Abstract
The evolutionary histories of complex traits are complicated because such traits are comprised of multiple integrated and interacting components, which may have different individual histories. Phylogenetic studies of complex trait evolution often do not take this into account, instead focusing only on the history of whole, integrated traits; for example, mapping eyes as simply present or absent through history. Using the biochemistry of animal vision as a model, we demonstrate how investigating the individual components of complex systems can aid in elucidating both the origins and diversification of such systems. Opsin-based phototransduction underlies all visual phenotypes in animals, using complex protein cascades that translate light information into changes in cyclic nucleotide gated (CNG) or canonical transient receptor potential (TRPC) ion-channel activity. Here we show that CNG ion channels play a role in cnidarian phototransduction. Transcripts of a CNG ion channel co-localize with opsin in specific cell types of the eyeless cnidarian Hydra magnipapillata. Further, the CNG inhibitor cis-diltiazem ablates a stereotypical photoresponse in the hydra. Our findings in the Cnidaria, the only non-bilaterian lineage to possess functional opsins, allow us to trace the history of CNG-based photosensitivity to the very origin of animal phototransduction. Our general analytical approach, based on explicit phylogenetic analysis of individual components, contrasts the deep evolutionary history of CNG-based phototransduction, today used in vertebrate vision, with the more recent assembly of TRPC-based systems that are common to protostome (e.g. fly and mollusc) vision.
Keywords: complex trait evolution, ancestral state, phototransduction, opsin, cnidarian, cyclic nucleotide gated ion channel
1. Introduction
Understanding the evolutionary origins of complexity is a central issue in biology. Studies of complex trait evolution are commonly conducted by mapping the presence or absence of whole, integrated traits on a phylogeny of related species. While such approaches allow important comparative inferences to be made on the timing and number of origins of the trait under investigation (Cunningham et al. 1998; Oakley & Cunningham 2002; Reznick et al. 2002; Avise 2006), they do not inform our understanding of the histories of its individual integrated components. Such individual components often vary over time and can shed light on the evolutionary patterns and processes of new trait origination (King et al. 2003; Edwards & Donoghue 2006; Plachetzki & Oakley 2007). Because the genes that specify individual components of complex traits are becoming increasingly well understood, hypotheses on the ancestral composition of such traits can now be tested by integrating studies on the evolution of their individual components.
Animal phototransduction cascades are prime examples of complex traits, with components that have been well characterized on functional, biochemical and genetic levels. Despite this progress, the evolutionary origins of these multi-component cascades remain little understood and have been the subject of both longstanding puzzlement among evolutionary biologists (Darwin 1859, 1987) and cynicism among others (Behe 1996). In particular, two types of ion channel delineate different modes of phototransduction known in animals. First, cyclic nucleotide gated (CNG) ion channels function in a ciliary phototransduction cascade, whereby CNG modulation effects a hyperpolarizing potential in the photoreceptor cell's response to light (Matulef & Zagotta 2003). Conversely, canonical transient receptor potential (TRPC) ion channels, members of a larger gene family of TRP channels (Venkatachalam & Montell 2007), function in a rhabdomeric pathway where activation leads to a depolarizing cell-physiological response to light (Hardie & Raghu 2001). Both CNG- and TRPC-modulated pathways are initiated by class-specific opsin paralogues (c-opsin and r-opsin, respectively), which are both present in protostomes and deuterostomes, and therefore predate the origin of bilaterian animals (Arendt et al. 2004; Velarde et al. 2005). In vertebrates, primary vision is mediated by CNG-based ciliary phototransduction, whereas in protostomes a TRPC-based rhabdomeric cascade is thought to serve this role (Hardie & Raghu 2001). Despite the centrality of these signalling pathways to vision and other photosensitivity phenotypes, an understanding of the origins of these cascades in animal evolution has evaded biologists, largely because accumulation of data from non-bilaterian animals like Cnidaria has lagged far behind the availability of data from bilaterians—especially flies, molluscs and vertebrates.
