Abstract
Amyloid fibrils are filamentous protein aggregates that accumulate in diseases such as Alzheimer’s or type II diabetes. The amyloid-forming protein is disease specific. Amyloids may also be formed in vitro from many other proteins, after first denaturing them. Unlike the diverse native folds of these proteins, their amyloids are fundamentally similar in being rigid, smooth-sided, and cross-β-structured, that is, with β strands running perpendicular to the fibril axis. In the absence of high-resolution fibril structures, increasingly credible models are being derived by integrating data from a crossfire of experimental techniques. Most current models of disease-related amyloids invoke “β arcades,” columnar structures produced by in-register stacking of “β arches.” A β arch is a strand-turn-strand motif in which the two β strands interact via their side chains, not via the polypeptide backbone as in a conventional β hairpin. Crystal structures of β-solenoids, a class of proteins with amyloid-like properties, offer insight into the β-arc turns found in β arches. General conformational and thermodynamic considerations suggest that complexes of 2 or more β arches may nucleate amyloid fibrillogenesis in vivo. The apparent prevalence of β arches and their components have implications for identifying amyloidogenic sequences, elucidating fibril polymorphisms, predicting the locations and conformations of β arcs within amyloid fibrils, and refining existing fibril models.—Kajava, A. V., Baxa, U., Steven, A. C. β arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils.
Keywords: amyloidogenicity, polymorphism, prion, misfolding, structural motif
It has long been known that certain diseases, particularly neurodegenerative diseases, are accompanied by the accumulation of amyloid fibrils: indigestible, cross-β-structured, polymers of normally innocuous proteins (usually of a proteolytic fragment of such a protein). About 20 different proteins have been identified as forming such fibrils, and their respective amyloidoses correlate specifically with different diseases (1). More recently, it has been discovered that amyloid fibrils may be produced in vitro from many other proteins by procedures that typically involve denaturing them and then removing the denaturant (2, 3). The latter systems are informative about the folding properties of polypeptide chains generally. However, the potential implications for human health have focused major interest on those amyloids that have demonstrated correlation with diseases, although we note that the nature of this correlation may differ between systems. In this context it is important to distinguish between pathogenicity and infectivity. For instance, there is evidence in some cases that the pathogenic form of a protein is not the amyloid fibril but a small oligomer (4), with the amyloid representing a passive by-product of the disease process. On the other hand, there is little doubt that, for the fungal prions, amyloid fibrils are indeed the infectious agent (5, 6).
Over the past decade, substantial progress has been made toward elucidating the structural arrangements in amyloid fibrils. Although these specimens remain refractory to the classical approach of X-ray crystallography, progress has stemmed largely from the application of experimental techniques such as solid state nuclear magnetic resonance, cryo-electron microscopy, scanning transmission electron microscopy mass measurements, and electron paramagnetic resonance spectroscopy of spin-labeled derivatives, as well as longer-established approaches such as X-ray fiber diffraction, conventional electron microscopy, and optical spectroscopy (7,8,9,10,11,12). At the same time, there has been progress in understanding the interactions of β strands in microcrystals of short amyloidogenic peptides (∼6 residues) (13, 14). Based on these and other experimental constraints, numerous structural models for amyloid and prion fibrils (prions are infectious amyloids) have been formulated (9). A major milestone has been the demonstration of parallel, in-register, arrangements of β strands in several kinds of amyloid fibrils (7, 8, 10, 15). An important additional insight has been the realization that there is a class of monomeric and oligomeric proteins, called β-solenoids, whose structural properties closely resemble those of amyloids in that they also have parallel, in-register, cross-β structures (with the qualification that the β strands stacked in register in β-solenoids tend to have similar rather than identical amino acid sequences, as they are in most amyloid fibrils). The growing set of high-resolution crystal structures of β-solenoid proteins affords a source of potential insight into the organization of amyloid fibrils (16). The status of the relationship between known β-solenoid structures and putative models of amyloid fibrils, and its implications, are central themes of this review. Analysis of these structures yields an enhanced understanding of their common structural determinants and sheds light on how they form and function. We also focus on the turn/loop regions that are necessary parts of amyloid fibrils assembled from longer peptides (>∼15 residues or so) and discuss their roles in stability, nucleation, and fibril polymorphism.
STRUCTURAL MODELS OF NATURALLY OCCURRING, DISEASE-RELATED, AMYLOID FIBRILS
Tightening experimental constraints have led to the formulation of progressively more precise structural models for amyloid fibrils linked to the most frequent and severe age-related and neurodegenerative diseases. Among them are Aβ fibrils (17, 18) and tau fibrils (19,20,21) related to Alzheimer’s disease; fibrils formed by glutamine expansion-containing proteins found in Huntington’s disease and other related diseases (22,23,24); fibrils of CA150 protein that are codeposited with huntingtin in Huntington’s disease (25); α-synuclein fibrils of Parkinson’s disease (26); and human amylin fibrils related to type 2 diabetes (27,28,29). Models have also been proposed for fibrils formed by the yeast prion proteins Ure2p and Sup35p (15, 23, 30, 31), the fungal HET-s prion protein (32, 33), and the human PrP protein, which is the main component of the infectious transmissible spongiform encephalopathy agent (34, 35). In addition, models have been suggested for amyloid fibrils of β2-microglobulin and insulin (9, 36, 37) formed in patients receiving hemodialysis and insulin injections, respectively.
