whole-cell lysates using β-actin as an internal standard, were approximately 0.74-, 2.56-, 0.24-, 3.87-, and 1.56-fold that of wild-type BCRP (Fig. 2, A and B). The expression levels of wild-type and mutant BCRP determined in whole-cell lysates and the respective plasma membrane preparations were comparable (Fig. 2C).
Fig. 2.
Immunoblotting analysis of the expression levels of wild-type and mutant BCRP. The expression levels of wild-type and mutant BCRP in stably transfected HEK cells were determined by immunoblotting and densitometric analysis as described. A, a representative immunoblot of whole-cell lysates for wild-type BCRP and the mutants K452A, K453D, H457A, R465A, and K473A. The numbers below the blot refer to the average protein expression levels of wild-type and mutant BCRP after normalization to β-actin. B, the relative protein levels of wild-type and mutant BCRP determined by immunoblotting of whole-cell lysates are presented as means ± S.D. of three independent experiments. C, a representative immunoblot of plasma membrane samples for wild-type and mutant BCRP. The numbers below the blots refer to the average levels of the mutants relative to wild-type BCRP after normalization to Na+/K+-ATPase. The experiments were repeated one to two times, and similar results were obtained.
Plasma Membrane Localization and Cell Surface Expression of Wild-Type and Mutant BCRP in HEK Cells.
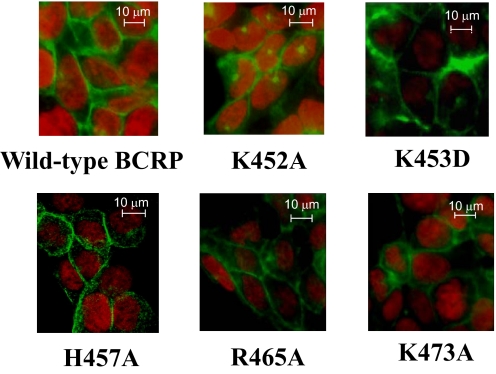
To explore whether plasma membrane localization of the mutants might be affected, the stably transfected HEK cells were examined by immunofluorescent confocal microscopy using the BCRP-specific mAb BXP-21. Strong plasma membrane staining of green fluorescence was observed in cells expressing wild-type or mutant BCRP, with little intracellular signal (Fig. 3), suggesting that all of the mutants were predominantly targeted to the plasma membrane of HEK cells and did not appear to affect BCRP folding and trafficking.
Fig. 3.
Confocal microscopy of HEK cells stably expressing wild-type and mutant BCRP. The cellular localization of wild-type and mutant BCRP in HEK cells was determined by immunofluorescence confocal microscopy using mAb BXP-21 as described. The staining of BCRP is indicated in green. Cell nuclei were stained with DAPI and are shown in red. No green fluorescence was detected in the vector control cells (data not shown). Selected areas of HEK cells expressing wild-type BCRP and the mutants K452A, K453D, H457A, R465A, and K473A are shown. Images have been enhanced for a suitable contrast between the black background and green fluorescence and are not intended for quantitative determination of BCRP expression.
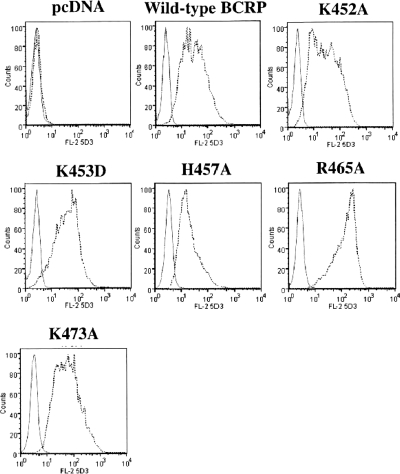
To further determine cell surface expression of wild-type and mutant BCRP, cells were stained with the phycoerythrin-conjugated BCRP-specific mAb 5D3, which recognizes a conformation-sensitive extracellular epitope (Ozvegy-Laczka et al., 2008), and the IgG2b control. Phycoerythrin fluorescence associated with cells was then determined by flow cytometry. As expected, the cells transfected with the empty vector did not exhibit any cell surface expression of BCRP (Fig. 4). Wild-type and mutant BCRP all showed significant cell surface expression that was generally consistent with the immunoblotting data (Fig. 2).
Fig. 4.
Cell surface expression of wild-type and mutant BCRP. The expression of wild-type and mutant BCRP on the surface of stably transfected HEK cells was detected by using mAb 5D3 as described. Shown are representative flow cytometry histograms showing cell surface expression of wild-type and mutant BCRP. The solid and dashed lines represent the phycoerythrin fluorescence associated with cells treated with the IgG2b negative control and 5D3 antibodies, respectively. No surface expression of BCRP was detected in the pcDNA vector control cells.
FTC-Inhibitable Efflux Activities of Wild-Type and Mutant BCRP.
We next examined whether mutations of these basic residues affect BCRP activity by using a flow cytometric efflux assay with three fluorescent substrates, MX, BODIPY-prazosin, and Hoechst33342. FTC, a specific inhibitor for BCRP, was used at 10 μM at which BCRP activity could be completely inhibited. This assay has been widely used to assess BCRP efflux activity (Robey et al., 2001; Vethanayagam et al., 2005). The intracellular fluorescence of the three substrates in cells expressing wild-type and mutant BCRP was increased by the addition of FTC; however, FTC did not significantly affect the intracellular fluorescence in the vector control cells (Table 1). This suggests that all five mutants were able to transport MX, BODIPY-prazosin, and Hoechst33342. After normalization to the BCRP levels of whole-cell lysates, statistically significant differences in efflux activities for all three substrates were noticed for K452A, K453D, H457A, and R465A compared with wild-type protein. Efflux activities of K473A were not significantly changed. Thus, the efflux activities of K452A and H457A were significantly increased 2- to 6-fold, whereas the activities of K453D and R465A were decreased by 40 to 60%, depending on substrate (Table 1). Notably, the activities of H457A for BODIPY-prazosin and Hoechst33342 were increased 3- to 6-fold, but its activity for MX was increased only 1.7-fold. We used the BCRP levels of whole-cell lysates, but not those of plasma membranes, in the calculations for three reasons. First, all of the mutants were expressed predominantly on the plasma membrane (Fig. 3). Second, unlike whole-cell lysates, plasma membranes were prepared through cell breakage and multiple centrifugation steps that could introduce more variations. Third, the BCRP levels of whole-cell lysates were validated by using a soluble protein β-actin as an internal standard.