Therefore, as the only non-bilaterian extant animal lineage known to possess phototransductive opsin proteins (Plachetzki et al. 2007; Suga et al. 2008), data from cnidarian taxa such as anemones, jellyfish and hydra can inform our understanding of the origin and early evolution of phototransduction. Elements of the phototransduction machinery that are shared between cnidarians and bilaterian animals may have been present at the origin of opsin-based photosensitivity phenotypes in animals. Here, we integrate studies of phylogenetics, gene expression, behavioural pharmacogenetics and character evolution to show that a CNG-based phototransduction system was the ancestral state of opsin-mediated photosensitivity and visual phenotypes in animal evolution. Our results highlight the ancient assembly of CNG-based ciliary phototransduction, which underlies function in vertebrate rods and cones, and the evolutionarily more recent assembly of the TRPC-based visual pathways that are common to the rhabdomeric photoreceptors of insects and other protostomes.
2. Material and methods
(a). Data mining
Publicly available gene models for all but the sponge Amphimedon queenslandica were compiled using BlastDB and subjected to tblastn (Altschul et al. 1997) searches using a wide range of bilaterian CNG and TRP loci as bait. For A. queenslandica, trace genome data were mined (http://www.ncbi.nlm.nih.gov/Traces).
For opsin phylogeny, we first obtained opsin sequences that could be specifically correlated with either CNG, TRP or, in the case of photoisomerases, no phototransductive activity (electronic supplementary material, table S3). These were then used as bait sequences in similarity searches using blastp (Altschul et al. 1997) of non-redundant protein databases curated by uniprot (http://www.uniprot.org/). For each opsin where the phototransductive ion channel was known, an additional two sequences from uniref90 and two sequences from uniref50 were retained. Redundant sequences obtained by this procedure were removed (see electronic supplementary material).
(b). Phylogenetic analyses
Amino acid sequences comprising CNG, TRP and opsin datasets were aligned under default parameters using TCOFFEE (Notredame et al. 2000) and alignment manipulations were done using Seaview (Galtier et al. 1996). Phylogenetic analyses were conducted using maximum likelihood (ML) as implemented in RaxML (Stamatakis et al. 2005) and Bayesian Markov Chain Monte Carlo (BMCMC) approaches using PhyloBayes (Lartillot & Philippe 2004). Support for internal nodes was assessed with 1000 bootstrap replicates for ML and posterior probability for BMCMC analyses. ML analysis assumed the best-fit model WAG + I + G determined by ProtTest (Abascal et al. 2005). BMCMC analyses were conducted under the default CAT approach. Convergence of BMCMC chains was determined to have occurred once the mean likelihood difference between chains fell below 0.01. Bayesian ancestral state reconstructions were done in SIMMAP using stochastic mutational mapping (Bollback 2006) and 20 000 trees sampled from the posterior (the consensus of which is given in electronic supplementary material, figure S6). Character states were reconstructed under a discrete three-state model with equal priors on character transition bias, and using branch length as rate. Mesquite (Maddison & Maddison 2004) was used for ancestral state reconstructions under ML.
(c). In situ hybridization
A partial transcript for hmCNG was cloned by reference to an expressed sequence tag (GenBank accession no. DT606755). The isolation of the hmOps2 clone has been described previously (Plachetzki et al. 2007). The expression of hmOps2 and hmCNG was studied using multi-channel in situ hybridization. RNA probes were synthesized using DIG (hmOps2) and fluorescein (hmCNG; Roche). Hybridization was conducted as described by Grens et al. (1995), with modifications, and co-localization data were collected using a Fluoview 500 confocal microscope (see electronic supplementary material, methods).
(d). Animal culture and photobehaviour assay
Cultures of Hydra magnipapillata were maintained using standard methods (Lenhoff 1982). Individual hydras were tested in 5 ml of hydra medium in 20 ml pyrex dishes. Subjects were staged for a minimum of 20 min in total darkness to allow dark-adaptation and recovery from any transportation shock. After 20 min, individual subjects were exposed to blue light of either 5 lux for dim light trials or 3500 lux for bright light trials. Light of a sharply defined spectrum peaking at 470 nm was emitted from a light-emitting diode (LED) array (SuperBright LEDs). Light intensity as given in lux was measured using a Smart Luxmeter (Aquatic Ecosystems, Inc.). Hydra were tested for the contraction response (Passano & McCullough 1962, 1965) in individual arenas so that only one individual was tested at a time. The time until first complete contraction was recorded and experiments were terminated after 20 min if the animal did not contract. For pharmacological treatments, cis-diltiazem was used at a concentration of 1 µM in standard hydra medium (see also electronic supplementary material, methods).
