Abstract
Duchenne muscular dystrophy (DMD) is a lethal muscle disease caused by dystrophin deficiency. In normal muscle, dystrophin helps maintain sarcolemmal stability. Dystrophin also recruits neuronal nitric oxide synthase (nNOS) to the sarcolemma. Failure to anchor nNOS to the membrane leads to functional ischemia and aggravates muscle disease in DMD. Over the past two decades, a great variety of therapeutic modalities have been explored to treat DMD. A particularly attractive approach is to increase utrophin expression. Utrophin shares considerable sequence, structural and functional similarity with dystrophin. Here, we test the hypothesis that utrophin also brings nNOS to the sarcolemma. Full-length utrophin cDNA was expressed in dystrophin-deficient mdx mice by gutted adenovirus or via transgenic overexpression. Subcellular nNOS localization was determined by immunofluorescence staining, in situ nNOS activity staining and microsomal preparation western blot. Despite supra-physiological utrophin expression, we did not detect nNOS at the sarcolemma. Furthermore, transgenic utrophin overexpression failed to protect mdx muscle from exercise-associated injury. Our results suggest that full-length utrophin cannot anchor nNOS to the sarcolemma. This finding might have important implications for the development of utrophin-based DMD therapies.
Keywords: Dystrophin, Utrophin, nNOS, Duchenne muscular dystrophy
Introduction
Duchenne muscular dystrophy (DMD) is the most common childhood lethal muscle disease. It is caused by mutations in the dystrophin gene (Kunkel, 2005). The dystrophin gene (DMD) encodes a 427 kDa multiple-domain cytosolic protein. The N-terminal domain of dystrophin interacts with cytosolic F-actin. The central rod domain contains 24 spectrin-like repeats and four hinges. The C-terminal domain carries the binding motifs for several cytosolic proteins such as syntrophin and dystrobrevin. A cysteine-rich domain sits between the central rod and the C-terminal domains and it connects dystrophin to the extracellular matrix via dystroglycan. The dystrophin-dystroglycan complex is further strengthened by the sarcoglycans and sarcospan. Together, dystrophin and its associated proteins protect the sarcolemma from contraction-induced injury (for a review, see Blake et al., 2002; Ervasti, 2007).
In DMD patients, dystrophin expression is abolished owing to gene mutation. As a result, dystrophin-associated proteins disassemble from the muscle membrane and the sarcolemma integrity is reduced. Although the loss of the physical support has certainly contributed to the muscle disease, recent studies have begun to appreciate other pathogenic factors (Heydemann et al., 2007). Among these, neuronal nitric oxide synthase (nNOS) is particularly interesting. In normal skeletal muscle, nNOS is recruited to the sarcolemma by dystrophin and syntrophin (Adams et al., 2000; Hillier et al., 1999; Kameya et al., 1999; Lai et al., 2009; Tochio et al., 1999). Membrane location of nNOS allows ready diffusion of nitric oxide to the nearby vasculature to counteract α-adrenergic vasoconstriction during muscle contraction. In the absence of dystrophin, sarcolemmal nNOS expression is lost. Consequently, the protective vessel relaxation mechanism is compromised (Brenman et al., 1995; Chang et al., 1996; Lai et al., 2009; Sander et al., 2000; Thomas et al., 1998). In this regard, contraction-associated ischemic injury has been recognized as one of the earliest pathological changes in DMD muscle (Mendell et al., 1971; Parker and Mendell, 1974). The physiological relevance of membrane-associated nNOS was further emphasized by several recent reports (Kobayashi et al., 2008; Lai et al., 2009; Percival et al., 2008). In these studies, investigators found that sarcolemmal nNOS prevented exercise-related fatigue and improved exercise performance in dystrophic subjects. In summary, restoring sarcolemmal nNOS could represent an important therapeutic endpoint.
Soon after the discovery of the dystrophin gene on the X-chromosome, the utrophin gene (UTRN) was identified as an autosomal paralog of the dystrophin gene (Khurana et al., 1990; Love et al., 1989; Tinsley et al., 1992). Similarly to dystrophin, utrophin also contains four major functional domains, including the N-terminal, central rod, cysteine-rich and C-terminal domains. The N-terminal, cysteine-rich and C-terminal domains are 80% identical to those of dystrophin (Tinsley et al., 1992). Because of the extraordinary sequence homology and structural resemblance, it is not surprising that utrophin stabilizes the sarcolemma by orchestrating dystrophin-associated proteins into a similar complex linking the extracellular matrix with the cytoskeleton (for a review, see Blake et al., 2002; Ervasti, 2007).
