Abstract
Understanding causal relationships between genotypes and phenotypes is a long-standing aim in genetics. In addition to high-throughput technologies that allow the measurement of many DNA variants it is possible to measure gene expression in specific tissues using array technology. “Systems genetics” is an emerging discipline that combines dense data on genotypes, gene expression, and outcome phenotypes to answer fundamental questions about causal pathways from genotype to phenotype. A recent paper by Chen et al. [Mol. Syst. Biol. 5, 310 (2009)] addressed the question of whether relative levels of mRNA expression help to elucidate causal paths from genotype to phenotype, using drug resistance in yeast as a model. The authors show that data on genetic markers and on gene expression, measured in a drug-free environment, can be combined to predict the growth of a yeast strain in the presence of a drug. They argue that their prediction can be used to identify causal pathways and for a subset of the genes used in prediction, the authors demonstrate that these genes cause an effect on drug sensitivity by deleting the gene or overexpressing it or swapping alleles between strains of yeast. This approach can also be applied to other species, including humans, and may become a tool in the study of personalized medicine.
Understanding the genetic basis of phenotypic differences between individuals in a population is a long-standing aim in genetics research with applications in medicine, evolutionary biology, and agriculture. For complex or quantitative traits, the phenotype depends on multiple genes and environmental factors. Traditionally, it has been difficult to identify specific polymorphisms causing variation in complex traits. Geneticists have estimated the proportion of phenotypic variance that is genetic (i.e., the heritability) by calculating the correlation between relatives and have predicted the phenotype of an individual from the phenotypes of its relatives (e.g., use of family history to assess one’s disease risk, Visscher et al., 2008). Phenotypes, such as future disease status, are also predicted from other conveniently measured phenotypes such as the use of serum cholesterol concentration to predict risk of heart disease. However, these studies do not tell us anything about the importance of specific genes or polymorphisms. With the advent of molecular markers it has become possible to map genes causing variation in a trait and even to identify the causal polymorphism.
Drug resistance is one example of a “complex trait” in that there are differences between individuals in the population in drug resistance and some of those differences are due to genetic factors. A better understanding of the genetic basis of drug resistance is important because it could be used to target and tailor drugs to specific genotypes, i.e., “personalized medicine.” For example in humans, the anticoagulant drug warfarin is used to reduce the risk of stroke, pulmonary embolism, and thrombosis but there is large variation between people in the dose needed for effective anticoagulation treatment. Nearly half of this dose variation is explained by common polymorphisms in three genes (VKORC1, CYP2C9, and CYP4F2, Takeuchi et al., 2009) so that genetic testing could aid in calibrating warfarin dose and thereby reduce the chance of serious illness or severe bleeding.
The extent of expression of a particular gene or transcript (mRNA abundance) is also a complex trait influenced by multiple polymorphisms and by environmental factors. Like other complex traits, it is possible to map genetic loci that explain some of the genetic variation in abundance of a particular transcript by using linkage or association analysis, to detect association within pedigrees or in the population, respectively, (Brem et al., 2002; Cheung et al., 2003a, 2003b, 2005; Jansen and Nap, 2001; Monks et al., 2004; Morley et al., 2004; Rockman and Kruglyak, 2006; Schadt et al., 2003; Stranger et al., 2005). It is also possible to study the correlation between gene expression and a conventional phenotype such as disease status. A number of investigators have gone one step further by studying the relationships between genetic markers, gene expression, and conventional phenotypes leading to gene networks where a polymorphism in one gene affects the expression of that same gene or other genes and this in turn affects a phenotype such as disease status (Schadt et al., 2005).
Chen et al. (2009) used a similar approach. They aim to predict a complex phenotype, the ability of yeast strains to grow in the presence of one of 94 drugs, using data on genetic markers and gene expression in a population of 104 recombinant strains derived from a cross of two widely divergent parents. Chen et al. specifically asked the question, “How useful are gene expression data collected in a drug-free environment to predict resistance to drugs when genotypes are challenged?”