TABLE 1.
FTC-inhibitable efflux activities of HEK cells expressing wild-type and mutant BCRP
FTC-inhibitable efflux activities of HEK cells expressing wild-type and mutant BCRP for mitoxantrone, BODIPY-prazosin, and Hoechst33342 are expressed as the differences in the median fluorescence (ΔF) between the FTC/efflux histograms and the respective efflux histograms. ΔF′ represents the differences in the median fluorescence after normalization for differences in BCRP expression level. Shown are means ± S.D. of three to six independent experiments. The relative efflux activities of the mutants as compared with wild-type BCRP after normalization to the BCRP expression levels are presented as ratios on which the activities of wild-type BCRP are set as 1. The relative BCRP levels determined by immunoblotting of whole-cell lysates shown in Fig. 2A were used for the calculations. The differences between the efflux activities of wild-type and mutant BCRP after normalization to the BCRP protein expression levels are statistically significant: *, P < 0.05 by Student's t test.
| Mitoxantrone |
BODIPY-Prazosin |
Hoechst33342 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔF | ΔF′ | Ratio | ΔF | ΔF′ | Ratio | ΔF | ΔF′ | Ratio | |
| pcDNA vector | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Wild-type BCRP | 11.1 ± 1.5 | 11.1 ± 1.5 | 1.0 | 49.9 ± 13.1 | 49.9 ± 13.1 | 1.0 | 976.5 ± 115.5 | 976.5 ± 115.5 | 1.0 |
| K452A | 22.0 ± 5.9 | 29.7 ± 7.9* | 2.7 | 76.6 ± 22.5 | 103.5 ± 30.4* | 2.1 | 1138.3 ± 134.7 | 1538.3 ± 182.1* | 1.6 |
| K453D | 19.1 ± 5.9 | 7.5 ± 3.3* | 0.7 | 57.7 ± 15.6 | 22.6 ± 6.1* | 0.4 | 1116.9 ± 132.2 | 436.3 ± 51.6* | 0.4 |
| H457A | 4.5 ± 2.1 | 18.7 ± 8.7* | 1.7 | 40.1 ± 9.7 | 167.0 ± 40.2* | 3.3 | 1259.5 ± 343.2 | 5247.9 ± 1429.9* | 5.4 |
| R465A | 26.1 ± 3.0 | 6.8 ± 0.8* | 0.6 | 96.5 ± 16.0 | 24.9 ± 4.1* | 0.5 | 2217.8 ± 255.2 | 573.1 ± 65.9* | 0.6 |
| K473A | 22.2 ± 5.0 | 14.3 ± 3.2 | 1.3 | 83.2 ± 17.6 | 53.3 ± 11.3 | 1.1 | 1411.5 ± 166.8 | 887.7 ± 104.9 | 0.9 |
Drug Resistance Conferred by Wild-Type and Mutant BCRP.
To further explore whether the mutations affect relative drug resistance of BCRP, we performed the cytotoxicity assay with MX, SN-38, Dox, and Rho123 on HEK cells expressing wild-type and mutant BCRP. The results are summarized in Table 2. Compared with the vector control cells, cells expressing wild-type and mutant BCRP were resistant to MX and SN-38, but not to Rho123 and Dox. These results are consistent with previous observations that Rho123 and Dox are not substrates of wild-type BCRP (Robey et al., 2003; Vethanayagam et al., 2005). After normalization to the BCRP levels, the IC50 values of cells expressing K452A, K453D, H457A, and R465A for MX and SN-38 were significantly different from those of cells expressing wild-type BCRP, whereas the IC50 values of cells expressing K473A and wild-type protein were comparable. Thus, the relative levels of resistance of K452A and H457A to MX were increased approximately 2- to 3-fold compared with wild-type protein, whereas those of K453D and R465A to MX were decreased by 30 to 70%. Likewise, the relative levels of resistance of H457A to SN-38 were increased approximately 3-fold, whereas those of K453D and R465A to SN-38 were decreased by 50 to 60%. However, the relative levels of resistance of K452A to SN-38 did not change much. We did not calculate the relative levels of resistance to Dox and Rho123 as all of the mutants did not confer any resistance to the two drugs. MX was used in both the efflux and drug resistance studies, and the results were in good agreement.
TABLE 2.
Relative drug resistance of HEK293 cells expressing wild-type and mutant BCRP
The resistance of HEK293 cells expressing wild-type and mutant BCRP to MX, SN-38, Dox, and Rho123 were determined as described under Materials and Methods. Relative resistance factors were obtained by dividing the IC50 values of cells expressing wild-type or mutant BCRP by the IC50 values of the vector control cells. The ratios represent the relative levels of resistance of the mutants compared with wild-type BCRP after normalization for differences in BCRP expression level. The relative BCRP expression levels determined by immunoblotting of whole-cell lysates shown in Fig. 2A were used for the calculations. Since wild-type and mutant BCRP did not confer resistance to Dox and Rho123, the ratios for the two compounds were not calculated. The IC50 values shown are means ± S.D. of nine independent experiments. The differences between the resistance factors of wild-type and mutant BCRP after normalization to the BCRP protein levels are statistically significant: *, p < 0.05 by Student's t test.