Considering the importance of sarcolemmal nNOS in DMD pathogenesis and therapy, we sought to determine whether utrophin was able to recruit nNOS to the sarcolemma. It has been well established that sarcolemmal nNOS anchoring is mediated by the syntrophin PDZ domain (Adams et al., 2001; Hillier et al., 1999; Tochio et al., 1999). We have recently shown that this process also requires dystrophin spectrin-like repeats 16 and 17 (R16/17) (Lai et al., 2009). Whereas dystrophin carries 24 spectrin-like repeats, utrophin contains 22 repeats. Although the repeats corresponding to dystrophin R16/17 appear to be preserved in utrophin, individual repeat units show considerable sequence divergence and it remains unclear whether utrophin repeats can interact with nNOS (Winder et al., 1995). Based on the known structural similarity, we hypothesized that utrophin could anchor nNOS to the sarcolemma. To test this hypothesis, we first overexpressed full-length utrophin using gutted adenoviral vectors in dystrophin-deficient mdx mouse muscles. Surprisingly, we did not detect sarcolemmal nNOS expression by immunofluorescence staining. To thoroughly address this issue, we compared nNOS expression in C57Bl/10 (BL10), mdx, utrophin-null, utrophin-dystrophin double knockout (u-dko) and full-length utrophin transgenic mdx mice. Through nNOS immunofluorescence staining, in situ activity staining and microsomal western analysis, we found that the loss of utrophin had a nominal effect on sarcolemmal nNOS localization. Furthermore, supra-physiological utrophin expression did not restore sarcolemmal nNOS expression. Importantly, chronic treadmill running resulted in apparent muscle degeneration-regeneration and reduction of specific muscle force in utrophin transgenic mdx mice, but not in normal control mice.
Results
Expression of gutted-adenovirus-mediated full-length dystrophin, but not utrophin, restores sarcolemmal nNOS
To determine whether utrophin recruits nNOS to the sarcolemma, we delivered full-length mouse utrophin to the anterior tibialis (TA) muscle in mdx mice using a gutted adenoviral vector. Gutted adenoviral vectors carrying full-length cDNA encoding human or mouse dystrophin were included in the study as the positive control. Two months after gene transfer, we examined nNOS expression by immunofluorescence staining. Robust utrophin and dystrophin expression was detected in mdx muscles injected with the respective adenoviral vectors (Fig. 1). Although sarcolemmal nNOS staining was restored following viral-mediated expression of full-length human (Fig. 1B) or mouse (data not shown) dystrophin, we did not observe membrane-associated nNOS immunoreactivity in mdx muscles infected by the full-length utrophin vector (Fig. 1B). As demonstrated before, some (but not all) revertant fibers showed membrane-associated nNOS expression (Lai et al., 2009; Wells et al., 2003).
Fig. 1.
Adenovirus-mediated full-length utrophin expression does not restore sarcolemmal nNOS. (A) Representative dystrophin and nNOS immunofluorescence staining photomicrographs from normal and mdx muscles. Some reverent fibers can restore nNOS (arrowhead) whereas others cannot (arrow). (B) Representative immunofluorescence staining photomicrographs at 2 months after mdx muscles were infected with gutted adenoviruses. Left panels are dystrophin and nNOS staining following human full-length dystrophin gutted adenovirus infection. Right panels are utrophin and nNOS staining following mouse full-length utrophin gutted adenovirus infection. gAd, gutted adenovirus; HDys, human dystrophin; Utr, utrophin. (C) Two examples of serial sections from full-length utrophin gutted adenovirus infected muscles stained with antibodies against utrophin (left panels) and α-syntrophin (right panels). Myofibers expressing high levels of utrophin at the sarcolemma also expressed α-syntrophin. Asterisks indicate the same myofiber in serial sections.
The absence of α-syntrophin compromises sarcolemmal nNOS localization (Adams et al., 2000; Kameya et al., 1999). If utrophin fails to recruit α-syntrophin, it might explain our finding. To test this hypothesis, we examined serial muscle sections from gutted adenoviral utrophin-injected muscles (Fig. 1C). Consistent with our previous studies (Odom et al., 2008), full-length utrophin delivered by gutted adenovirus restored sarcolemmal α-syntrophin expression (Fig. 1C).