3. Results
We first identified homologues of ion channel genes, with particular focus on non-bilaterian genomes. Bioinformatic screens recovered loci belonging to both the CNG and TRPC ion-channel gene families from most of 22 animal genomes included in the study, with the exception that TRPC loci were not recovered from the genomes of the sponge A. queenslandica or the placozoan Trichoplax adhaerens (a full listing is given in the electronic supplementary material, tables S1 and S2). A single CNG locus (hmCNG) with membership in the CNG A clade, which also includes representatives that function in vertebrate phototransduction, was recovered from the genome sequence of the hydra (electronic supplementary material, figures S1 and S2). A single putative but truncated TRPC locus was also identified in the hydra genome, but this gene is neither found in the hundreds of thousands of publicly available hydra EST sequences, nor could it be amplified from our cDNA preps. In addition, this sequence lacks protein-coding regions present in gene homologues from other species and it failed a reciprocal best-hit test of orthology with known TRPC genes. For these reasons we concluded that this cnidarian TRPC locus is a pseudogene (for details, see electronic supplementary material, methods and figures S3 and S4).
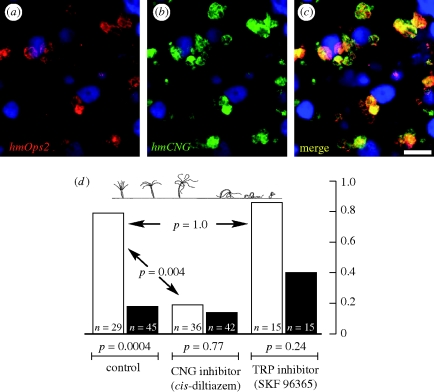
Because intermediary enzymes (Kozmik et al. 2008) and second messengers (Koyanagi et al. 2008) associated with the CNG-activating, ciliary mode of phototransduction have recently been identified in the camera eyes of cubozoan jellyfish, we hypothesized that hmCNG plays a role in cnidarian phototransduction. Supporting this hypothesis, hydra hmCNG is specifically co-expressed with a previously described (Plachetzki et al. 2007) member of the cnidops clade of cnidarian opsins, hmOps2. Cells expressing both hmOps2 and hmCNG are distributed in neurons throughout the animal, and are especially concentrated in the oral hypostome and in cells associated with the cells of cnidarians known as nematocytes, which include stinging and other ejective cells (figure 1a–c).
Figure 1.
Studies of gene expression and behavioural pharmacogenetics suggest the involvement of a CNG ion channel in cnidarian phototransduction. (a–c) Transcripts of cnidarian opsin and CNG ion channels co-localize to the same cell types. (a) hmOps2, red; (b) hmCNG, green; (c) merge; nuclei, blue. Confocal optical section = 0.2 µm. Scale bar = 50 µm. (d) The contraction response (top) in the hydra is a photobehaviour that is perturbed by a CNG inhibitor but not a TRPC inhibitor. Animal subjects were adapted in total darkness prior to presentation with bright light (see §2). For each treatment, behavioural trials were conducted under conditions of blue light (3500 lux, 470 nm; white bars) or dim conditions (5 lux, 470 nm; black bars). Under control conditions a significant difference exists in the behaviour of animal subjects, indicating a direct response to light. Under treatment with the CNG inhibitor cis-diltiazem (5 µM), the frequency of the response to bright light is decreased significantly. However, no significant difference exists between bright light trials conducted in the presence of the TRPC inhibitor SKF 96395 (1 µM). Animals treated with either drug were responsive to mechanical stimuli following trials. Drugs were used at the highest non-lethal concentrations. Contraction response illustration is from Passano & McCullough (1963).