| MX |
SN-38 |
Dox |
Rho123 |
|||||
|---|---|---|---|---|---|---|---|---|
| IC50 | Relative Resistance (Ratio) | IC50 | Relative Resistance (Ratio) | IC50 | Relative Resistance | IC50 | Relative Resistance | |
| nM | nM | nM | μM | |||||
| pcDNA vector | 24.0 ± 3.5 | 2.4 ± 0.3 | 24.0 ± 8.7 | 7.26 ± 1.15 | ||||
| Wild-type BCRP | 145.1 ± 52.8 | 6.0 (1.0) | 125.3 ± 6.1 | 52.2 (1.0) | 31.5 ± 12.6 | 1.3 | 10.97 ± 1.84 | 1.5 |
| K452A | 354.8 ± 68.6 | 14.8 (3.3)* | 103.0 ± 12.5 | 42.9 (1.1)* | 24.2 ± 4.4 | 1.0 | 9.27 ± 1.75 | 1.3 |
| K453D | 244.0 ± 99.9 | 10.2 (0.7)* | 136.2 ± 9.9 | 56.8 (0.4)* | 34.0 ± 6.7 | 1.4 | 8.22 ± 0.97 | 1.1 |
| H457A | 71.9 ± 12.8 | 3.0 (2.1)* | 90.6 ± 4.6 | 37.8 (3.0)* | 21.8 ± 16.6 | 0.9 | 7.61 ± 1.29 | 1.0 |
| R465A | 169.1 ± 49.0 | 7.0 (0.3)* | 224.2 ± 39.7 | 93.4 (0.5)* | 37.9 ± 17.5 | 1.6 | 14.65 ± 1.26 | 2.0 |
| K473A | 243.1 ± 114.0 | 10.1 (1.1) | 188.0 ± 19.2 | 78.3 (0.9) | 36.0 ± 10.1 | 1.5 | 15.11 ± 1.43 | 2.1 |
ATPase Activities of Wild-Type and Mutant BCRP.
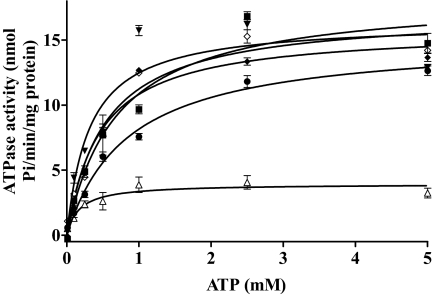
We then examined whether the altered transport properties of the mutants might possibly be caused by changes in ATP hydrolysis by these mutants. We first determined vanadate-sensitive ATPase activities of the plasma membrane samples of wild-type and mutant BCRP or the vector control. The ATPase activities attributable to wild-type and mutant BCRP were then calculated by subtracting the background of vanadate-sensitive ATPase activities in the vector control. ATP concentration-dependent vanadate-sensitive ATPase activities of wild-type and mutant BCRP are shown in Fig. 5. The kinetic parameters Km and Vmax of wild-type and mutant BCRP for ATP hydrolysis were estimated by fitting to the Michaelis-Menten equation to the data points and are shown in Table 3. The Km values of K453D, R465A, and K473A were comparable with that of wild-type protein; however, the Km values of K452A and H457A were decreased by approximately 50 and 70%, respectively, suggesting that these two mutations, particularly the one at position 457, increased the binding affinity of ATP to BCRP. The Km value of wild-type BCRP was comparable with that reported previously (Pozza et al., 2006). After normalization to the BCRP levels, the Vmax values of K453D and R465A were approximately 60% lower than that of wild-type BCRP, whereas the Vmax values of other mutants did not change. As a result, the Vmax/Km values of K452A and H457A were increased approximately 2- and 4-fold compared with wild-type BCRP, respectively, suggesting that these two mutants catalyzed ATP hydrolysis more efficiently than wild-type protein. In contrast, the Vmax/Km values of K453D and R465A were decreased by approximately 40 to 70%. The Vmax/Km value of K473A was comparable with that of wild-type protein.
Fig. 5.
ATP hydrolysis profiles of wild-type and mutant BCRP. Vanadate-sensitive ATPase activities of wild-type and mutant BCRP were measured with plasma membrane preparations over an ATP concentration range of 0 to 5 mM as described. Shown are means ± S.D. of three independent experiments for wild-type BCRP (■), K452A (▾), K453D (●), H457A (▵), R465A (♦), and K473A (♢).
TABLE 3.
Kinetic parameters of ATP hydrolysis by wild-type and mutant BCRP
ATP hydrolysis activities of plasma membrane preparations from HEK293 cells expressing wild-type and mutant BCRP were determined as described under Materials and Methods. The kinetic parameters of ATP hydrolysis were calculated by fitting the Michaelis-Menten equation to the data points by nonlinear regression as described under Materials and Methods. The values of Vmax were normalized to the BCRP expression level. The BCRP expression levels determined by immunoblotting of plasma membrane preparations shown in Fig. 2C were used for the calculations. Shown are means ± S.D. of three independent experiments.
| Wild-type BCRP | K452A | K453D | H457A | R465A | K473A | |
|---|---|---|---|---|---|---|
| Vmax (nmol Pi/min/mg protein) | 18.4 ± 1.8 | 16.4 ± 1.9 | 15.1 ± 1.4 | 3.94 ± 0.07 | 15.8 ± 2.6 | 17.1 ± 1.3 |
| Vmax normalized to BCRP level (nmol Pi/min/mg protein) | 18.4 ± 1.8 | 18.4 ± 2.1 | 7.4 ± 0.7 | 19.6 ± 0.3 | 6.7 ± 1.1 | 17.6 ± 1.3 |
| Km for ATP (mM) | 0.69 ± 0.21 | 0.32 ± 0.15 | 0.85 ± 0.12 | 0.17 ± 0.07 | 0.46 ± 0.11 | 0.52 ± 0.13 |
| Vmax/Km (nmol Pi/min/mg protein/mM) | 26.7 | 57.5 | 8.7 | 115.3 | 14.6 | 33.8 |
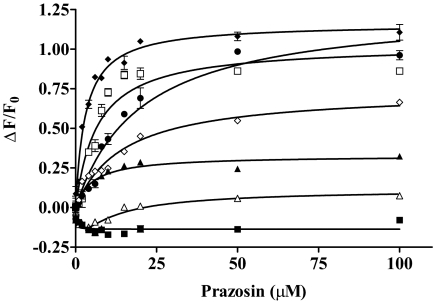
Concentration-Dependent Effects of Prazosin on the Binding of 5D3 to Wild-Type and Mutant BCRP.