Transgenic overexpression of full-length utrophin fails to recover sarcolemmal nNOS
To further extend the intriguing finding seen with the gutted adenovirus, we examined nNOS localization in utrophin-deficient mice (utrophin knockout and u-dko) and full-length utrophin transgenic mdx mice. Immunostaining was performed on serial TA muscle sections for dystrophin, utrophin, syntrophin and nNOS. In the case of syntrophin, a pan-syntrophin antibody was used to detect all syntrophin isoforms. To corroborate immunostaining results, we also stained for nNOS activity in adjacent muscle sections. In BL10 mice, we observed uniform dystrophin, syntrophin and nNOS expression at the sarcolemma. Consistent with previous reports (Blake et al., 2002; Nguyen et al., 1991; Rivier et al., 1997), utrophin was detected in the microvasculature in normal muscle (Fig. 2A). In dystrophin-deficient mdx mice, utrophin was upregulated and elevated utrophin resulted in low-level syntrophin staining at the sarcolemma. However, neither nNOS immunostaining nor nNOS activity staining generated positive signals (Fig. 2B). Eliminating utrophin alone had minimal influence on sarcolemmal nNOS expression (Fig. 2C). As expected, u-dko mice displayed much severe muscle disease (Deconinck et al., 1997; Grady et al., 1997). We observed substantial sarcolemmal damage in u-dko mice, as illustrated by extensive immunoglobulin infiltration (Lai et al., 2005). Nonetheless, the nNOS expression pattern was not altered (Fig. 2D). In the Fiona line of full-length utrophin transgenic mdx mice (Fiona mice) (Tinsley et al., 1998), strong utrophin expression was detected in every myofiber (Fig. 2E). Transgenic full-length utrophin overexpression also reduced central nucleation and inflammation (Fig. 2E) (Tinsley et al., 1998). Consistent with the results of the adenoviral vector experiment (Fig. 1), supra-physiological utrophin expression in transgenic mice did not restore nNOS to the sarcolemma (Fig. 2E).
Fig. 2.
In situ evaluation of muscle histology and dystrophin, utrophin, syntrophin and nNOS expression. Serial muscle sections from normal BL10 (A), mdx (B), utrophin knockout (C), u-dko (D) and Fiona strain of utrophin transgenic mdx mice (E) were stained for general histology with hematoxylin and eosin (HE), and for dystrophin, utrophin, syntrophin, nNOS and nNOS activity. Representative photomicrographs from each mouse model are presented. nNOS (pAb), immunofluorescence staining for nNOS; Asterisks indicate the same myofiber in serial sections; empty arrowheads in C and E indicate neuromuscular junctions; arrows in D show damaged myofibers with infiltrated mouse immunoglobulin; filled arrowheads in E indicate myofibers with centrally located nucleus.
To confirm immunostaining results, we performed western analysis with total muscle lysates and membrane-enriched microsomal preparations. Using whole-muscle lysates, dystrophin was detected only in BL10 and utrophin-null muscles. Normal muscle showed little utrophin expression. Utrophin was completely eliminated in utrophin-null and u-dko muscles. Although utrophin levels were moderately increased in mdx muscles, the highest utrophin expression was observed in Fiona mouse muscles (Fig. 3A). Total cellular nNOS levels were substantially reduced in mdx and u-dko muscles, but not in utrophin-null and Fiona mouse muscles (Fig. 3A).
Fig. 3.
Western blot analysis of nNOS expression in whole-muscle lysate and microsomal preparation. (A) Representative western blot of whole-muscle lysate. (B) Representative microsomal western blot results. Rapid blue staining of a duplicated gel was included as the loading control for microsomal preparation western blot.
Membrane-associated syntrophin and nNOS were evaluated by western blot analysis of muscle microsomes. Similarly to the immunostaining results, syntrophin levels were normalized in utrophin transgenic muscles. However, supra-physiological utrophin expression did not restore nNOS to the sarcolemma (Fig. 3B). In summary, utrophin overexpression or elimination had a nominal effect on subcellular nNOS distribution (Figs 1, 2 and 3).
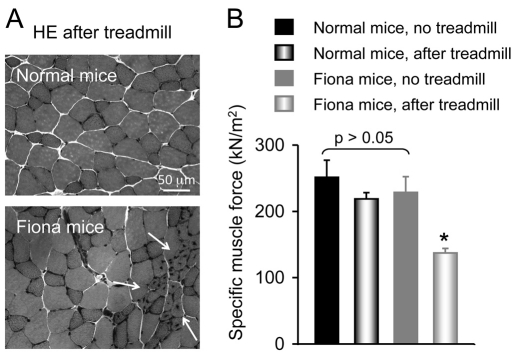
Chronic treadmill exercise leads to focal degeneration-regeneration and force reduction in utrophin transgenic Fiona mice
To evaluate the physiological consequences, we challenged 4-week-old Fiona and BL10 mice with treadmill exercise for 8 weeks (Grounds et al., 2008). Non-exercised mice were included as controls. At the end of the study, we isolated the extensor digitorium longus muscle and compared histology and force between sedentary and treadmill-challenged mice.
In non-exercised Fiona mice, centrally nucleated myofibers were occasionally observed (Fig. 2E). This might reflect accumulated functional ischemic damage from daily activity. Consistent with our previous publication (Lai et al., 2009), focal muscle degeneration-regeneration was substantially aggravated in Fiona mice after chronic treadmill running (Fig. 4A). Large patches of myofibers now showed centrally located nuclei (Fig. 4A). Force measurement further suggested muscle damage in exercised Fiona mice (Fig. 4B). Although treadmill challenge did not influence force generation in BL10 mice, specific tetanic force was significantly reduced in Fiona mice after continuous treadmill running (Fig. 4B).
Fig. 4.