These models vary in their level of plausibility, depending on the number and stringency of the experimental constraints that have been established. Although a quantitative measure cannot be given for the confidence to be placed on these models, they can be approximately ranked. For example, the fungal HET-s prion (32) and variants of Aβ fibrils (17, 18) can be considered as the most rigorous models currently available, but the other models listed above are supported by enough data to merit serious consideration.
All the above-listed fibrils are known to have cross-β structures. However, some disease-related fibrils are formed by stacks of native or refolded globular structures and do not necessarily exhibit cross-β structure. For example, the fibrils of transthyretin (38, 39), cystatin C (40), and mutants of superoxide dismutase (41) consist of stacks of globular dimers. Here we will focus on cross-β fibrils and will not consider further fibrils formed by globular protomers, which have different structures and mechanisms of fibrillogenesis.
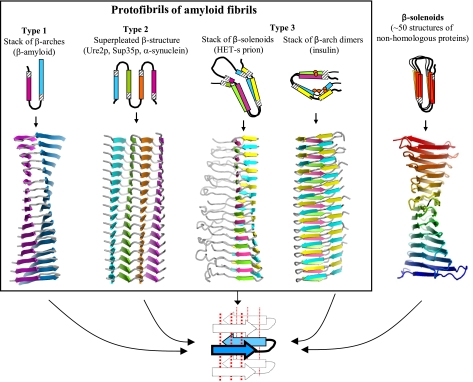
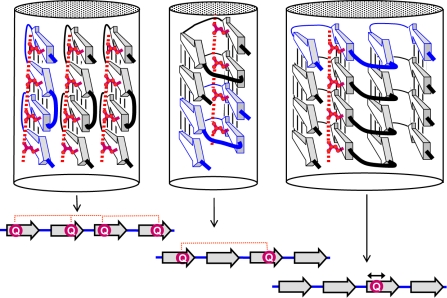
One characteristic common to many cross-β models is their parallel and in-register nature (8). Despite this commonality, the models look quite different. The envisaged amyloids vary in the number of protofibrils that are assembled into a fibril and in the structure of their individual protofibrils. Topologically, the protofibril models can be classified into 3 types (Fig. 1).
Figure 1.
Three types of cross-β models for amyloid protofibrils. Top: axial views of the repetitive structural units (rectangles represent β strands); bottom: lateral views of protofibrils formed by stacking the corresponding repetitive units. Orange circles in the insulin model show Cys residues forming disulfide bonds. Beneath, schematic diagram of a β arcade, considered as a structural motif common to all 3 types of models. One β arch is colored in blue, with depth cuing; arrows indicate β strands; dotted lines show H bonds. Right: crystal structure of the SufD dimer (69), shown as an illustrative β-solenoid with a stacking of β arches similar to that envisaged in the amyloid models.
The first type includes models proposed for fibrils of Aβ and of the K3 fragment of β2-microglobulin, one kind of fibril formed by human amylin, and fibrils of CA150 protein (17, 18, 25, 29, 37). These structures are formed by axial stacking of identical structural elements consisting of 2 long β strands connected by a turn. The repetitive elements, which are β arches (16, 42), are H bonded along the fibril axis and form a double layer of parallel β sheets.
Models of the second type, which we call parallel superpleated β structures, have been proposed for fibrils of Ure2p, Sup35p, α-synuclein, poly-Gln tracts, amylin (3-stranded fibrils), tau, and the B1 domain of IgG binding protein G (19, 23, 26, 28, 43). In these models, each polypeptide chain has several β strands connected by loops, and zigzags in a planar serpentine fold. These serpentines are stacked axially, in register, generating an array of parallel β sheets (Fig. 1). The number of strands, Ns (which equals the number of sheets), may, in principle, vary. If Ns = 2, this structure simplifies to a type 1 model. The example shown in Fig. 1 has Ns = 4.
The third type of model, applicable to fibrils of the fungal HET-s prion, is based on axial stacking of 2-coil β-solenoids (32). The sequence motifs of the 2 coils are similar but not identical. Each coil has 2 long and 2 short β strands (Fig. 1). In principle, this model may be generalized by stacking β-solenoids that have more than 2 coils. Insulin fibrils may be also considered as of this type (Fig. 1). In the latter case, the repetitive module is a dimer of similar but not identical peptides, covalently connected by 2 disulfide bonds. Within the dimer, these peptides form 2 coils that interact with each other via β-structural H bonding. In turn, the dimers stack axially, forming parallel cross-β structures in which the 2 peptides alternate along the fibril axis.
STACKING OF β ARCHES AS A COMMON STRUCTURAL MOTIF
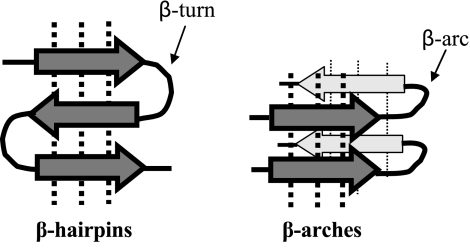
These models all invoke stacks of β arches (Fig. 1). The β arch is a strand-loop-strand motif that attracted our attention when surveying several dozen β-solenoid structures that had been solved by X-ray crystallography (16, 42). In the vast majority of proteins with known globular structures, strand-loop-strand motifs are primarily β hairpins in which the 2 strands form an antiparallel β sheet (Fig. 2). In the β arch, however, each strand is relatively rotated by ∼90° around its axis so that they interact via the side chains, not via the polypeptide backbones. When a β arch is stacked into a β arcade, its 2 strands are integrated into 2 different β sheets. Unlike conventional globular proteins in which β arches are rare (44), β-solenoids are built almost entirely of these motifs, whereby β arches stack in register to form β arcades.