It has been shown that the binding of the 5D3 antibody to BCRP is sensitive to conformational changes that result in increased binding of the antibody in the presence of substrates or inhibitors (Ozvegy-Laczka et al., 2008). We speculate that such substrate or inhibitor binding induced conformational changes may be affected by the mutations, thus contributing to the altered transport and/or ATPase activity. Therefore, the binding of 5D3 to wild-type and mutant BCRP was measured in the absence and presence of varying concentrations of prazosin, a model substrate of BCRP. When cells expressing wild-type BCRP were incubated with the IgG2b control antibody, prazosin at up to 100 μM did not affect the phycoerythrin fluorescence (data not shown). In addition, when the pcDNA vector control cells were incubated with 5D3, the addition of prazosin did not change the phycoerythrin fluorescence much (Fig. 6). This suggests that prazosin itself had no effect on phycoerythrin fluorescence. However, the addition of prazosin differentially increased the binding of 5D3 to wild-type BCRP, K452A, K453D, H457A, and R465A in a concentration-dependent manner (Fig. 6), suggesting that the binding equilibrium between prazosin and BCRP could be monitored by measuring the binding of 5D3 to the transporter. Thus, the apparent dissociation constants of the prazosin complex with wild-type or mutant BCRP were estimated to be 5.3 ± 1.1, 14.7 ± 2.3, 3.1 ± 0.4, 20.1 ± 4.0, and 6.7 ± 1.8 μM for wild-type BCRP, K452A, K453D, H457A, and R465A, respectively. In contrast, prazosin only slightly decreased, rather than increased, the binding of 5D3 to K473A at low concentrations. These data suggest that the interactions of prazosin with BCRP and the consequent substrate binding induced conformational changes can be differentially affected by replacing these basic residues.
Fig. 6.
Effects of prazosin on the binding of 5D3 to wild-type and mutant BCRP. The concentration-dependent effects of prazosin on the binding of 5D3 to wild-type and mutant BCRP over a concentration range of 0 to 100 μM were determined by using flow cytometry as described. Shown are means ± S.D. of three independent experiments for the pcDNA control (■), wild-type BCRP (▴), K452A (♢), K453D (♦), H457A (●), R465A (□), and K473A (▵).
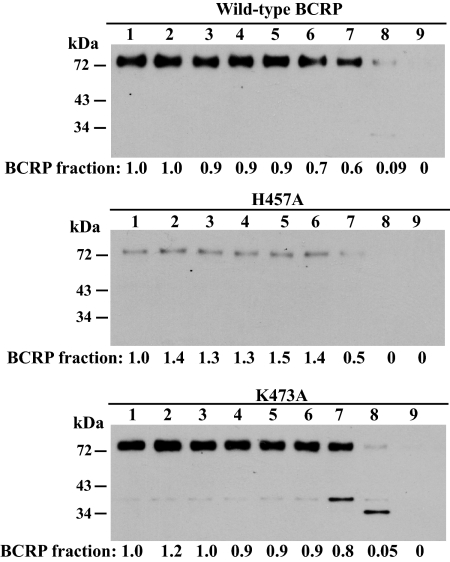
Limited Trypsin Digestion of Wild-Type BCRP and the Mutants H457A and K473A.
It has previously been shown that conformational changes in MRP1 induced by point mutations (Leslie et al., 2003) or in P-gp caused by single-nucleotide polymorphisms (Kimchi-Sarfaty et al., 2007) could be detected by limited trypsin digestion of membrane preparations caused by changes in exposure of trypsin cleavage sites. Therefore, to provide additional evidence of conformational changes in the BCRP mutants, we performed limited trypsin digestion of plasma membrane preparations of wild-type BCRP and two representative mutants H457A and K473A that showed significant changes in the pattern of 5D3 binding (Fig. 6). We compared their relative susceptibility to trypsin cleavage. We found that significant trypsin cleavage of wild-type BCRP began to occur at a trypsin/protein ratio of 1:250 because the BCRP fraction at this ratio decreased to 0.7 (Fig. 7A). In contrast, significant trypsin cleavage of both H457A and K473A began to occur at a trypsin/protein ratio of 1:100 (Fig. 7, B and C). These results suggest that the mutants are more resistant to trypsin digestion than wild-type BCRP by a factor of approximately two to three. These results again imply that there are small differences in tertiary structure between the mutants and wild-type BCRP. Interestingly, increased resistance to trypsin digestion has also been reported for MRP1 mutants (Leslie et al., 2003) and P-gp single-nucleotide polymorphisms (Kimchi-Sarfaty et al., 2007).
Fig. 7.
Trypsin digestion of wild-type BCRP, H457A, and K473A. Plasma membrane preparations expressing wild-type BCRP, H457A, or K473A (2 μg of protein each lane) were subjected to limited trypsin digestion and immunoblotting as described under Materials and Methods. Trypsin/protein ratios (w/w) used are indicated above the blots as follows: lane 1, 0 (the control with no trypsin added); lane 2, 1:10,000; lane 3, 1:5000; lane 4, 1:1000; lane 5, 1:500; lane 6, 1:250; lane 7, 1:100; lane 8, 1:50; and lane 9, 1:10. The extent of trypsin digestion of BCRP at different trypsin/protein ratios normalized to the control is shown as BCRP fraction under each blot. Molecular mass markers are indicated on the left.
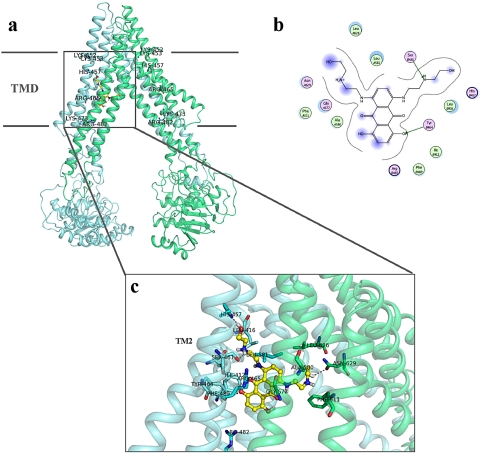
Structural Models of the BCRP–Substrate Complexes.