Continuous treadmill running leads to aggravated degeneration-regeneration and specific force reduction in the extensor digitorum longus muscle of Fiona mice. Experimental mice were divided into running and no-running groups. In the running group, mice were challenged with treadmill exercise for 8 weeks. (A) Representative photomicrographs of HE-stained muscle cross-sections from exercised mice. Arrows indicate areas of ischemic damage. (B) Specific muscle force (mean ± s.e.m., n=6 per group). Asterisk indicates a significantly lower value compared with other groups.
Discussion
The sequence homology and the structural similarity between utrophin and dystrophin have stirred a tremendous interest in developing utrophin-based therapies for DMD (for reviews, see Khurana and Davies, 2003; Miura and Jasmin, 2006; Perkins and Davies, 2002; Tinsley and Davies, 1993). Numerous studies have further established functional redundancy between the two proteins (for reviews, see Blake et al., 1996; Blake et al., 2002; Ervasti, 2007). These studies suggest that the utrophin and dystrophin genes are derived from a common evolutionary ancestor. Except for some spatial and/or temporal differences in the expression pattern and a minor difference in the rod-domain length, utrophin seems sufficient to correct virtually all the cellular defects caused by dystrophin deficiency (Tinsley et al., 1998). Encouraging results from gene therapy, protein therapy and pharmacological interventions have further substantiated the therapeutic promise of utrophin (Cerletti et al., 2003; Deol et al., 2007; Khurana and Davies, 2003; Miura and Jasmin, 2006; Odom et al., 2008; Sonnemann et al., 2009).
In an effort to better understand the basic biology of utrophin-mediated DMD therapy, here we examined whether utrophin could recover sarcolemmal nNOS. Based on the striking similarity between dystrophin and utrophin, we initially hypothesized that utrophin could recruit nNOS to the sarcolemma. Our data however, show the hypothesis not to be true. For example, utrophin is upregulated in mdx muscles (Fig. 2B and Fig. 3A), but nNOS is not observed at the mdx sarcolemma (Fig. 1A and Fig. 2B). One possible explanation is that utrophin has a lower affinity for nNOS binding. If this were the case, the amounts of sarcolemmal nNOS in mdx muscle would be too low and beyond the detection threshold. To thoroughly test our hypothesis, we examined whether sarcolemmal nNOS expression could be restored when utrophin was overexpressed. Two different approaches were used to increase utrophin expression in mdx muscles, injection of gutted adenoviral vectors and the use of transgenic mice. We did not detect nNOS at the sarcolemma by immunofluorescence staining, despite robust full-length utrophin expression from the gutted adenoviral vectors (Fig. 1). Three independent approaches (immunofluorescence staining, in situ activity staining and microsomal preparation western blot) were then applied to determine membrane-associated nNOS expression in transgenic mice. Several lines of utrophin transgenic mdx mice have been described (Tinsley et al., 1998). Among these, the Fiona line showed the highest utrophin expression (~10-fold higher than the endogenous level) (Perkins and Davies, 2002). Importantly, morphological and physiological defects were almost completely corrected in Fiona mice (Fig. 2E) (Perkins and Davies, 2002; Tinsley et al., 1998). Despite the supra-physiological utrophin levels, we were not able to detect nNOS at the sarcolemma (Figs 2 and 3). To gain more insight into the role utrophin might have, additional studies were performed in utrophin-null and u-dko mice. Subcellular nNOS localization was not altered in these mice (Fig. 2C,D and Fig. 3B).
We have recently shown that dystrophin-mediated sarcolemmal nNOS localization prevents functional ischemia and enhances exercise performance in dystrophin-deficient mice (Lai et al., 2009). Specifically, focal ischemic damage (such as degeneration and regeneration) was found in transgenic mdx mice expressing the ΔH2-R19 mini-dystrophin gene (this minigene cannot restore nNOS) but not in ΔH2-R15 mini-dystrophin transgenic mdx mice, which contain sarcolemmal nNOS (Lai et al., 2009). In sedentary Fiona muscle, we also observed sporadic focal degeneration and regeneration (Fig. 2E, arrowhead). This might reflect accumulated functional ischemic damage from daily activity. To further evaluate the physiological alterations that might occur with utrophin-overexpression therapy, mice were challenged with treadmill running for 8 weeks. Consistent with our previous observation (Lai et al., 2009), treadmill exercise induced significant degeneration-regeneration and compromised muscle force generation in Fiona mice, but not in normal mice (Fig. 4).
Previous studies suggest that some quantitative functional differences between dystrophin and utrophin might exist. For example, dystrophin binds actin via two separate contact sites whereas utrophin interacts with actin through a single continuous unit (for a review, see Ervasti, 2007). Furthermore, utrophin shows a twofold lower affinity for β-dystroglycan (Ishikawa-Sakurai et al., 2004). Nevertheless, both proteins are fully capable of interacting with F-actin and β-dystroglycan, and each alone is sufficient to preserve the sarcolemmal integrity. It appears that dystrophin and utrophin are functionally exchangeable. Our results identify for the first time a significant difference in the functional capacity of utrophin and dystrophin. Dystrophin can recruit nNOS to the sarcolemma, but utrophin cannot.