DESIGN AND ANALYSIS
The drug response data were growth yields in the presence of 94 different chemicals, including well-known drugs such as Resveratrol, Clotrimazole, and Tamoxifen, where 526 genetic markers were used, spread throughout the genome. The gene expression data were from 854 candidate genes. Importantly, gene expression data were generated in the absence of the chemicals. All these data were generated previously by Brem and Kruglyak (2005), Brem et al. (2002), and Perlstein et al. (2007).
The challenge is to use these different sources of data to predict the phenotype and drug resistance in this case. One method is to correlate the genetic markers with outcome, using linkage analysis. A strong correlation implies that somewhere in the region of the genome that is linked to the marker there are one or more genetic polymorphisms that cause the observed outcome. A disadvantage of this approach is the resolution because the segment of chromo-some that is correlated can harbor many genes. An alternative approach is to correlate gene expression with drug resistance. The advantage of this is resolution since a specific gene is implicated. The disadvantage is that a correlation is not evidence of causation. A prediction of drug resistance based on either genetic markers or gene expression might be satisfactory even if neither had a causal relationship to drug resistance but the authors argue, reasonably, that the prediction will be more robust if the correlations are causal. In addition, the discovery of the causes of phenotypic variation may be useful for purposes other than prediction including identification of new drug targets and understanding of the biological system. The authors combine the marker and gene expression data in a sophisticated model fitting procedure they call causal modeling with expression linkage for complex traits (Camelot). The aim of the study is to simultaneously construct the best predictor for outcome and identify causal mechanisms.
The authors present evidence that Camelot is able to select a good predictor and that the addition of gene expression can improve the prediction of drug resistance dramatically. For example, for the drug Haloperidol, the classification accuracy (a measure of how well the prediction works) is 0.45 when only genetic markers are used and 0.72 when both markers and gene expression is used.
Developing a prediction equation based on 526 markers and expression of 854 transcripts using 104 data points is inherently difficult because it is always possible to find a prediction equation that works in the data where it is estimated (the training data set) but often such equations fail when applied to new data (the test data set). How does Camelot work? The main reason for success seems to be the judicious choice of the variables or “features” to include in the prediction equation. Once the variables for use are chosen, their effects are estimated by linear regression. There appear to be at least three mechanisms by which the variables are chosen. The statistical analysis uses various methods to make the choice of variables robust such as the nonparametric bootstrap. We would expect that this is advantageous when the data (i.e., growth in response to a drug) is highly non-normally distributed. The accuracy of the final prediction is evaluated using a tenfold cross-validation scheme to avoid biasing the estimate of accuracy. This means that the prediction equation is estimated using 90% of the yeast strains and tested in the other 10%. There is a little concern that Camelot may have been developed in the same data and therefore there may still be some bias in the estimates of accuracy. It would be desirable to test Camelot in a completely new data set. However, the authors test a number of their predictions of causality with new experimental data and this supports the functional role of the single nucleotide polymorphisms (SNPs) and transcripts used in the prediction equations (see below).
The selection of the SNPs to include in the prediction equation is aided by what the authors call “zoom-in.” Many SNPs in a chromosome segment are likely to be correlated with the phenotype due to linkage. However, only a SNP in the gene containing the causal polymorphism is likely to affect the expression of its own transcript and to show a correlation between the transcript abundance and the phenotype. Therefore, Camelot selects SNPs showing these characteristics for inclusion in the prediction equation and it also favors genes that are highly conserved across yeast species on the grounds that mutations in highly conserved genes are more likely to have an effect.
The choice of transcripts to use in the prediction equation depends on several types of data. First, the 854 transcripts considered were chosen from the full set of 6189 on the basis of the known function of the gene and its likelihood of affecting drug resistance. Second, to be included, transcripts had to pass a “triangle test.” This appears to be a conditional test to ensure that transcripts remain significant when added to a prediction that already contained the chosen SNPs.