Owing to the lack of high-resolution crystal structures, we built a BCRP homology model by using the crystal structure of P-gp (Aller et al., 2009) as a template. Because BCRP shares a low sequence similarity with P-gp, the sequence alignment of the MSD between BCRP and P-gp was essentially based on the topology structure of BCRP we recently determined (Wang et al., 2008). The BCRP model generated represents the substrate-bound and nucleotide-free inward-facing conformation of the transporter (Fig. 8A).
Fig. 8.
Three-dimensional model of the BCRP–mitoxantrone complex. A, a side view of the three-dimensional model of the BCRP–MX complex in the inward-facing form of BCRP. Only the complex of BCRP with one MX molecule is presented. Individual BCRP monomer in the BCRP dimer is shown in green or light green. The putative boundary of the transmembrane domain (TMD) is indicated by straight lines. The approximate locations of Lys452, Lys453, His457, Arg465, Lys473, and Arg482 are indicated by LYS-452, LYS-453, HIS-457, ARG-465, LYS-473, and ARG-482, respectively. MX is presented in yellow and red. B, a schematic representation of hydrogen bonds and hydrophobic interactions between MX and BCRP. Arrows and lines indicate potential hydrogen bonds and hydrophobic interactions between MX and BCRP, respectively. Basic (His457 and Arg465), polar (Ser461 and Tyr464), and hydrophobic (Ile412, Leu416, Phe489, Phe511, Ala580, Leu581, and Leu626) residues potentially involved in interactions between MX and BCRP are presented. A and B in the cycles indicate residues from different BCRP monomers in the dimer. Pink and green indicate polar and hydrophobic residues, respectively. C, an expanded view of residues potentially involved in interactions with MX in the model. Only the complex of BCRP with one MX molecule is presented, and only TM2 of one BCRP monomer is indicated. MX is shown in yellow and red in stick format. Residues involved in potential interactions with MX are indicated in blue and green in stick format.
Based on the BCRP model, there is a large central cavity in which all the TMs appear to participate (Fig. 8A). To understand whether the residues analyzed in this study might be directly involved in BCRP–substrate interaction, we performed docking calculations of MX, prazosin, SN-38, and Hoechst33342 to the BCRP model. The four substrates may share highly overlapping, but distinct, binding sites (Fig. 8C and Supplements 1–3). The putative binding sites all are located in the central cavity in the middle of lipid bilayer, and apparently at least two substrate molecules could bind simultaneously based on the size of the central cavity. Similarly, the crystal structures of P-gp complexed with two large ligands indicate that two substrates can bind simultaneously to P-gp at the drug-binding sites (Aller et al., 2009). The best-ranked calculated docking poses demonstrate interactions of the four substrates with residues from all of the TM α-helices. Of the five basic residues, His457 and Arg465, but not Lys452, Lys453, and Lys473, seem to directly participate in the binding of all four substrates. Arg482, which is known to be crucial for substrate specificity and transport activity, may interact with MX and Hoechst33342 through the side chain (Fig. 8C and Supplement 3); however, Arg482 seems unlikely to directly interact with prazosin and SN-38 in the inward-facing form of BCRP as revealed in the docked models (Supplements 1 and 2). This suggests that the functional importance of Arg482 might be substrate-dependent, which is consistent with the results from previous studies showing that resistance to MX was increased, but resistance to SN-38 or efflux of prazosin was not significantly affected, by the mutations of Arg482 (Miwa et al., 2003; Robey et al., 2003). This also appears to be in good agreement with the findings that prazosin binds to a pharmacologically distinct site relative to other BCRP substrates such as MX and Hoechst33342 (Clark et al., 2006), and the binding of a prazosin derivative to BCRP was relatively unaffected by the mutations of Arg482 (Ejendal et al., 2006).
Based on the BCRP model, side chains of Lys453, His457, and Arg465 in TM2 are facing toward the central cavity, which is consistent with the helical wheel analysis (Fig. 1C). His457 and Arg465 may form hydrogen bonds or electrostatic interactions with substrates (Fig. 8, B and C, and Supplements 1–3). Lys453 seems unlikely to be directly involved in substrate binding in the docked models; however, it may form a salt bridge with Glu585 of the opposite BCRP molecule in the dimer. Lys452 and Lys473 are located outside of the central cavity with Lys452 on the top and Lys473 at the bottom (Fig. 8A).
Discussion
In the present study, we analyzed the effects of mutations of Lys452, Lys453, His457, Arg465, and Lys473 within or near the highly amphiphatic TM2 on BCRP expression and activity. All of the mutants were expressed as a mature form, properly targeted to the plasma membrane, and functional in drug transport and resistance, suggesting that amino acid substitutions of these basic residues do not cause a major alteration in BCRP structure that could affect proper folding, plasma membrane targeting, and biogenesis of the transporter. In contrast, the basic residue Arg383 in the cytosolic region near TM1 seems to be crucial for BCRP biogenesis (Polgar et al., 2009).
Instead, mutations of some of these basic residues altered BCRP activity. Notably, replacing Lys452 or His457 with Ala (K452A or H457A) markedly increased the efflux of MX, BODIPY-prazosin, and Hoechst33342 (Table 1). Likewise, K452A and H457A conferred significantly increased resistance to MX and SN-38 compared with wild-type BCRP (Table 2). Such a “gain of function” also occurred with mutations of other residues within TMs. For example, R482G and R482T exhibited markedly increased resistance to MX (Miwa et al., 2003; Robey et al., 2003). The natural variant of bovine ABCG2, Y581S, at position 581 within TM5 also showed a 1.4-fold increase in MX efflux (Merino et al., 2009). It is worth noting that K452A and H457A are not selected during evolution. In contrast, substitutions of Lys453 and Arg465 with Asp (K453D) or Ala (R465A) caused a nonselective global reduction in BCRP activity (Tables 1 and 2). Similarly, one earlier study showed that mutation of His630 within TM6 to Glu or Leu reduced resistance of BCRP to MX and SN-38 by 50% (Miwa et al., 2003). Conversion of Lys473 to Ala had no significant effect on BCRP activity. These results suggest that certain basic residues within or near TM2 are important for BCRP activity.