Despite intensive investigation, the molecular mechanism(s) underlying sarcolemmal nNOS localization remains incompletely understood. The current model suggests the involvement of syntrophin and dystrophin R16/17. The syntrophin family consists of five members, including α-, β1-, β2-, γ1- and γ2-syntrophin (Albrecht and Froehner, 2002). Only α-syntrophin has been conclusively shown to interact with nNOS (Adams et al., 2000; Adams et al., 2001; Hillier et al., 1999; Kameya et al., 1999; Tochio et al., 1999). Based on this model, one possible explanation for our results might relate to the specific syntrophin isoform(s) recruited by utrophin.
It has been shown in wild-type muscle that dystrophin primarily interacts with α-syntrophin and β1-syntrophin whereas utrophin primarily interacts with β1-syntrophin and β2-syntrophin (Peters et al., 1997). However, utrophin has also been shown to recruit α-syntrophin to the sarcolemma at places where utrophin is highly expressed (such as the neuromuscular junction and regenerating myofibers) (Peters et al., 1997). We have recently shown that a synthetic micro-utrophin protein also interacts with α-syntrophin (Odom et al., 2008). To determine whether the failure to recruit α-syntrophin underlies our observation, we examined α-syntrophin expression in mdx muscles that have been transduced with gutted adenoviral vectors expressing full-length mouse utrophin (Fig. 1C). Immunofluorescence staining was performed with an α-syntrophin-specific antibody (Adams et al., 2000; Peters et al., 1997). As shown in Fig. 1C, we observed robust sarcolemmal α-syntrophin expression in myofibers that were transduced by the adenoviral vector.
The finding that utrophin cannot recover sarcolemmal nNOS might also reflect a poor interaction between nNOS and utrophin. Dystrophin R16/17 constitutes the nNOS binding domain. Utrophin spectrin-like repeats 15 and 16 (R15/16) appear to be the corresponding repeats of dystrophin R16/17 (Winder et al., 1995). Our result offers a platform to dissect the molecular interaction between nNOS and dystrophin R16/17 or utrophin R15/16. Since the majority of the amino acid residues are highly conserved in these repeats, reciprocal mutagenesis could help to identify residues crucial for dystrophin-nNOS interaction.
Taken together, our results reveal a qualitative difference between utrophin and dystrophin. Dystrophin can anchor nNOS to the sarcolemma and maintain blood perfusion in contracting muscle. However, utrophin cannot recover membrane-associated nNOS expression. The absence of sarcolemmal nNOS is known to result in functional ischemia during muscle contraction. To enhance efficacy of utrophin-based therapies, innovative means are needed to maintain muscle perfusion in exercise.
Materials and Methods
Animal studies
Animal experiments were performed in accordance with the NIH and institutional guidelines of the University of Missouri, the University of Washington and Oxford University. C57Bl/6 (this strain was used as the normal control in adenoviral experiments), BL10 and mdx mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The original breeding pair of u-dko mice was kindly provided by Mark Grady (Washington University, St Louis, MO) (Grady et al., 1997). U-dko mice were subsequently backcrossed with mdx mice to the BL10 background (Yue et al., 2006). Experimental u-dko mice were all on the BL10 background. Utrophin-null mice were generated by two rounds crossing of BL10 mice with BL10 background u-dko mice. Fiona strain full-length utrophin transgenic mdx mice was reported before (Tinsley et al., 1998). Treadmill exercise was performed according to a published protocol (Grounds et al., 2008). Briefly, mice were run on a horizontal treadmill at a speed of 12 m/minute, twice a week for a total of 8 weeks. At the beginning of each session, mice received 10 minute warm-up exercise at the speed of 8 m/minute.
Gutted adenoviral vector
The full-length dystrophin and utrophin gutted adenoviral vectors have been described before (DelloRusso et al., 2002; Scott et al., 2002). High-titer gutted adenoviral vectors was grown and purified as described (Hartigan-O'Connor et al., 2002). Local muscle injection was performed as reported previously to the TA muscle in adult mdx mice using 4×1010 particles of vector per muscle (DelloRusso et al., 2002). Muscles were analyzed for expression 2 months after injection.
Immunofluorescence staining and in situ nNOS activity staining
Dystrophin was detected with a mouse monoclonal antibody against the C-terminal domain (Dys-2, 1:30, Novocastra, Newcastle, UK). Utrophin was examined with a mouse monoclonal antibody against the utrophin N-terminal domain (1:20; Vector Laboratories, Burlingame, CA). Syntrophin was revealed with two different antibodies including a polyclonal α-syntrophin specific antibody (1:200, a gift from Stanley Froehner, University of Washington, Seattle, WA) and a pan-syntrophin mouse monoclonal antibody that recognizes the PDZ domain (1:200; Abcam, Cambridge, MA) (Adams et al., 2000; Peters et al., 1997). nNOS was detected with a polyclonal antibody (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA). Immunofluorescence staining was performed using protocols we described before (Lai et al., 2009; Lai et al., 2005; Yue et al., 2003). In situ nNOS activity staining was performed according to a published protocol (Lai et al., 2009).