The co-localization of hmOps2 and hmCNG suggested a possible functional relationship in phototransduction, which we tested using a behavioural assay. A diversity of photobehaviours has been described in cnidarian taxa that range from diurnal migration in pelagic taxa (Mackie 1999) to phototaxis in benthic species (Ewer 1947). Because of its reproducibility in an experimental setting, we used one previously described photobehaviour in hydra—the contraction response—to further test the possible role of CNG in cnidarian phototransduction. Dark-adapted hydras display a series of predictable and repeatable postures that culminate in the tight retraction of the animal into its most condensed state after presentation with bright light (Passano & McCullough 1962, 1965; Rushforth 1973; figure 1d). We assayed the ability of the CNG inhibitor cis-diltiazem (Haynes 1992) to ablate this contraction response in dark-adapted hydra. Upon stimulation with a bright blue light generated by an LED array, most (23 out of 29) individually assayed hydra subjects displayed the contraction response within 20 min. However, under dim light conditions only eight out of 45 trials resulted in a response. The results from these control experiments (bright light versus dim light) are significantly different (Fisher's exact test, p = 4 × 10−4), confirming the long-known behavioural response to light. However, in the presence of the CNG inhibitor the contraction response to stimulation with bright light rarely occurred (7 responses in 36 trials). The difference between the control (no drug) and experimental (drug) bright light trials is also significant (p = 4 × 10−3), consistent with a pharmacological inhibition of the light-induced contraction behaviour. In addition, data resulting from two dim light trials (drug and no drug) were not significantly different (p = 0.77), thus demonstrating the dependence of CNG-mediated behaviour on light. Even though we found no evidence for a functional TRPC gene in the hydra genome (discussed in §4), we also assayed the efficacy of SKF 96365, a TRPC inhibitor (Merritt et al. 1990), to ablate the light-activated contraction response, and found this agent to have no significant effect (figure 1d). In all experimental trials, animals responded to mechanical stimulation directly following the experiments, thus eliminating the possibility that drug treatments ablated total nervous system function. Together with our gene expression data and previous indirect evidence (Koyanagi et al. 2008; Kozmik et al. 2008), our behavioural–pharmacological results strongly implicate CNG function in cnidarian phototransduction.
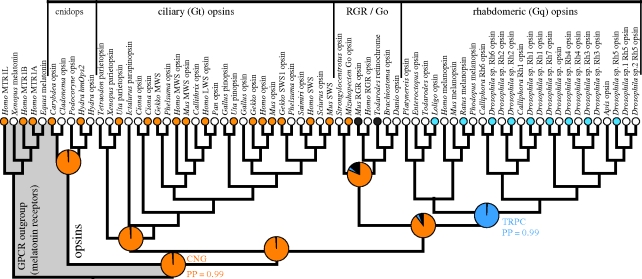
In order to assess the ancestral and intervening states of ion channel components during the origin and diversification of animal phototransduction cascades, we mapped data reported here, together with the published data (electronic supplementary material, table S2), onto a phylogeny for animal opsins under an explicit statistical model of character evolution. Our results clearly support the hypothesis that the ancestral animal phototransduction cascade used a CNG ion channel for signalling (posterior probability and proportion of likelihood both = 0.99; figure 2). Our results also indicate with strong support that the ancestral rhabdomeric pathway, denoted in our analysis by a monophyletic clade of r-opsins, used TRPC, not CNG, in signalling, and therefore represents a functional transition from a CNG- to a TRPC-based cascade. This transition must have occurred along the branch leading to the rhabdomeric opsins, prior to the origin of bilaterians but after cnidarians and bilaterians diverged. Importantly, posterior probabilities for these reconstructed ancestral states were determined by integrating across all possible trees in proportion to the inferred posterior probability that each tree is correct given our model for molecular evolution (see §2). Therefore, our conclusions from Bayesian analyses are robust to alternative rooting hypotheses for animal opsins, which are difficult to distinguish with certainty. For example, although we illustrate the ML opsin phylogeny in figure 1 with cnidops as the sister group to all other opsins, this root placement is not robust in Bayesian analyses, and the totality of evidence favours root placement between ciliary and all other opsins (Plachetzki et al. 2007).
Figure 2.
Ancestral state reconstruction supports the hypothesis that CNG ion channels functioned in the ancestral phototransduction cascade. A phylogeny for animal opsins represents the evolutionary history of the phototransduction cascades. Outgroups (grey) were selected based on a previous study of GPCR evolution (Fredriksson et al. 2003). Functional relationships between specific opsin sequences and their ion channel types were coded based on new data reported here and those abstracted from the literature (orange, CNG; blue, TRPC; black, no phototransduction as per photoisomerases). See electronic supplementary material, table S2 for justification of functional data. Additional sequences (white) mined from public databases (see §2). Reconstruction of ancestral states using character mapping reveals, with a strong proportion of likelihood (pie chart), that the ancestral animal phototransduction cascade used a CNG ion channel (orange; proportion of likelihood = 0.99). The node joining the rhabdomeric opsin is also reconstructed, with a high proportion of likelihood, to have used a TRPC ion channel (blue; proportion of likelihood = 0.99). Additional analyses using Bayesian methodology strongly support these findings. Outgroup rooted ML tree is shown (see electronic supplementary material, figure S5 for support indices). Posterior probabilities (PP) of reconstructed states from Bayesian analyses integrate over all topologies sampled from the posterior (electronic supplementary material, figure S6) and allow multiple character state transitions to occur along branches.