The exact mechanism by which these mutations affect BCRP activity is not known. Docking calculations of the substrates tested to the BCRP model suggest that His457 and Arg465 may be directly involved in substrate binding (Fig. 8 and Supplements 1–3). Hence, mutations of His457 and Arg465 might affect BCRP activity by directly altering the interaction mode of substrates with the transporter. According to the docked models, Lys453 does not participate in BCRP–substrate interaction; instead, it may form a salt bridge with Glu585 of the opposite BCRP molecule. Salt bridges may provide protein stability, promote interactions with substrates, and/or facilitate α-helix formation (Popot and Engelman, 2000). Because Glu585 is located at the extracellular membrane interface of TM5, this potential salt bridge might play an important role in the linkage of TM2 of one BCRP monomer with TM5 of the opposite monomer, leading to formation of the inward-facing form of the BCRP dimer that is closed from the extracellular side. Substitution of Lys453 with Asp would abolish the salt bridge and affect the shape of substrate binding sites and the conformation of BCRP, leading to an overall loss of transport activity. We will test this possibility in future work. Lys452 and Lys473 are located at the membrane interfaces of TM2 and not directed toward the central cavity, and thus do not appear to participate in substrate binding (Fig. 8 and Supplements 1–3). Collectively, Lys452, Lys453, His457, and Arg465 might be directly involved in or indirectly affect BCRP–substrate interaction. Mutations of Lys452 and Lys453 showed opposite effects on BCRP activity (Tables 1–3) even though they are next to each other in protein sequence. According to the BCRP model, the side chains of Lys452 and Lys453 point toward the outside and inside of the drug translocation pathway, respectively. Thus, the two Lys residues do not seem to have a strong interaction with each other.
K452A and H457A were associated with a 50 to 70% decrease in Km and a 2- to 5-fold increase in Vmax/Km for ATP hydrolysis, whereas the Vmax/Km values of K453D and R465A were decreased by 40 to 70%, and the Km values of K453D and R465A did not change much (Table 3). This suggests that ATP binding affinity and/or the efficiency of ATP hydrolysis are increased for K452A and H457A, but decreased for K453D and R465A, thus affecting BCRP activity accordingly. It is not uncommon that mutations in a MSD of an ABC transporter could affect interaction with ATP and thus transport activity. For example, replacing a Pro residue in the third MSD of MRP1 with Ala caused an increase in transporting 17β-estradiol 17β-d-glucuronide that was associated with a 4-fold decrease in Km for ATP (Koike et al., 2004). More importantly, the Km and Vmax/Km values for basal ATPase activity of R482T with a similar gain of function have also been shown to be decreased and increased, respectively (Pozza et al., 2006). As to why mutations of these basic residues affect ATPase activity is not known. By analyzing projection structures of BCRP from two-dimensional crystals, we have provided direct evidence that the presence of a BCRP substrate can induce conformational changes (M. F. Rosenberg, Z. Bikadi, J. Chan, X. Liu, Z. Ni, X. Cai, R. C. Ford, and Q. Mao, submitted for publication). Substrate binding may set BCRP to an intermediate state in the transport cycle, which might be critical for its overall transport activity. It might be possible that, in addition to the direct effects on BCRP–substrate interactions at the drug binding sites as discussed above, mutations of the basic residues cause subtle conformational changes in BCRP at a distance. If this is the case, such changes could alter substrate binding-induced state transition of BCRP in the transport cycle, thus contributing to the altered drug binding, transport, and/or ATPase activity. In support of this hypothesis, we demonstrated concentration-dependent effects of prazosin on 5D3 binding to wild-type and mutant BCRP. The 5D3 antibody recognizes a conformation-sensitive extracellular epitope in BCRP. Prazosin differentially increased 5D3 binding to wild-type BCRP, K452A, K453D, H457A, and R465A, but had little effect on 5D3 binding to K473A (Fig. 6). Intriguingly, the apparent dissociation constant of the prazosin complex with K452A or H457A was increased approximately 3- to 4-fold compared with wild-type BCRP, suggesting that the association rate of prazosin to BCRP may be decreased and/or prazosin could dissociate from the BCRP–prazosin complex more readily. Given the nature of gain of function associated with K452A and H457A, a similar observation was reported in which R482T was shown to be much less intensively photolabeled by a photoactive analog of Rho123 than wild-type BCRP (Alqawi et al., 2004), indicating the binding affinity of the photoactive substrate to R482T was decreased even though R482T can effectively transport Rho123, but wild-type BCRP cannot. It is worth noting that subtle conformational changes may occur even in mutants that do not show apparent functional changes. K473A appears to be the case that revealed a completely different pattern of 5D3 binding (Fig. 6) and increased resistance to trypsin digestion (Fig. 7) compared with wild-type protein, but showed no activity changes. At present, it is impossible to describe exactly what conformational changes may be caused by mutations of these basic residues.
In general, the experimental data of this study appear to be in good agreement with the results of homology modeling and docking calculations. Docking calculations provided additional insight into BCRP–substrate interactions. In addition to His457 and Arg465, other polar residues, Ser461 and Tyr464 of TM2, and Asn629 of TM6, might form hydrogen bonds with MX (Fig. 8, B and C). Other substrates were calculated to form hydrogen bonds with different combinations of polar residues such as Arg482 of TM3 and Ser622 and Trp627 within or near TM6 (Supplements 1–3). This might be caused by the central binding cavity that is large enough to allow individually distinct, but partially overlapping, binding interactions for different substrates. Similarly, recent crystal structures of P-gp have shown that two different ligands can bind simultaneously to P-gp at partially overlapping, but distinct, sites in the same binding pocket (Aller et al., 2009). The existence of a similar large drug binding pocket in a BCRP dimer may explain why the mutation of His457 revealed the obviously differential effects on the efflux of different substrates (Table 1) and the earlier observation that two different substrates did not reciprocally inhibit the efflux of the other (Nakanishi et al., 2003). In addition, the involvement of a large number of hydrophobic residues in substrate interactions (e.g., Ile412, Leu416, Phe489, Phe511, Leu581, and Leu626 for MX) suggests that the nature of substrate binding may not be solely based on hydrophilic interactions (Fig. 8, B and C). We realize that the results of docking calculations should be interpreted with caution when a homology model possessing a low sequence identity to the template is used. Clearly, more extensive mutagenesis studies are required in future work to validate the functional importance of the residues predicted in docking calculations.