Western blot
Whole-muscle lysate was extracted from limb muscle and western blot was performed according to a previously published protocol (Lai et al., 2009; Li et al., 2009; Li et al., 2008). Membrane-enriched microsomal preparations were produced from limb muscle according to a previously described protocol (Ervasti and Campbell, 1991; Lai et al., 2009). Proteins were resolved on a 6% SDS-polyacrylamide gel and transferred to a PVDF membrane. Dystrophin was detected with the anti-Dys-2 antibody (1:100; Novocastra). Utrophin was detected with a mouse monoclonal antibody (1:200; BD Biosciences, San Jose, CA). Syntrophin was revealed with a mouse monoclonal antibody (1:2000; Abcam). nNOS was detected with a rabbit anti-nNOS polyclonal antibody (1:2000; Upstate, Lake Placid, NY). α-tubulin (1:3000; Sigma, St Louis, MO) was used as the loading control in whole-muscle-lysate western blot. Rapid blue staining (Geno Technology, St Louis, MO) of a duplicated gel was used as the loading control in microsomal preparation western blot.
Acknowledgments
This work was supported by grants from the National Institutes of Health (AR-49419, D.D; AR-44533, J.S.C.), the Muscular Dystrophy Association (D.D. and J.S.C.), the Clarendon Fund and Kokil Pathak scholarship (A.B.). The authors (D.L., Y.Y., Y.L. and D.D.) thank Robert J. McDonald, Jr, for the generous support to Duchenne muscular dystrophy research in the Duan lab. We thank Stanley Froehner for providing the polyclonal anti-α-syntrophin antibody. We also thank Sarah Squire, Brian Bostick, Chun Long, Dave Powell and Yadong Zhang for technical help. Deposited in PMC for release after 12 months.
References
- Adams M. E., Kramarcy N., Krall S. P., Rossi S. G., Rotundo R. L., Sealock R., Froehner S. C. (2000). Absence of alpha-syntrophin leads to structurally aberrant neuromuscular synapses deficient in utrophin. J. Cell Biol. 150, 1385-1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. E., Mueller H. A., Froehner S. C. (2001). In vivo requirement of the alpha-syntrophin PDZ domain for the sarcolemmal localization of nNOS and aquaporin-4. J. Cell Biol. 155, 113-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht D. E., Froehner S. C. (2002). Syntrophins and dystrobrevins: defining the dystrophin scaffold at synapses. Neurosignals 11, 123-129 [DOI] [PubMed] [Google Scholar]
- Blake D. J., Tinsley J. M., Davies K. E. (1996). Utrophin: a structural and functional comparison to dystrophin. Brain Pathol. 6, 37-47 [DOI] [PubMed] [Google Scholar]
- Blake D. J., Weir A., Newey S. E., Davies K. E. (2002). Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol. Rev. 82, 291-329 [DOI] [PubMed] [Google Scholar]
- Brenman J. E., Chao D. S., Xia H., Aldape K., Bredt D. S. (1995). Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell 82, 743-752 [DOI] [PubMed] [Google Scholar]
- Cerletti M., Negri T., Cozzi F., Colpo R., Andreetta F., Croci D., Davies K. E., Cornelio F., Pozza O., Karpati G., et al. (2003). Dystrophic phenotype of canine X-linked muscular dystrophy is mitigated by adenovirus-mediated utrophin gene transfer. Gene Ther. 10, 750-757 [DOI] [PubMed] [Google Scholar]
- Chang W. J., Iannaccone S. T., Lau K. S., Masters B. S., McCabe T. J., McMillan K., Padre R. C., Spencer M. J., Tidball J. G., Stull J. T. (1996). Neuronal nitric oxide synthase and dystrophin-deficient muscular dystrophy. Proc. Natl. Acad. Sci. USA 93, 9142-9147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deconinck A. E., Rafael J. A., Skinner J. A., Brown S. C., Potter A. C., Metzinger L., Watt D. J., Dickson J. G., Tinsley J. M., Davies K. E. (1997). Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell 90, 717-727 [DOI] [PubMed] [Google Scholar]
- DelloRusso C., Scott J. M., Hartigan-O'Connor D., Salvatori G., Barjot C., Robinson A. S., Crawford R. W., Brooks S. V., Chamberlain J. S. (2002). Functional correction of adult mdx mouse muscle using gutted adenoviral vectors expressing full-length dystrophin. Proc. Natl. Acad. Sci. USA 99, 12979-12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deol J. R., Danialou G., Larochelle N., Bourget M., Moon J. S., Liu A. B., Gilbert R., Petrof B. J., Nalbantoglu J., Karpati G. (2007). Successful compensation for dystrophin deficiency by a helper-dependent adenovirus expressing full-length utrophin. Mol. Ther. 15, 1767-1774 [DOI] [PubMed] [Google Scholar]