4. Discussion
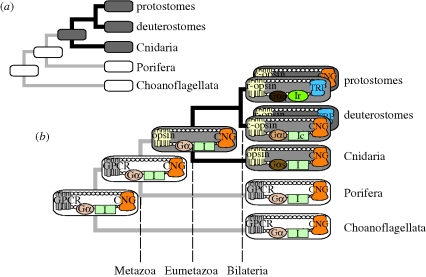
This study exemplifies how tracing the evolutionary histories of individual components of integrated systems can provide a framework for understanding how complex traits originate during evolution (figure 3). Our results indicate that the TRPC-based phototransduction cascade used in the rhabdomeric visual systems of many modern invertebrates probably originated from within a CNG-based radiation of cascades. This transition probably involved an exchange of multiple components because the transition we report here, from CNG to TRPC ion channel, is also associated with a change to Gα-q as the G-protein component of rhabdomeric phototransduction (Hardie & Raghu 2001). Such a G-protein transition, occurring prior to the bilaterian ancestor, may have been caused by amino acid changes in the third cytoplasmic loop of opsin, which activate Gα-q (Marin et al. 2000; Arendt et al. 2004; Plachetzki et al. 2007) and lead to modulation of TRPC ion channels. While we infer that this transition gave rise to a novel light-gated cell physiology, which also correlates temporally with the origin of a new morphological class of photoreceptors (Eakin 1979; Arendt 2003), other alterations in Gα subunit usage that did not precipitate shifts in ion channel usage must have also occurred. Our previous attempts to reconstruct the ancestral G-protein complement in animal phototransduction proved equivocal (Plachetzki et al. 2007), because as many as four different Gα paralogues are presently known to participate in various modes of animal vision, including Gαi in vertebrates (Franke et al. 1990), Gαo in molluscs (Gomez & Nasi 2000), Gαs in cubozoan cnidarians (Koyanagi et al. 2008) and Gαq in many protostomes (Hardie & Raghu 2001). While our current analyses suggest that the ion channel components of animal phototransduction cascades remain deeply conserved in evolution, the Gα components of these systems appear to be less conserved.
Figure 3.
Examining the histories of their individual components can illuminate the origination of complex traits. (a) Illustrated is a traditional view of animal phylogeny (Philippe et al. 2009) with the presence (grey) and absence (white) of opsin-based phototransduction mapped onto the tree using parsimony. This approach illustrates that cnidarians, including Hydra magnipapillata, are in a key phylogenetic position to inform our view of the origins of this complex trait. (b) Analysing the individual components of phototransduction in a phylogenetic context yields deeper insights into its origin and evolution. Round-cornered rectangles represent phototransduction (grey) or other G-protein-coupled receptor (GPCR) pathways (white), with individual components inside. Our results indicate that CNG ion channels are involved in hydra phototransduction (figure 1) as well as the ancestral phototransduction cascade (figure 2). Because CNG is present outside eumetazoan animals and is involved in non-opsin GPCR pathways (Stumpf et al. 2009), we infer that—like G-proteins and other phototransduction components (Suga et al. 1999; Plachetzki et al. 2007)—CNG ion channels predate phototransduction. Therefore, the origin of phototransduction may have involved the gain of light sensitivity in an ancient CNG–GPCR pathway. Our results also indicate that TRPC-based rhabdomeric phototransduction originated by changing multiple components. Protostomes and deuterostomes are illustrated to have both ciliary and rhabdomeric pathways (Arendt 2003), with the pathway dominant in vision positioned on top. Dashed lines indicate major clades in metazoan evolution. Additional abbreviations: c-opsin, ciliary opsin; r-opsin, rhabdomeric opsin; I, intermediary molecules; Ic, ciliary intermediaries such as phosphodiesterase (PDE) and guanylate cyclase (GC); Ir, rhabdomeric intermediaries such as phospholipase C (PLC), DAG (diacylglycerol) and PIP2 (phosphatidylinositol-4,5-biphosphate).