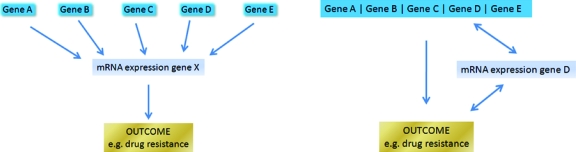
The phenotypes recorded were the average of a strain and presumably the differences between strains are genetic not environmental. Consequently, one might expect that the SNPs, which cover the whole genome, could completely predict the phenotype without the need to include the gene expression data. The authors offer two explanations why gene expression gives additional information over and above the value of genotypic information. The first one is that gene expression as a phenotype may capture the accumulated effects of many genetic variants that influence it. The effect of a specific genetic polymorphism on the outcome trait (drug response) may be through a change in the amount of mRNA expression at one or more genes. The polymorphism may have too small an effect on the phenotype to be detected in the data set of 104 yeast strains. However, if many such polymorphisms effect the expression of the same transcript and also affect the phenotype, the transcript abundance may integrate the effects of many polymorphisms and therefore be a more useful predictor than the individual polymorphisms. Obviously, this could occur if the effect of the SNPs on drug resistance is mediated by the expression level of this transcript (Fig. 1). The second mechanism by which the gene expression data improves the prediction is through the zoom-in method described above for selecting SNPs.
Figure 1. Two models to explain the added value of gene expression.
(Left panel) Variants in multiple (unlinked) genes (A)–(E) affect mRNA expression in a particular gene (X) and this affects outcome. (Right panel) Zoom-in: gene expression in gene D helps to identify which of the linked genes (A)–(E) contains a DNA variant that affects outcome.
PREDICTION AND CAUSALITY
The intention of Camelot is that it selects features for the prediction equation that have a causal relationship to the phenotype. Chen et al. tested this in a subset of cases. The expression of DHH1 was negatively correlated with growth in the presence of hydrogen peroxide. To show that this was causal, the authors deleted the DHH1 gene and observed an increase in growth. The prediction equation for growth in hydrogen peroxide included a SNP in the gene ERG6 and Chen et al. showed that this gene had a causal role by overexpressing ERG6 and observing a decrease in growth. A SNP in the gene PHO84 was selected by the zoom-in method of Camelot to predict resistance to many drugs. Chen et al. confirmed that this gene is causal by swapping the PHO84 alleles between the two parent strains and showing that this changed resistance to the appropriate drugs. This is a surprising result because the difference between the alleles is one amino acid, which should not directly affect the expression of the gene. However, Chen et al. showed that the change in amino acid sequence of the protein affects the function of the protein causing a feedback loop to alter the gene expression. Because they can identify causal features, Chen et al. are able to identify pathways by which the drugs inhibit growth and also new drug targets. For instance, hydrogen peroxide and several other drugs seem to work through mitochondrial function.
SIMILAR STUDIES SHOW SIMILAR RESULTS
The results from a very similar study that used the same yeast strains were published in PLoS One (Ruderfer et al., 2009). These authors came to very similar conclusions as Chen et al. in that (1) drug response can be predicted from transcript levels measured in the absence of drugs; (2) drug response can be predicted from marker data; and (3) combining marker and transcript abundance increases prediction accuracy. Quantitatively, most of the prediction power came from genetic markers in the study of Ruderfer et al. andthe authors conclude that in the absence of environmental perturbations, genotype determines both expression levels and drug response so that most information is contained in the genotype information.