Figure 2.
β hairpins and β arches. Arrows indicate β strands, dotted lines show H bonds.
Of the 3 types distinguished in Fig. 1, fibrils of types 1 and 2 are formed by stacking identical β arches. In type 3 fibrils, 2 or more β arches with differing but related sequences alternate along the fibril.
Each β arcade has a double-layer structure in which 2 parallel in-register β sheets face each other (Fig. 2). The stabilizing effect of sheet-on-sheet packing was already recognized in models formulated for Aβ and Ure2p fibrils (17, 18, 23, 45) and in crystal structures in which short peptides engage in amyloid-like interactions (13). The latter structures revealed details of side-chain interactions inside the double β sheet, also called a cross-β spine (13, 14). The side chains protruding from apposing β sheets form a tight interdigitated packing, also called a steric zipper. Additional stabilization may come from H-bonded axial stacking in “ladders” of Gln or Asn side chains. It is noteworthy that in crystallites of short peptides (13), Gln side chains also form H bonds with the backbones of the apposing β sheet. As a result, the interior Gln side chains knit the double β sheets together via networks of H bonds, both axial and lateral (intersheet). In terms of its spatial network of H bonds, this structure resembles polyglycine, which also forms stable insoluble aggregates (46, 47). Models of Aβ fibrils also suggest that oppositely charged side chains can stabilize the double β sheet by intersheet ionic bonds (17, 18). In this scenario, “knobs-into-holes” packing of the sheets, supplemented by H bonds and ionic bonds formed by interior polar side chains, imparts the high stability characteristic of amyloid fibrils.
β ARCS CAN STABILIZE PARALLEL, IN-REGISTER, CROSS-β STRUCTURES
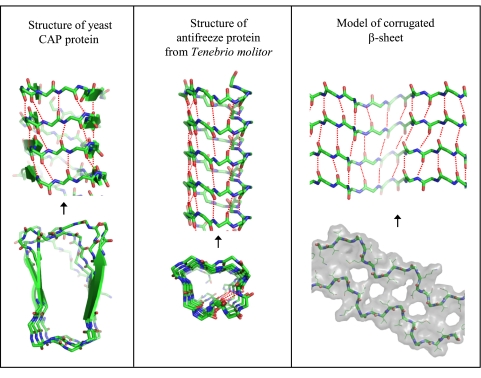
In a β arcade, one may distinguish two principal structural elements: a double β sheet and a stack of β arcs. The importance of the double β sheet for fibril stability is discussed above. Less is known about the interactions of β arcs, as structural data on fibrils are not yet sufficiently detailed to define the arc conformations. The most reliable source of information on these conformations comes from crystal structures of β-solenoids (16, 42, 48, 49). Analysis of these structures has revealed that β arcs assume a limited number of preferred conformations, which are coded by different sequence motifs (42). Of these, U-turn β arcs, which are the most relevant to amyloid fibrils, have 3 to 7 residues. Moreover, it was shown that in β-arc conformations, turns can be completed without interrupting the axial pattern of backbone H bonding (Fig. 3). This observation implies that stacks of β arcs can enhance the stability of β arcades rather than detracting from it. In line with this inference, of 20 axially H-bonded peptide groups in the recently determined structure of HET-s fibrils, 7 residues belong to β arcs (32).
Figure 3.
Lateral and axial views of some structures containing inter-β-arc H bonding. Left: crystal structure of yeast CAP protein (70); middle: crystal structure of antifreeze protein from Tenebrio molitor (50); right: model of corrugated β-structure of Aβ (unpublished results) built with reference to cryo-electron microscopy data (11). Only the polypeptide backbone is shown (except in the axial view of the corrugated structure). Dotted lines show H bonds.
Carried to the extreme, the tenet that β arcs can be stabilizing elements of cross-β structures predicts that amyloid-like fibrils may, in principle, be formed by stacking polypeptides whose folds are made up of β arcs without any β strands. The crystal structure of antifreeze protein from Tenebrio molitor (50) provides a good example of such a strandless β-solenoid. As for amyloid fibrils, a form of Aβ fibril with paired “corrugated β sheets“ has been recently described on the basis of cryo-electron microscopy showing the fibril’s axial profile at 8-Å resolution (11); molecular modeling suggests that this structure might correspond to a tandem run of several alternately inverted β arcs (Fig. 3, right).