Although the origin of rhabdomeric phototransduction involved multiple components (including TRPC, Gαq and other intermediates associated with this pathway; see figure 3b), the origin of phototransduction in animals may have involved the alteration of only a single component—the receptor—allowing gain of light sensitivity in an ancient CNG-based GPCR pathway. We infer that the CNG ion channel component of the ancestral phototransduction pathway was present prior to the origin of animal opsins. CNG loci were recovered from taxa outside Eumetazoa (electronic supplementary material, figures S1 and S2) and, based on comparison of opsin-mediated pathways with related outgroup GPCR pathways like human melatonin (Stumpf et al. 2009), probably functioned in other GPCR pathways prior to the origin of functional opsins. In addition, earlier studies showed that G-proteins and other components of animal phototransduction cascades diversified prior to Metazoa (Suga et al. 1999). Of the components studied thus far, only the origin of opsin from non-opsin GPCR genes (by definition) corresponds with the origin of animal phototransduction (Plachetzki & Oakley 2007). Animal opsins originated prior to Eumetazoa, but opsins are unknown from non-animals, including fungi and choanoflagellates, and non-Eumetazoan animal lineages, such as poriferans and placozoans (Plachetzki et al. 2007; Suga et al. 2008). Further, type-I opsins common to bacteria and other taxa are not homologous to animal opsins (Spudich et al. 2000; Larusso et al. 2008).
Taken together, these data support a view that the origination of animal phototransduction, and by extension of animal vision itself, involved the evolution of an existing CNG-based cascade into a functionally new cascade, founded by a novel opsin lineage of light-sensitive G-protein-coupled receptors. Our inferences that CNG and G-proteins predate animals are robust to alternative views of animal phylogeny, but the specific timing of opsin origin depends on these assumptions. If the traditional view that sponges are the sister group to other animals is assumed (Philippe et al. 2009), then the functional transition to light sensitivity occurred during the phylogenetic interval separating Eumetazoa from earlier animal lineages at least 600 Myr ago (Peterson et al. 2004). A second major transition, from CNG- to TRPC-based phototransduction, would have occurred much later in animal evolution predating the bilaterian clade and giving rise to the rhabdomeric system.
Acknowledgements
We thank John Robson and two anonymous reviewers for their helpful comments on this manuscript. Funding was provided by NSF DEB-0710406 to D.C.P and T.H.O, and by NSF 0643840 to T.H.O.
References
- Abascal F., Zardoya R., Posada D.2005ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (doi:10.1093/bioinformatics/bti263) [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J.1997Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 25, 3389–3402 (doi:10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D.2003Evolution of eyes and photoreceptor cell types. Int. J. Dev. Biol. 47, 563–571 [PubMed] [Google Scholar]
- Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A. W., Wittbrodt J.2004Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869–871 (doi:10.1126/science.1099955) [DOI] [PubMed] [Google Scholar]
- Avise J. C.2006Evolutionary pathways in nature: a phylogenetic approach Cambrige, UK: Cambrige University Press [Google Scholar]
- Behe M. J.1996Darwin's black box New York, NY: Free Press [Google Scholar]
- Bollback J. P.2006SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinform. 7, 88 (doi:10.1186/1471-2105-7-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C. W., Omland K. E., Oakley T. H.1998Reconstructing ancestral character states: a critical reappraisal. Trends Ecol. Evol. 13, 361–366 (doi:10.1016/S0169-5347(98)01382-2) [DOI] [PubMed] [Google Scholar]