In summary, this study demonstrates that certain basic resides within or near TM2 of BCRP may play an important role in BCRP–substrate interaction. TM2 of BCRP, along with other TMs, might form the large central drug binding cavity and the substrate translocation pathway as predicted by homology modeling and docking calculations. Further mutagenesis studies are needed to verify the predicted features.
Supplementary Material
Acknowledgments
We thank Dr. Susan E. Bates (National Cancer Institute, Bethesda, MD) for the pcDNA3.1 plasmid containing full-length human BCRP cDNA and the Drug Synthesis and Chemistry Branch, National Cancer Institute, National Institutes of Health (Bethesda, MD) for FTC.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM073715].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.109.163493.
 The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- BCRP
- breast cancer resistance protein
- MRP
- multidrug resistance protein
- P-gp
- P-glycoprotein
- HEK
- human embryonic kidney
- FTC
- fumitremorgin C
- MEM
- Eagle's minimum essential medium
- PBS
- phosphate-buffered saline
- FBS
- fetal bovine serum
- TM
- transmembrane
- MSD
- membrane-spanning domain
- MX
- mitoxantrone
- Dox
- doxorubucin
- Rho123
- rhodamine 123 (2′-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-5′-bi-1H-benzimidazole)
- mAb
- monoclonal antibody
- DAPI
- 4′,6-diamidino-2-phenylindole
- SN-38
- 7-ethyl-10-hydroxy-camptothecin
- ABC
- ATP-binding cassette
- BODIPY
- boron-dipyrromethene.
References
- Aller et al., 2009.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, et al. (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323:1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqawi et al., 2004.Alqawi O, Bates S, Georges E. (2004) Arginine482 to threonine mutation in the breast cancer resistance protein ABCG2 inhibits rhodamine 123 transport while increasing binding. Biochem J 382:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikadi and Hazai, 2009.Bikadi Z, Hazai E. (2009) Application of the PM6 semiempirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark et al., 2006.Clark R, Kerr ID, Callaghan R. (2006) Multiple drug binding sites on the R482G isoform of the ABCG2 transporter. Br J Pharmacol 149:506–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conseil et al., 2006.Conseil G, Deeley RG, Cole SP. (2006) Functional importance of three basic residues clustered at the cytosolic interface of transmembrane helix 15 in the multidrug and organic anion transporter MRP1 (ABCC1). J Biol Chem 281:43–50 [DOI] [PubMed] [Google Scholar]
- Doyle et al., 1998.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. (1998) A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA 95:15665–15670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejendal et al., 2006.Ejendal KF, Diop NK, Schweiger LC, Hrycyna CA. (2006) The nature of amino acid 482 of human ABCG2 affects substrate transport and ATP hydrolysis but not substrate binding. Protein Sci 15:1597–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo et al., 2001.Honjo Y, Hrycyna CA, Yan QW, Medina-Pérez WY, Robey RW, van de Laar A, Litman T, Dean M, Bates SE. (2001) Acquired mutations in the MXR/BCRP/ABCP gene alter substrate specificity in MXR/BCRP/ABCP-overexpressing cells. Cancer Res 61:6635–6639 [PubMed] [Google Scholar]
- Huey et al., 2007.Huey R, Morris GM, Olson AJ, Goodsell DS. (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152 [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty et al., 2007.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525–528 [DOI] [PubMed] [Google Scholar]
- Koike et al., 2004.Koike K, Conseil G, Leslie EM, Deeley RG, Cole SP. (2004) Identification of proline residues in the core cytoplasmic and transmembrane regions of multidrug resistance protein 1 (MRP1/ABCC1) important for transport function, substrate specificity, and nucleotide interactions. J Biol Chem 279:12325–12336 [DOI] [PubMed] [Google Scholar]
- Leslie et al., 2003.Leslie EM, Létourneau IJ, Deeley RG, Cole SP. (2003) Functional and structural consequences of cysteine substitutions in the NH2 proximal region of the human multidrug resistance protein 1 (MRP1/ABCC1). Biochemistry 42:5214–5224 [DOI] [PubMed] [Google Scholar]
- Maliepaard et al., 2001.Maliepaard M, Scheffer GL, Faneyte IF, van Gastelen MA, Pijnenborg AC, Schinkel AH, van De Vijver MJ, Scheper RJ, Schellens JH. (2001) Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Res 61:3458–3464 [PubMed] [Google Scholar]
- Mao et al., 2004.Mao Q, Conseil G, Gupta A, Cole SP, Unadkat JD. (2004) Functional expression of the human breast cancer resistance protein in Pichia pastoris. Biochem Biophys Res Commun 320:730–737 [DOI] [PubMed] [Google Scholar]
- Mao and Unadkat, 2005.Mao Q, Unadkat JD. (2005) Role of the breast cancer resistance protein (ABCG2) in drug transport. Aaps J 7:E118–E133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt et al., 2006.McDevitt CA, Collins RF, Conway M, Modok S, Storm J, Kerr ID, Ford RC, Callaghan R. (2006) Purification and 3D structural analysis of oligomeric human multidrug transporter ABCG2. Structure 14:1623–1632 [DOI] [PubMed] [Google Scholar]
- Merino et al., 2009.Merino G, Real R, Baro MF, Gonzalez-Lobato L, Prieto JG, Alvarez AI, Marques MM. (2009) Natural allelic variants of bovine ATP-binding cassette transporter ABCG2: increased activity of the Ser581 variant and development of tools for the discovery of new ABCG2 inhibitors. Drug Metab Dispos 37:5–9 [DOI] [PubMed] [Google Scholar]