β ARCHES MAY FORM NUCLEATION COMPLEXES FOR AMYLOID FIBRILLATION
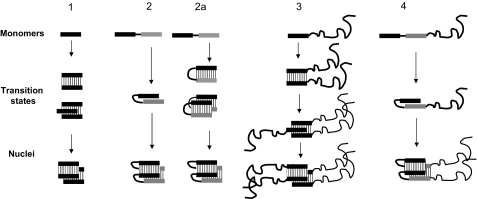
Monomers of amyloid-forming proteins tend to remain in solution for extended periods before assembling rapidly into fibrils (51,52,53). The lag period indicates that nucleation poses the major energy barrier on the pathway from unstructured monomers to well-ordered fibrils. For short peptides (∼6 residues), it has been suggested that the transition-state structures, which determine the kinetic barrier to fibril formation, are either 2 molecules in a single sheet or this pair interacting with 1 molecule from the apposing sheet, leading to a nucleus consisting of 3–4 molecules, 2 in one sheet interdigitated with 1–2 in the apposing sheet (13) (Fig. 4, pathways 1 and 3). It was also pointed out that in their initial transient state, unfolded peptides are well exposed to solvent and therefore cannot provide a significant change in the enthalpy of the system; however, they can pose a substantial entropic barrier (54). Fibrils formed by longer peptides (>∼15 residues), which fully engage in amyloid structures, that is, without protruding flexible tails, should have at least 1 turn, that is, a β arch, suggesting a different scenario. An intramolecular conformational change such as formation of a β arch or β hairpin (Fig. 4, pathways 2 and 2a) is independent of monomer concentration and therefore is energetically more favorable, because of the entropic component, than intermolecular associations of 2 or more peptides (Fig. 4, pathways 1 and 3). Given that a single β arch or β hairpin can also contribute favorable van der Waals, H bond, and ionic bond interactions to the enthalpy of the system, these structures are plausible intermediate states on the fibrillation pathways with the lowest transition states. The nucleus may be provided by the stacking of 2 β arches (Fig. 4, pathway 2). If, instead, one were to imagine that the early intermediate structure is not a β arch but a β hairpin, the step of intermolecular interaction could be accompanied by reorientation of peptide groups in the β hairpin, leading to a 2-β-arch nucleus (Fig. 4, pathway 2a).
Figure 4.
Hypothetical pathways of amyloid fibril assembly. Bars represent β strands; curved lines represent other parts of the polypeptide chains. In some cases, two similar but nonidentical strand-forming peptides are shown in different shades. Thin lines between bars represent H bonds. Pathway 1: short amyloid-forming peptides (<15 residues or so) form dimers, trimers (transition state), and then tetrameric double-β-sheet nucleus (13). Pathway 2: long peptides (>15 residues or so) form a β-arch (transition state) and then a double-β-sheet nucleus. Pathway 2a: long peptides (>15 residues or so) form β hairpins, and then 2 β hairpins side by side (transition state), which transforms into a double-β-sheet nucleus. Pathway 3: short amyloid-forming peptides with nonamyloidogenic flanking regions follow a pathway similar to pathway 1. Pathway 4: long peptides with nonamyloidogenic flanking regions follow a pathway similar to those of pathway 2 or 2a.
Evidently, the location of a β arc within a given peptide should depend on the amino acid sequence. β-arch nuclei could be formed by β-arc-favoring sequence motifs (42) or typical β-turn motifs (44, 52, 55) and by favorable van der Waals, H-bond (Asn and Gln), and ionic interactions of side chains between adjacent β strands (13, 23, 24, 45, 56). Among energetically equivalent β arches, those with fewer residues in their β arcs pose lower entropic barriers (the decrease is proportional to the logarithm of number of residues in the loop; ref. 57) and therefore represent the most likely fibril-forming structures. Moreover, as noted above, peptide groups in “tight” standard β arcs (42) can form interarc H bonds of the same strength as those in β strands. Thus, it is plausible that in at least some cases, the nuclei for fibrillation are short stacks of β arcs; consistent with this idea, some mutations in putative turn regions have been found to affect amyloid fibril formation (52).
Both short and long amyloidogenic peptides form fibrils in vitro at concentrations >1 μM; however, it does not necessarily follow that these peptides will also fibrillize equally well under in vivo conditions. Due to the bi- and trimolecular transition states of short peptides, the increase in entropic barrier to fibrillation that accompanies decreasing monomer concentration will be stronger (proportional to at least the logarithm of concentration) than the barrier to monomolecular β-arch formation. Therefore, longer β-arch-forming peptides should retain their ability to fibrillize in vivo better than short peptides. In fact, peptides of <∼15 residues rarely reach fibril-forming concentrations in human cells because once they are produced, they are rapidly degraded by endogenous proteases (58). A short fibril-forming region may also occur within a longer polypeptide chain. As with the short peptide alone, fibrillation of this protein via the short peptide pathway with a nucleus of 3 to 4 molecules (Fig. 4, pathway 3) should diminish with decreasing concentration. The flanking regions could inhibit this process in 2 ways: by steric impedance or interactions with the amyloid-forming region, and by the increased entropic barrier due to motion in the flanking region when in the transition state structure (59). In disordered flanking regions, this additional increase of the kinetic barrier should be proportional to the logarithm of the number of residues. In contrast to short peptides, β-arch-containing fibrillizing peptides should be less sensitive to inhibitory effects from flanking regions, because the β-arch transition state consists of fewer molecules and therefore has fewer inhibiting flanking regions (Fig. 4, pathways 3 vs. 4).
These general considerations suggest that β arches may provide nuclei for fibrillogenesis under conditions characterized by low concentrations of monomers and interference from flanking regions. Several observations are consistent with this hypothesis. First, all known naturally occurring amyloid-forming proteins have amyloidogenic regions that are longer than 15 residues. Second, although it has been shown that fusing short amyloidogenic peptides with soluble proteins can trigger fibrillation in vitro at high concentrations of 300–600 μM (60, 61), the same results were not observed at lower in vivo-like concentrations.