- Darwin C.1859On the origin of the species by means of natural selection, or the preservation of favoured races in the struggle for life London, UK: John Murray; [PMC free article] [PubMed] [Google Scholar]
- Darwin C.1987Transmutation notebooks 1836–1844. In Charles Darwin's notebooks (ed. Kohn D. B.-E., transcriber). Ithaca, NY: Cornell University Press [Google Scholar]
- Eakin R. M.1979Evolutionary significance of photoreceptors: in retrospect. Am. Zool. 19, 647–653 [Google Scholar]
- Edwards E. J., Donoghue M. J.2006Pereskia and the origin of the cactus life-form. Am. Nat. 167 (doi:10.1086/504605) [DOI] [PubMed] [Google Scholar]
- Ewer R. F.1947On the functions and mode of action of the nematocytes of hydra. Proc. Zool. Soc. Lond. 117, 365–376 [Google Scholar]
- Franke R. R., Konig B., Sakmar T. P., Khorana H. G., Hofmann K. P.1990Rhodopsin mutants that bind but fail to activate transducin. Science 250, 123–125 (doi:10.1126/science.2218504) [DOI] [PubMed] [Google Scholar]
- Fredriksson R., Lagerstrom M. C., Lundin L. G., Schioth H. B.2003The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 (doi:10.1124/mol.63.6.1256) [DOI] [PubMed] [Google Scholar]
- Galtier N., Gouy M., Gautier C.1996SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12, 543–548 [DOI] [PubMed] [Google Scholar]
- Gomez M. P., Nasi E.2000Light transduction in invertebrate hyperpolarizing photoreceptors: possible involvement of a Go-regulated guanylate cyclase. J. Neurosci. 20, 5254–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grens A., Mason E., Marsh J. L., Bode H. R.1995Evolutionary conservation of a cell fate specification gene: the Hydra achaete-scute homolog has proneural activity in Drosophila. Development 121, 4027–4035 [DOI] [PubMed] [Google Scholar]
- Hardie R. C., Raghu P.2001Visual transduction in Drosophila. Nature 413, 186–193 (doi:10.1038/35093002) [DOI] [PubMed] [Google Scholar]
- Haynes L. W.1992Block of the cyclic GMP-gated channel of vertebrate rod and cone photoreceptors by l-cis-diltiazem. J. Gen. Physiol. 100, 783–801 (doi:10.1085/jgp.100.5.783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N., Hittinger C. T., Carroll S. B.2003Evolution of key cell signaling and adhesion protein families predates animal origins. Science 301, 361–363 (doi:10.1126/science.1083853) [DOI] [PubMed] [Google Scholar]
- Koyanagi M., Takano K., Tsukamoto H., Ohtsu K., Tokunaga F., Terakita A.2008Jellyfish vision starts with cAMP signaling mediated by opsin-G(s) cascade. Proc. Natl Acad. Sci. USA 105, 15 576–15 580 (doi:10.1073/pnas.0806215105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z., et al. 2008Assembly of the cnidarian camera-type eye from vertebrate-like components. Proc. Natl Acad. Sci. USA 105, 8989–8993 (doi:10.1073/pnas.0800388105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N., Philippe H.2004A Bayesian mixture model for across-site heterogeneities in the amino acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (doi:10.1093/molbev/msh112) [DOI] [PubMed] [Google Scholar]
- Larusso N. D., Ruttenberg B. E., Singh A. K., Oakley T. H.2008Type II opsins: evolutionary origin by internal domain duplication? J. Mol. Evol. 66, 417–423 (doi:10.1007/s00239-008-9076-6) [DOI] [PubMed] [Google Scholar]
- Lenhoff H.1982Water, culture solutions and buffers. In Hydra research methods (ed. Lenhoff H.), pp. 19–28 New York, NY: Plenum [Google Scholar]
- Mackie G. O.1999Colenterate organs. Mar. Freshw. Behav. Physiol. 32, 113–127 (doi:10.1080/10236249909379043) [Google Scholar]
- Maddison W. P., Maddison D. R.2004Mesquite: a modular system for evolutionary analysis. Version 2.72. http://mesquiteproject.org [Google Scholar]
- Marin E. P., Krishna A. G., Zvyaga T. A., Isele J., Siebert F., Sakmar T. P.2000The amino terminus of the fourth cytoplasmic loop of rhodopsin modulates rhodopsin–transducin interaction. J. Biol. Chem. 275, 1930–1936 (doi:10.1074/jbc.275.3.1930) [DOI] [PubMed] [Google Scholar]
- Matulef K., Zagotta W. N.2003Cyclic nucleotide-gated ion channels. Annu. Rev. Cell. Dev. Biol. 19, 23–44 (doi:10.1146/annurev.cellbio.19.110701.154854) [DOI] [PubMed] [Google Scholar]