- Ervasti J. M. (2007). Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim. Biophys. Acta 1772, 108-117 [DOI] [PubMed] [Google Scholar]
- Ervasti J. M., Campbell K. P. (1991). Membrane organization of the dystrophin-glycoprotein complex. Cell 66, 1121-1131 [DOI] [PubMed] [Google Scholar]
- Grady R. M., Teng H., Nichol M. C., Cunningham J. C., Wilkinson R. S., Sanes J. R. (1997). Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell 90, 729-738 [DOI] [PubMed] [Google Scholar]
- Grounds M. D., Radley H. G., Lynch G. S., Nagaraju K., De Luca A. (2008). Towards developing standard operating procedures for pre-clinical testing in the mdx mouse model of Duchenne muscular dystrophy. Neurobiol. Dis. 31, 1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan-O'Connor D., Barjot C., Salvatori G., Chamberlain J. S. (2002). Generation and growth of gutted adenoviral vectors. Methods Enzymol. 346, 224-246 [DOI] [PubMed] [Google Scholar]
- Heydemann A., Doherty K. R., McNally E. M. (2007). Genetic modifiers of muscular dystrophy: Implications for therapy. Biochim. Biophys. Acta 172, 216-228 [DOI] [PubMed] [Google Scholar]
- Hillier B. J., Christopherson K. S., Prehoda K. E., Bredt D. S., Lim W. A. (1999). Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science 284, 812-815 [PubMed] [Google Scholar]
- Ishikawa-Sakurai M., Yoshida M., Imamura M., Davies K. E., Ozawa E. (2004). ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to beta-dystroglycan. Hum. Mol. Genet. 13, 693-702 [DOI] [PubMed] [Google Scholar]
- Kameya S., Miyagoe Y., Nonaka I., Ikemoto T., Endo M., Hanaoka K., Nabeshima Y., Takeda S. (1999). alpha1-syntrophin gene disruption results in the absence of neuronal-type nitric-oxide synthase at the sarcolemma but does not induce muscle degeneration. J. Biol. Chem. 274, 2193-2200 [DOI] [PubMed] [Google Scholar]
- Khurana T. S., Davies K. E. (2003). Pharmacological strategies for muscular dystrophy. Nat. Rev. Drug Discov. 2, 379-390 [DOI] [PubMed] [Google Scholar]
- Khurana T. S., Hoffman E. P., Kunkel L. M. (1990). Identification of a chromosome 6-encoded dystrophin-related protein. J. Biol. Chem. 265, 16717-16720 [PubMed] [Google Scholar]
- Kobayashi Y. M., Rader E. P., Crawford R. W., Iyengar N. K., Thedens D. R., Faulkner J. A., Parikh S. V., Weiss R. M., Chamberlain J. S., Moore S. A., et al. (2008). Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature 456, 511-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel L. M. (2005). 2004 William Allan Award address. Cloning of the DMD gene. Am. J. Hum. Genet. 76, 205-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Yue Y., Liu M., Ghosh A., Engelhardt J. F., Chamberlain J. S., Duan D. (2005). Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat. Biotechnol. 23, 1435-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Thomas G. D., Yue Y., Yang H. T., Li D., Long C., Judge L., Bostick B., Chamberlain J. S., Terjung R. L., et al. (2009). Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J. Clin. Invest. 119, 624-635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Yue Y., Duan D. (2008). Preservation of muscle force in mdx3cv mice correlates with low-level expression of a near full-length dystrophin protein. Am. J. Pathol. 172, 1332-1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Long C., Yue Y., Duan D. (2009). Sub-physiological sarcoglycan expression contributes to compensatory muscle protection in mdx mice. Hum. Mol. Genet. 18, 1209-1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love D. R., Hill D. F., Dickson G., Spurr N. K., Byth B. C., Marsden R. F., Walsh F. S., Edwards Y. H., Davies K. E. (1989). An autosomal transcript in skeletal muscle with homology to dystrophin. Nature 339, 55-58 [DOI] [PubMed] [Google Scholar]
- Mendell J. R., Engel W. K., Derrer E. C. (1971). Duchenne muscular dystrophy: functional ischemia reproduces its characteristic lesions. Science 172, 1143-1145 [DOI] [PubMed] [Google Scholar]
- Miura P., Jasmin B. J. (2006). Utrophin upregulation for treating Duchenne or Becker muscular dystrophy: how close are we? Trends Mol. Med. 12, 122-129 [DOI] [PubMed] [Google Scholar]