POLYMORPHISM OF AMYLOID FIBRILS AND OF β-ARCADE STRUCTURES
β arcades are stacked columns of identical β arches. In such arrangements, the same side chains recur in the same positions and are axially nearest neighbors (Fig. 5). The interior of the fibril may accommodate both apolar residues and uncharged polar Asn and Gln engaged in H-bonded ladders, as well as Ser, Thr, and Cys, interacting with each other and/or with the polypeptide backbone as in known β-solenoid structures (16, 42). The ability of β arches to accommodate both polar and apolar internal residues increases the number of peptide sequences with the potential to form β arcades. Furthermore, placing the β arcs at all possible positions in an amyloidogenic sequence results in many different β-arcade structures, while preserving the in-register arrangement. Accordingly, the axial stacks of both apolar side chains and H-bonded Asn or Gln side chains are not disrupted. These considerations imply that one amyloidogenic peptide may form several different β-arcade structures with similar energies. Such a peptide may, in principle, assume different β arches when generated from different nucleators. Once a β-arch nucleus is formed, it should impose the same structure on other like peptides recruited into the growing fibril. Unlike parallel in-register arrangements (Fig. 5, middle and right), an antiparallel cross-β structure (Fig. 5, left) requires a particular periodic pattern of Asn and/or Gln residues in the sequence to have stabilizing H-bond ladders within the fibril. This restriction makes antiparallel β structures less likely for amyloid fibrils. Nevertheless, peptides with a certain sequence periodicity of Gln and/or Asn residues or poly-Asn and poly-Gln peptides can form fibrils with antiparallel stacking of β arches (24, 62). Such a model has recently been suggested for fibrils of a particular mutant of the Aβ peptide (62).
Figure 5.
Distinct cross-β structures. Left: a fibril formed from antiparallel cross β-structures, in this case β meanders packed side by side; middle: fibril formed by a stacking of β-solenoids (one shown with black turns; one with blue turns); right: fibril formed from in-register stacking of β arches (parallel superpleated structure). In these examples, each monomer has 4 β strands (arrows). Gln side chains in H-bonded ladders located inside the fibrils are shown in red. Bottom: red circles indicate locations of Gln residues (Q) within the corresponding peptides that form H-bonded ladders. Models are arranged from left to right in order of progressive relaxation of the requirement for specific sequence motifs. In the model at right, a Gln in any position produces a ladder. Ladders based on differing serpentine folds of the same polypeptide chain are, in principle, possible, which would provide a structural basis for fibril polymorphism. The other two structures require a specific distance between the Glns for a ladder to be formed, and this constraint reduces the number of possible structures. We consider that antiparallel β-structured fibrils formed from peptides that are rich in Gln and/or Asn and whose stability depends on ladders of these residues are likely to be rare.
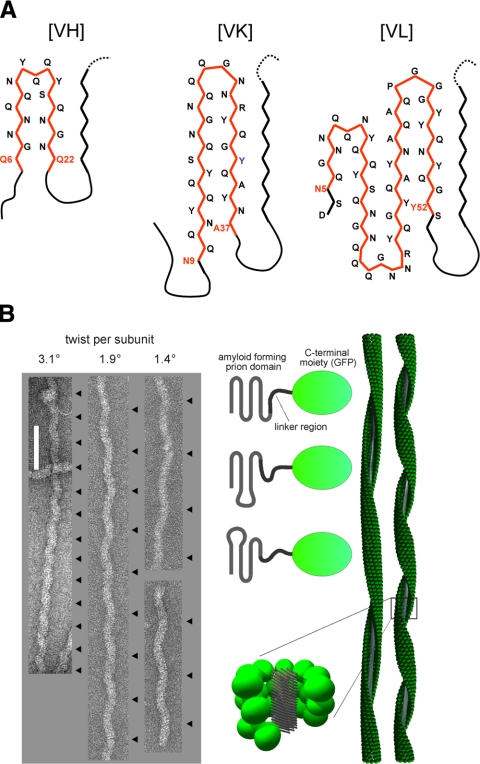
The adaptability ascribed above to arrangements of axially stacked polypeptides offers an explanation for the observed polymorphism of amyloid fibrils (63,64,65). Polymorphism may be expressed either by structural differences in the elementary protofibril and/or by different numbers or different associations of essentially the same protofibril. Polymorphisms of the former kind are difficult to characterize by electron microscopy, as protofibrils tend to be smooth surfaced and hence generate little contrast; moreover, the axial step per subunit tends to be short (4.7 Å or small multiples thereof) and therefore difficult to detect at normal operating resolutions. However, several different morphological types have been distinguished for single-protofibril filaments of the yeast prion protein Sup35 and correlated with different strains, although this is not a one-to-one correlation (63) (Fig. 6A). Similarly, single-protofibril filaments of the Ure2p prion domain fused with GFP express their polymorphism in terms of “corkscrew” morphologies whose axial repeats vary from ∼50 to ∼200 nm but are consistently maintained along a given filament (15, 66) (Fig. 6B). Much evidence supports the view that these filaments have amyloid fibril backbones surrounded by the globular GFP moieties (67, 68) and an axial step of 4.7 Å per subunit (67). Thus, the differing axial repeats correspond to small rotations of 0.8–3.4° per axial step. These properties underlie the working hypothesis that differences in the shape of the serpentine fold (type 2 model; Fig. 1) result in differing rotation angles and hence in the differing axial repeats that are observed (Fig. 6B).