APPLICATIONS TO HUMANS
How could these methods be applied to other species, for example, humans? In humans the map resolution from marker-trait association studies is much better than for experimental crosses (such as the one in yeast used in Chenet al.) because the effective population size is larger. Therefore, the confidence interval for the causal polymorphism is smaller and may contain only a few genes. However, the zoom-in method that is used in the study of Chen et al. could still be used to predict which of these genes contains the causal polymorphism. Obtaining the relevant mRNA levels is more problematic in humans because often the best tissue cannot be sampled and researchers use blood samples or cell lines to measure gene expression (Cheung et al., 2003a; Monks et al., 2004; Morley et al., 2004; Stranger et al., 2005). The results of Chen et al. implied that it may not be necessary to measure gene expression in the correct tissue and physiological state since they used gene expression data in the absence of any drug to predict drug resistance. How this applies to multicellular organisms remains to be seen.
The results of Chen et al. implied that few genes and pathways are needed to explain growth in the presence of a drug. This seems to be at odds with the data on most complex traits in mammals where many genes typically affect a complex trait (Donnelly, 2008). This would imply that much larger sample sizes would be needed to achieve the same accuracy of prediction in humans and other mammals. The existence of some polymorphisms of large effect on drug resistance may occur because natural selection has not eliminated either allele because in the absence of the drug, the polymorphism is nearly neutral. Similarly, in humans, it is unlikely that there has been strong evolutionary pressure on the response to drugs so common variants with large effects may exist, as exemplified by the genes affecting variation in warfarin dose response (Takeuchi et al., 2009).
The “system genetics” (Jansen and Nap, 2001) approach taken by Chen et al. could in principle be taken further by adding other levels of relevant data. For example, proteomic and epigenetic data on specific genes may help both to make the prediction more accurate and to elucidate causal pathways.
CONCLUSION
Systems genetics is an emerging discipline in which several levels of biological data, often characterized by high volumes through the use of omics technologies, are measuredto elucidate causal pathways. It is characterized by having a genetically informative design (for example, yeast segregants in the Chen et al. and Ruderfer et al. papers) in which intermediate “phenotypes” such as gene expression, gene methylation, protein abundance, or metabolites are measured to understand and predict the relationship between genetic information and outcome phenotypes such as drug resistance, disease susceptibility, and quantitative traits. It is a logical step forward from the “genetical genomics” approaches suggested by Jansen and Nap (2001) when it became clear that transcript abundance has a strong genetic basis.
Chen et al. (2009) and Ruderfer et al. (2009) combined the experimental data on genetic markers and gene expression in yeast with data from the public domain (sequence conservation and annotation of gene function) to predict drug resistance for individual genotypes. They show that adding gene expression improves the accuracy of prediction and facilitates the identification of causal pathways. Extending this work to humans could be an important step to fulfill the promise of personalized medicine.
References
- Brem, R B, and Kruglyak, L (2005). “The landscape of genetic complexity across 5,700 gene expression traits in yeast.” Proc. Natl. Acad. Sci. U.S.A. 102, 1572–1577. 10.1073/pnas.0408709102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R B, Yvert, G, Clinton, R, and Kruglyak, L (2002). “Genetic dissection of transcriptional regulation in budding yeast.” Science 296, 752–755. 10.1126/science.1069516 [DOI] [PubMed] [Google Scholar]