- Merritt J. E., et al. 1990SK&F 96365, a novel inhibitor of receptor-mediated calcium entry. Biochem. J. 271, 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J.2000T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (doi:10.1006/jmbi.2000.4042) [DOI] [PubMed] [Google Scholar]
- Oakley T. H., Cunningham C. W.2002Molecular phylogenetic evidence for the independent evolutionary origin of an arthropod compound eye. Proc. Natl Acad. Sci. USA 99, 1426–1430 (doi:10.1073/pnas.032483599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passano L. M., McCullough C. B.1962The light response and the rhythmic potentials of hydra. Proc. Natl Acad. Sci. USA 48, 1376–1382 (doi:10.1073/pnas.48.8.1376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passano L. M., McCullough C. B.1963Pacemaker hierarchies controlling the behaviour of hydras. Nature 199, 1174–1175 (doi:10.1038/1991174a0) [DOI] [PubMed] [Google Scholar]
- Passano L. M., McCullough C. B.1965Co-ordinating systems and behaviour in hydra. II. The rhythmic potential system. J. Exp. Biol. 42, 205–231 [DOI] [PubMed] [Google Scholar]
- Peterson K. J., Lyons J. B., Nowak K. S., Takacs C. M., Wargo M. J., McPeek M. A.2004Estimating metazoan divergence times with a molecular clock. Proc. Natl Acad. Sci. USA 101, 6536–6541 (doi:10.1073/pnas.0401670101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H., et al. 2009Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712 (doi:10.1016/j.cub.2009.02.052) [DOI] [PubMed] [Google Scholar]
- Plachetzki D., Oakley T. H.2007Key transitions during the evolution of animal phototransduction: novelty, ‘tree-thinking,’ co-option, and co-duplication. Integr. Comp. Biol. 47, 759–769 (doi:10.1093/icb/icm050) [DOI] [PubMed] [Google Scholar]
- Plachetzki D. C., Degnan B. M., Oakley T. H.2007The origins of novel protein interactions during animal opsin evolution. PLoS ONE 2, e1054 (doi:10.1371/journal.pone.0001054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick D. N., Mateos M., Springer M. S.2002Independent origins and rapid evolution of the placenta in the fish genus Poeciliopsis. Science 298, 1018–1020 (doi:10.1126/science.1076018) [DOI] [PubMed] [Google Scholar]
- Rushforth N. B.1973Behavioral modifications in Colenterates. In Invertebrate learning, vol. 1 (eds Corning W., Dyal J., Willows A.), pp. 123–269 New York, NY: Plenum [Google Scholar]
- Spudich J. L., Yang C. S., Jung K. H., Spudich E. N.2000Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell. Dev. Biol. 16, 365–392 (doi:10.1146/annurev.cellbio.16.1.365) [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Ludwig T., Meier H.2005RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21, 456–463 (doi:10.1093/bioinformatics/bti191) [DOI] [PubMed] [Google Scholar]
- Stumpf I., Bazwinsky I., Peschke E.2009Modulation of the cGMP signaling pathway by melatonin in pancreatic beta-cells. J. Pineal Res. 46, 140–147 (doi:10.1111/j.1600-079X.2008.00638.x) [DOI] [PubMed] [Google Scholar]
- Suga H., Koyanagi M., Hoshiyama D., Ono K., Iwabe N., Kuma K., Miyata T.1999Extensive gene duplication in the early evolution of animals before the parazoan–eumetazoan split demonstrated by G proteins and protein tyrosine kinases from sponge and hydra. J. Mol. Evol. 48, 646–653 (doi:10.1007/PL00006508) [DOI] [PubMed] [Google Scholar]
- Suga H., Schmid V., Gehring W. J.2008Evolution and functional diversity of jellyfish opsins. Curr. Biol. 18, 51–55 (doi:10.1016/j.cub.2007.11.059) [DOI] [PubMed] [Google Scholar]
- Velarde R. A., Sauer C. D., Walden K. K., Fahrbach S. E., Robertson H. M.2005Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem. Mol. Biol. 35, 1367–1377 (doi:10.1016/j.ibmb.2005.09.001) [DOI] [PubMed] [Google Scholar]
- Venkatachalam K., Montell C.2007TRP channels. Annu. Rev. Biochem. 76, 387–417 (doi:10.1146/annurev.biochem.75.103004.142819) [DOI] [PMC free article] [PubMed] [Google Scholar]