Figure 6.
A) Polymorphism of Sup35p fibrils. Cross-sections of hypothetical models of Sup35 related to the strains of the yeast prion [PSI]: VH strain requires residues 7–21; VK strain requires residues 9–37; VL strain requires residues 5–52 (63). Each model envisages a parallel in-register stacking of the corresponding serpentine. Primary β arches are outlined in red; secondary pleats are in black. B) Axial repeat length polymorphism in Ure2p-derived filaments. Left: electron micrographs of negatively stained specimens of Ure2p (1–80)-GFP filaments that exhibit different axial repeat lengths (15). Repeats are indexed with black arrowheads at right of each filament. Scale bar = 100 nm. Right: schematic representation of 3 different variants that lead to different repeat lengths. We hypothesize that this polymorphism arises from different serpentine folds of the prion domain that result in slightly differing axial twists per subunit when the subunits are stacked in parallel in-register arrangements. Specifics of the envisaged folds are unknown, and the examples shown are simply for illustrative purposes. The corkscrew appearance of the filaments arises from clusters of globular GFP moieties precessing around the amyloid fibril backbone (middle, bottom).
To exemplify the way in which parallel in-register stacks of β arches could account for the observed polymorphism, we show hypothetical serpentine folds for 3 different variants of Sup35p that have been characterized extensively by King and coworkers (63) (Fig. 6A). Amyloidogenic sequences of >30 residues may polymerize into “superpleated” β structures (type 2 model, Fig. 1) formed by axial stacking of planar serpentine folds, involving >1 β arcade (23). The β arcades of a given superpleated β structure may observe a hierarchy whereby a primary arcade is first formed with 2 β sheets, to which additional sheets then join, forming secondary pleats (Fig. 6). We conjecture that a primary β arch should have a short β arc with a tight conformation and favorable interstrand interactions, whereas the β arcs of secondary arcades may be longer and have looser folds.
PERSPECTIVE
The recent efflorescence of information on the 3-dimensional structures of β-structured fibrous proteins and of amyloid fibrils stems from data gleaned from emerging experimental techniques. On this basis, we posit that stacks of β arches play a central role in the formation of at least some disease-related amyloid fibrils. While further and more precise experimental data are certainly acquired, this hypothesis has the potential to lead to future applications in such areas as improved prediction of amyloidogenic regions in amino acid sequences. Ultimately this may lead to patient-specific assessment of risk to develop age-related and neurodegenerative diseases. In experimental biology and biotechnology, sequence-based identification of aggregation-prone regions can contribute to improved design of soluble recombinant protein constructs. An improved appreciation of the relationship between solved β-arch structures and the corresponding amino acid sequences should facilitate identification of β-arch formers and prediction of their conformations and, eventually, to identification of the most appropriate sites for amyloidogenesis-suppressing drugs. Apart from its biological and biomedical implications, enhanced knowledge of amyloid fibril structures should open new avenues in the growing field of nanoscale engineering of biomaterials and the structure-based design and functionalization of nanofibers.
Acknowledgments
The authors thank Drs. Oleg Borisov and Christian Ligoure for discussion and suggestions. This work was supported in part by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases at the National Institutes of Health, Bethesda, MD, USA.
References
- Pepys M B. Amyloidosis. Annu Rev Med. 2006;57:223–241. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson C M. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci U S A. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani M, Dobson C M. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- Lashuel H A, Lansbury P T., Jr Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q Rev Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- King C Y, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- Brachmann A, Baxa U, Wickner R B. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzutkin O N, Balbach J J, Leapman R D, Rizzo N W, Reed J, Tycko R. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of beta-sheets in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q Rev Biophys. 2008;41:265–297. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- Benzinger T L, Gregory D M, Burkoth T S, Miller-Auer H, Lynn D G, Botto R E, Meredith S C. Propagating structure of Alzheimer’s beta-amyloid(10–35) is parallel beta-sheet with residues in exact register. Proc Natl Acad Sci U S A. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse C, Fandrich M, Grigorieff N. Paired beta-sheet structure of an Abeta(1–40) amyloid fibril revealed by electron microscopy. Proc Natl Acad Sci U S A. 2008;105:7462–7466. doi: 10.1073/pnas.0712290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Shinchuk L M, Inouye H, Wetzel R, Kirschner D A. Polyglutamine homopolymers having 8–45 residues form slablike beta-crystallite assemblies. Proteins. 2005;61:398–411. doi: 10.1002/prot.20602. [DOI] [PubMed] [Google Scholar]