- Mitomo et al., 2003.Mitomo H, Kato R, Ito A, Kasamatsu S, Ikegami Y, Kii I, Kudo A, Kobatake E, Sumino Y, Ishikawa T. (2003) A functional study on polymorphism of the ATP-binding cassette transporter ABCG2: critical role of arginine-482 in methotrexate transport. Biochem J 373:767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa et al., 2003.Miwa M, Tsukahara S, Ishikawa E, Asada S, Imai Y, Sugimoto Y. (2003) Single amino acid substitutions in the transmembrane domains of breast cancer resistance protein (BCRP) alter cross-resistance patterns in transfectants. Int J Cancer 107:757–763 [DOI] [PubMed] [Google Scholar]
- Miyake et al., 1999.Miyake K, Mickley L, Litman T, Zhan Z, Robey R, Cristensen B, Brangi M, Greenberger L, Dean M, Fojo T, et al. (1999) Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: demonstration of homology to ABC transport genes. Cancer Res 59:8–13 [PubMed] [Google Scholar]
- Nakanishi et al., 2003.Nakanishi T, Doyle LA, Hassel B, Wei Y, Bauer KS, Wu S, Pumplin DW, Fang HB, Ross DD. (2003) Functional characterization of human breast cancer resistance protein (BCRP, ABCG2) expressed in the oocytes of Xenopus laevis. Mol Pharmacol 64:1452–1462 [DOI] [PubMed] [Google Scholar]
- Ozvegy-Laczka et al., 2005.Ozvegy-Laczka C, Köblös G, Sarkadi B, Váradi A. (2005) Single amino acid (482) variants of the ABCG2 multidrug transporter: major differences in transport capacity and substrate recognition. Biochim Biophys Acta 1668:53–63 [DOI] [PubMed] [Google Scholar]
- Ozvegy-Laczka et al., 2008.Ozvegy-Laczka C, Laczkó R, Hegedus C, Litman T, Várady G, Goda K, Hegedus T, Dokholyan NV, Sorrentino BP, Váradi A, et al. (2008) Interaction with the 5D3 monoclonal antibody is regulated by intramolecular rearrangements but not by covalent dimer formation of the human ABCG2 multidrug transporter. J Biol Chem 283:26059–26070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar et al., 2009.Polgar O, Ediriwickrema LS, Robey RW, Sharma A, Hegde RS, Li Y, Xia D, Ward Y, Dean M, Ozvegy-Laczka C, et al. (2009) Arginine 383 is a crucial residue in ABCG2 biogenesis. Biochim Biophys Acta 1788:1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar et al., 2006.Polgar O, Ozvegy-Laczka C, Robey RW, Morisaki K, Okada M, Tamaki A, Koblos G, Elkind NB, Ward Y, Dean M, et al. (2006) Mutational studies of G553 in TM5 of ABCG2: a residue potentially involved in dimerization. Biochemistry 45:5251–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popot and Engelman, 2000.Popot JL, Engelman DM. (2000) Helical membrane protein folding, stability, and evolution. Annu Rev Biochem 69:881–922 [DOI] [PubMed] [Google Scholar]
- Pozza et al., 2006.Pozza A, Perez-Victoria JM, Sardo A, Ahmed-Belkacem A, Di Pietro A. (2006) Purification of breast cancer resistance protein ABCG2 and role of arginine-482. Cell Mol Life Sci 63:1912–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey et al., 2003.Robey RW, Honjo Y, Morisaki K, Nadjem TA, Runge S, Risbood M, Poruchynsky MS, Bates SE. (2003) Mutations at amino acid 482 in the ABCG2 gene affect substrate and antagonist specificity. Br J Cancer 89:1971–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey et al., 2001.Robey RW, Honjo Y, van de Laar A, Miyake K, Regis JT, Litman T, Bates SE. (2001) A functional assay for detection of the mitoxantrone resistance protein, MXR (ABCG2). Biochim Biophys Acta 1512:171–182 [DOI] [PubMed] [Google Scholar]
- Robey et al., 2009.Robey RW, To KK, Polgar O, Dohse M, Fetsch P, Dean M, Bates SE. (2009) ABCG2: a perspective. Adv Drug Deliv Rev 61:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu et al., 2000.Ryu S, Kawabe T, Nada S, Yamaguchi A. (2000) Identification of basic residues involved in drug export function of human multidrug resistance-associated protein 2. J Biol Chem 275:39617–39624 [DOI] [PubMed] [Google Scholar]
- Stewart, 2009.Stewart JJ. (2009) Application of the PM6 method to modeling proteins. J Mol Model 15:765–805 [DOI] [PubMed] [Google Scholar]
- Taguchi et al., 1997.Taguchi Y, Kino K, Morishima M, Komano T, Kane SE, Ueda K. (1997) Alteration of substrate specificity by mutations at the His61 position in predicted transmembrane domain 1 of human MDR1/P-glycoprotein. Biochemistry 36:8883–8889 [DOI] [PubMed] [Google Scholar]
- Vethanayagam et al., 2005.Vethanayagam RR, Wang H, Gupta A, Zhang Y, Lewis F, Unadkat JD, Mao Q. (2005) Functional analysis of the human variants of breast cancer resistance protein: I206L, N590Y, and D620N. Drug Metab Dispos 33:697–705 [DOI] [PubMed] [Google Scholar]
- Wang et al., 2008.Wang H, Lee EW, Cai X, Ni Z, Zhou L, Mao Q. (2008) Membrane topology of the human breast cancer resistance protein (BCRP/ABCG2) determined by epitope insertion and immunofluorescence. Biochemistry 47:13778–13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu et al., 2004.Xu J, Liu Y, Yang Y, Bates S, Zhang JT. (2004) Characterization of oligomeric human half-ABC transporter ATP-binding cassette G2. J Biol Chem 279:19781–19789 [DOI] [PubMed] [Google Scholar]
- Xu et al., 2007.Xu J, Peng H, Chen Q, Liu Y, Dong Z, Zhang JT. (2007) Oligomerization domain of the multidrug resistance-associated transporter ABCG2 and its dominant inhibitory activity. Cancer Res 67:4373–4381 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.