- Nguyen T. M., Ellis J. M., Love D. R., Davies K. E., Gatter K. C., Dickson G., Morris G. E. (1991). Localization of the DMDL gene-encoded dystrophin-related protein using a panel of nineteen monoclonal antibodies: presence at neuromuscular junctions, in the sarcolemma of dystrophic skeletal muscle, in vascular and other smooth muscles, and in proliferating brain cell lines. J. Cell Biol. 115, 1695-1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom G. L., Gregorevic P., Allen J. M., Finn E., Chamberlain J. S. (2008). Microutrophin delivery through rAAV6 increases lifespan and improves muscle function in dystrophic dystrophin/utrophin-deficient mice. Mol. Ther. 16, 1539-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. M., Mendell J. R. (1974). Proximal myopathy induced by 5-HT-imipramine simulates Duchenne dystrophy. Nature 247, 103-104 [DOI] [PubMed] [Google Scholar]
- Percival J. M., Anderson K. N., Gregorevic P., Chamberlain J. S., Froehner S. C. (2008). Functional deficits in nNOSmu-deficient skeletal muscle: myopathy in nNOS knockout mice. PLoS ONE 3, e3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins K. J., Davies K. E. (2002). The role of utrophin in the potential therapy of Duchenne muscular dystrophy. Neuromuscul. Disord. 12Suppl. 1, S78-S89 [DOI] [PubMed] [Google Scholar]
- Peters M. F., Adams M. E., Froehner S. C. (1997). Differential association of syntrophin pairs with the dystrophin complex. J. Cell Biol. 138, 81-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier F., Robert A., Hugon G., Mornet D. (1997). Different utrophin and dystrophin properties related to their vascular smooth muscle distributions. FEBS Lett. 408, 94-98 [DOI] [PubMed] [Google Scholar]
- Sander M., Chavoshan B., Harris S. A., Iannaccone S. T., Stull J. T., Thomas G. D., Victor R. G. (2000). Functional muscle ischemia in neuronal nitric oxide synthase-deficient skeletal muscle of children with Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 97, 13818-13823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J., Li S., Harper S., Welikson R., Bourque D., DelloRusso C., Hauschka S., Chamberlain J. (2002). Viral vectors for gene transfer of micro-, mini-, or full-length dystrophin. Neuromuscul. Disord. 12Suppl, S23 [DOI] [PubMed] [Google Scholar]
- Sonnemann K. J., Heun-Johnson H., Turner A. J., Baltgalvis K. A., Lowe D. A., Ervasti J. M. (2009). Functional substitution by TAT-utrophin in dystrophin-deficient mice. PLoS Med. 6, e1000083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. D., Sander M., Lau K. S., Huang P. L., Stull J. T., Victor R. G. (1998). Impaired metabolic modulation of alpha-adrenergic vasoconstriction in dystrophin-deficient skeletal muscle. Proc. Natl. Acad. Sci. USA 95, 15090-15095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley J. M., Davies K. E. (1993). Utrophin: a potential replacement for dystrophin? Neuromuscul. Disord. 3, 537-539 [DOI] [PubMed] [Google Scholar]
- Tinsley J. M., Blake D. J., Roche A., Fairbrother U., Riss J., Byth B. C., Knight A. E., Kendrick-Jones J., Suthers G. K., Love D. R., et al. (1992). Primary structure of dystrophin-related protein. Nature 360, 591-593 [DOI] [PubMed] [Google Scholar]
- Tinsley J., Deconinck N., Fisher R., Kahn D., Phelps S., Gillis J. M., Davies K. (1998). Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med. 4, 1441-1444 [DOI] [PubMed] [Google Scholar]
- Tochio H., Zhang Q., Mandal P., Li M., Zhang M. (1999). Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide. Nat. Struct. Biol. 6, 417-421 [DOI] [PubMed] [Google Scholar]
- Wells K. E., Torelli S., Lu Q., Brown S. C., Partridge T., Muntoni F., Wells D. J. (2003). Relocalization of neuronal nitric oxide synthase (nNOS) as a marker for complete restoration of the dystrophin associated protein complex in skeletal muscle. Neuromuscul. Disord. 13, 21-31 [DOI] [PubMed] [Google Scholar]
- Winder S. J., Gibson T. J., Kendrick-Jones J. (1995). Dystrophin and utrophin: the missing links! FEBS Lett. 369, 27-33 [DOI] [PubMed] [Google Scholar]
- Yue Y., Li Z., Harper S. Q., Davisson R. L., Chamberlain J. S., Duan D. (2003). Microdystrophin gene therapy of cardiomyopathy restores dystrophin-glycoprotein complex and improves sarcolemma integrity in the Mdx mouse heart. Circulation 108, 1626-1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Liu M., Duan D. (2006). C-terminal truncated microdystrophin recruits dystrobrevin and syntrophin to the dystrophin-associated glycoprotein complex and reduces muscular dystrophy in symptomatic utrophin/dystrophin double knock-out mice. Mol. Ther. 14, 79-87 [DOI] [PMC free article] [PubMed] [Google Scholar]