- Nelson R, Sawaya M R, Balbirnie M, Madsen A O, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya M R, Sambashivan S, Nelson R, Ivanova M I, Sievers S A, Apostol M I, Thompson M J, Balbirnie M, Wiltzius J J, McFarlane H T, Madsen A O, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Baxa U, Cassese T, Kajava A V, Steven A C. Structure, function, and amyloidogenesis of fungal prions: filament polymorphism and prion variants. Adv Protein Chem. 2006;73:125–180. doi: 10.1016/S0065-3233(06)73005-4. [DOI] [PubMed] [Google Scholar]
- Kajava A V, Steven A C. Beta-rolls, beta-helices, and other beta-solenoid proteins. Adv Protein Chem. 2006;73:55–96. doi: 10.1016/S0065-3233(06)73003-0. [DOI] [PubMed] [Google Scholar]
- Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-beta(1–42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova A T, Yau W M, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai M, Langen R. Template-assisted filament growth by parallel stacking of tau. Proc Natl Acad Sci U S A. 2004;101:10278–10283. doi: 10.1073/pnas.0401911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronesi O C, von Bergen M, Biernat J, Seidel K, Griesinger C, Mandelkow E, Baldus M. Characterization of Alzheimer’s-like paired helical filaments from the core domain of tau protein using solid-state NMR spectroscopy. J Am Chem Soc. 2008;130:5922–5928. doi: 10.1021/ja7100517. [DOI] [PubMed] [Google Scholar]
- Jeganathan S, von Bergen M, Mandelkow E M, Mandelkow E. The natively unfolded character of tau and its aggregation to Alzheimer-like paired helical filaments. Biochemistry. 2008;47:10526–10539. doi: 10.1021/bi800783d. [DOI] [PubMed] [Google Scholar]
- Thakur A K, Wetzel R. Mutational analysis of the structural organization of polyglutamine aggregates. Proc Natl Acad Sci U S A. 2002;99:17014–17019. doi: 10.1073/pnas.252523899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava A V, Baxa U, Wickner R B, Steven A C. A model for Ure2p prion filaments and other amyloids: the parallel superpleated beta-structure. Proc Natl Acad Sci U S A. 2004;101:7885–7890. doi: 10.1073/pnas.0402427101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorsky P, Atkins E. New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules. 2005;6:425–432. doi: 10.1021/bm0494388. [DOI] [PubMed] [Google Scholar]
- Ferguson N, Becker J, Tidow H, Tremmel S, Sharpe T D, Krause G, Flinders J, Petrovich M, Berriman J, Oschkinat H, Fersht A R. General structural motifs of amyloid protofilaments. Proc Natl Acad Sci U S A. 2006;103:16248–16253. doi: 10.1073/pnas.0607815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der-Sarkissian A, Jao C C, Chen J, Langen R. Structural organization of alpha-synuclein fibrils studied by site-directed spin labeling. J Biol Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- Wiltzius J J, Sievers S A, Sawaya M R, Cascio D, Popov D, Riekel C, Eisenberg D. Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin) Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava A V, Aebi U, Steven A C. The parallel superpleated beta-structure as a model for amyloid fibrils of human amylin. J Mol Biol. 2005;348:247–252. doi: 10.1016/j.jmb.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Luca S, Yau W M, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Lindquist S L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Wickner R B, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci U S A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer A B, Riek R, Meier B H. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- Ritter C, Maddelein M L, Siemer A B, Luhrs T, Ernst M, Meier B H, Saupe S J, Riek R. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts C, Wille H, Prusiner S B, Cohen F E. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci U S A. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye H, Kirschner D A. X-ray fiber and powder diffraction of PrP prion peptides. Adv Protein Chem. 2006;73:181–215. doi: 10.1016/S0065-3233(06)73006-6. [DOI] [PubMed] [Google Scholar]
- Jimenez J L, Nettleton E J, Bouchard M, Robinson C V, Dobson C M, Saibil H R. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci U S A. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata K, Fujiwara T, Matsuki Y, Akutsu H, Takahashi S, Naiki H, Goto Y. 3D structure of amyloid protofilaments of beta2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci U S A. 2006;103:18119–18124. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M J. Transthyretin mutations in health and disease. Hum Mutat. 1995;5:191–196. doi: 10.1002/humu.1380050302. [DOI] [PubMed] [Google Scholar]
- Westermark P, Sletten K, Johansson B, Cornwell G G., 3rd Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci U S A. 1990;87:2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A, Jeremy Craven C, Higgins L D, Giannini S, Conroy M J, Hounslow A M, Waltho J P, Staniforth R A. Cystatin forms a tetramer through structural rearrangement of domain-swapped dimers prior to amyloidogenesis. J Mol Biol. 2004;336:165–178. doi: 10.1016/j.jmb.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Elam J S, Taylor A B, Strange R, Antonyuk S, Doucette P A, Rodriguez J A, Hasnain S S, Hayward L J, Valentine J S, Yeates T O, Hart P J. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- Hennetin J, Jullian B, Steven A C, Kajava A V. Standard conformations of beta-arches in beta-solenoid proteins. J Mol Biol. 2006;358:1094–1105. doi: 10.1016/j.jmb.2006.02.039. [DOI] [PubMed] [Google Scholar]