- Chen, B J, Causton, H C, Mancenido, D, Goddard, N L, Perlstein, E O, and Pe’er, D (2009). “Harnessing gene expression to identify the genetic basis of drug resistance.” Mol. Syst. Biol. 5, 310. 10.1038/msb.2009.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, V G, Conlin, L K, Weber, T M, Arcaro, M, Jen, K Y, Morley, M, and Spielman, R S (2003a). “Natural variation in human gene expression assessed in lymphoblastoid cells.” Nat. Genet. 33, 422–425. 10.1038/ng1094 [DOI] [PubMed] [Google Scholar]
- Cheung, V G, Jen, K Y, Weber, T, Morley, M, Devlin, J L Ewens, K G, and Spielman, R S (2003b). “Genetics of quantitative variation in human gene expression.” Cold Spring Harbor Symp. Quant. Biol. 68, 403–407. [DOI] [PubMed] [Google Scholar]
- Cheung, V G, Spielman, R S, Ewens, K G, Weber, T M, Morley, M, and Burdick, J T (2005). “Mapping determinants of human gene expression by regional and genome-wide association.” Nature (London) 437, 1365–1369. 10.1038/nature04244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly, P (2008). “Progress and challenges in genome-wide association studies in humans.” Nature (London) 456, 728–731. 10.1038/nature07631 [DOI] [PubMed] [Google Scholar]
- Jansen, R C, and Nap, J P (2001). “Genetical genomics: the added value from segregation.” Trends Genet. 17, 388–391. 10.1016/S0168-9525(01)02310-1 [DOI] [PubMed] [Google Scholar]
- Monks, S A, Leonardson, A, Zhu, H, Cundiff, P, and Pietrusiak, P (2004). “Genetic inheritance of gene expression in human cell lines.” Am J. Hum. Genet. 75, 1094–1105. 10.1086/426461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, M, Molony, C M, Weber, T M, Devlin, J L, Ewens, K G, Spielman, R S, and Cheung, V G (2004). “Genetic analysis of genome-wide variation in human gene expression.” Nature (London) 430, 743–747. 10.1038/nature02797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlstein, E O, Ruderfer, D M, Roberts, D C, Schreiber, S L, and Kruglyak, L (2007). “Genetic basis of individual differences in the response to small-molecule drugs in yeast.” Nat. Genet. 39, 496–502. 10.1038/ng1991 [DOI] [PubMed] [Google Scholar]
- Rockman, M V, and Kruglyak, L (2006). “Genetics of global gene expression.” Nat. Rev. Genet. 7, 862–872. 10.1038/nrg1964 [DOI] [PubMed] [Google Scholar]
- Ruderfer, D M, Roberts, D C, Schreiber, S L, Perlstein, E O, and Kruglyak, L (2009). “Using expression and genotype to predict drug response in yeast.” PLoS ONE 4, e6907. 10.1371/journal.pone.0006907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt, E E, Monks, S A, Drake, T A, Lusis, A J, Che, N, Colinayo, V, Ruff, T G, Milligan, S B, Lamb, J R, Cavet, G, Linsley, P S, Mao, M, Stoughton, R B, and Friend, S H (2003). “Genetics of gene expression surveyed in maize, mouse and man.” Nature (London) 422, 297–302. 10.1038/nature01434 [DOI] [PubMed] [Google Scholar]
- Schadt, E E, Lamb, J, Yang, X, Zhu, J, Edwards, S, Guhathakurta, D, Sieberts, S K, Monks, S, Reitman, M, Zhang, C, Lum, P Y, Leonardson, A, Thieringer, R, Metzger, J M, Yang, L, Castle, J, Zhu, H, Kash, S F, Drake, T A, Sachs, Aand Lusis, A J(2005). “An integrative genomics approach to infer causal associations between gene expression and disease.” Nat. Genet. 37, 710–717. 10.1038/ng1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger, B E, Forrest, M S, Clark, A G, Minichiello, M J, Deutsch, S, Lyle, R, Hunt, S, Kahl, B, Antonarakis, S E, Tavare, S, Deloukas, P, and Dermitzakis, E T (2005). “Genome-wide associations of gene expression variation in humans.” PLoS Genet. 1, e78. 10.1371/journal.pgen.0010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, F, McGinnis, R, Bourgeois, S, Barnes, C, Eriksson, N, Soranzo, N, Whittaker, P, Ranganath, V, Kumanduri, V, McLaren, W, Holm, L, Lindh, J, Rane, A, Wadelius, M, and Deloukas, P (2009). “A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose.” PLoS Genet. 5, e1000433. 10.1371/journal.pgen.1000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher, P M, Hill, W G, and Wray, N R (2008). “Heritability in the genomics era—concepts and misconceptions.” Nat. Rev. Genet. 9, 255–266. 10.1038/nrg2322 [DOI] [PubMed] [Google Scholar]