- Wang J, Gulich S, Bradford C, Ramirez-Alvarado M, Regan L. A twisted four-sheeted model for an amyloid fibril. Structure. 2005;13:1279–1288. doi: 10.1016/j.str.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Efimov A V. Pseudo-homology of protein standard structures formed by two consecutive beta-strands. FEBS Lett. 1987;224:372–376. [Google Scholar]
- Ma B, Nussinov R. Stabilities and conformations of Alzheimer’s beta-amyloid peptide oligomers (Abeta 16–22, Abeta 16–35, and Abeta 10–35): sequence effects. Proc Natl Acad Sci U S A. 2002;99:14126–14131. doi: 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F H, Rich A. Structure of polyglycine II. Nature. 1955;176:780–781. doi: 10.1038/176780a0. [DOI] [PubMed] [Google Scholar]
- Kajava A V. Dimorphism of polyglycine I: structural models for crystal modifications. Acta Crystallographica D. 1999;55:436–442. doi: 10.1107/s0907444998012438. [DOI] [PubMed] [Google Scholar]
- Iengar P, Joshi N V, Balaram P. Conformational and sequence signatures in beta helix proteins. Structure. 2006;14:529–542. doi: 10.1016/j.str.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Tsai H H, Gunasekaran K, Nussinov R. Sequence and structure analysis of parallel beta helices: implication for constructing amyloid structural models. Structure. 2006;14:1059–1072. doi: 10.1016/j.str.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Liou Y C, Tocilj A, Davies P L, Jia Z. Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein. Nature. 2000;406:322–324. doi: 10.1038/35018604. [DOI] [PubMed] [Google Scholar]
- Kelly J W. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- Frieden C. Protein aggregation processes: in search of the mechanism. Protein Sci. 2007;16:2334–2344. doi: 10.1110/ps.073164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson C M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Wesson M, Yamashita M. Interpretation of protein folding and binding with atomic solvation parameters. Chemica Scripta. 1989;29A:217–221. [Google Scholar]
- Grant M A, Lazo N D, Lomakin A, Condron M M, Arai H, Yamin G, Rigby A C, Teplow D B. Familial Alzheimer’s disease mutations alter the stability of the amyloid beta-protein monomer folding nucleus. Proc Natl Acad Sci U S A. 2007;104:16522–16527. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova A T, Ishii Y, Balbach J J, Antzutkin O N, Leapman R D, Delaglio F, Tycko R. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H X. Loops, linkages, rings, catenanes, cages, and crowders: entropy-based strategies for stabilizing proteins. Acc Chem Res. 2004;37:123–130. doi: 10.1021/ar0302282. [DOI] [PubMed] [Google Scholar]
- Saveanu L, Fruci D, van Endert P. Beyond the proteasome: trimming, degradation and generation of MHC class I ligands by auxiliary proteases. Mol Immunol. 2002;39:203–215. doi: 10.1016/s0161-5890(02)00102-5. [DOI] [PubMed] [Google Scholar]
- Rubinstein M, Colby R H. Oxford, UK: Oxford University Press; Polymer Physics. 2003 [Google Scholar]
- Guo Z, Eisenberg D. The structure of a fibril-forming sequence, NNQQNY, in the context of a globular fold. Protein Sci. 2008;17:1617–1623. doi: 10.1110/ps.036368.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteras-Chopo A, Serrano L, Lopez de la Paz M. The amyloid stretch hypothesis: recruiting proteins toward the dark side. Proc Natl Acad Sci U S A. 2005;102:16672–16677. doi: 10.1073/pnas.0505905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R, Sciarretta K L, Orgel J P, Meredith S C. Evidence for novel beta-sheet structures in Iowa mutant beta-amyloid fibrils. Biochemistry. 2009;48:6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H Y, Lin J Y, Lee H C, Wang H L, King C Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc Natl Acad Sci U S A. 2008;105:13345–13350. doi: 10.1073/pnas.0802215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Weissman J S, DePace A H. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- Diaz-Avalos R, King C Y, Wall J, Simon M, Caspar D L. Strain-specific morphologies of yeast prion amyloid fibrils. Proc Natl Acad Sci U S A. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Speransky V, Steven A C, Wickner R B. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci U S A. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Taylor K L, Wall J S, Simon M N, Cheng N, Wickner R B, Steven A C. Architecture of Ure2p prion filaments: the N-terminal domains form a central core fiber. J Biol Chem. 2003;278:43717–43727. doi: 10.1074/jbc.M306004200. [DOI] [PubMed] [Google Scholar]
- Baxa U, Cheng N, Winkler D C, Chiu T K, Davies D R, Sharma D, Inouye H, Kirschner D A, Wickner R B, Steven A C. Filaments of the Ure2p prion protein have a cross-beta core structure. J Struct Biol. 2005;150:170–179. doi: 10.1016/j.jsb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Badger J, Sauder J M, Adams J M, Antonysamy S, Bain K, Bergseid M G, Buchanan S G, Buchanan M D, Batiyenko Y, Christopher J A, Emtage S, Eroshkina A, Feil I, Furlong E B, Gajiwala K S, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis H A, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh C D, Miller I, Molinari J, Muller-Dieckmann H J, Newman J M, Noland B W, Pagarigan B, Park F, Peat T S, Post K W, Radojicic S, Ramos A, Romero R, Rutter M E, Sanderson W E, Schwinn K D, Tresser J, Winhoven J, Wright T A, Wu L, Xu J, Harris T J. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- Dodatko T, Fedorov A A, Grynberg M, Patskovsky Y, Rozwarski D A, Jaroszewski L, Aronoff-Spencer E, Kondraskina E, Irving T, Godzik A, Almo S C. Crystal structure of the actin binding domain of the cyclase-associated protein. Biochemistry. 2004;43:10628–10641. doi: 10.1021/bi049071r. [DOI] [PubMed] [Google Scholar]