Abstract
Recent outbreaks of highly pathogenic avian influenza (HPAI) in poultry have raised interest in the interplay between avian influenza (AI) viruses and their wild hosts. Studies linking virus ecology to host ecology are still scarce, particularly for non-duck species. Here, we link capture–resighting data of greater white-fronted geese Anser albifrons albifrons with the AI virus infection data collected during capture in The Netherlands in four consecutive winters. We ask what factors are related to AI virus prevalence and whether there are ecological consequences associated with AI virus infection in staging white-fronted geese. Mean seasonal (low pathogenic) AI virus prevalence ranged between 2.5 and 10.7 per cent, among the highest reported values for non-duck species, and occurred in distinct peaks with near-zero prevalence before and after. Throat samples had a 2.4 times higher detection frequency than cloacal samples. AI virus infection was significantly related to age and body mass in some but not other winters. AI virus infection was not related to resighting probability, nor to maximum distance travelled, which was at least 191 km during the short infectious lifespan of an AI virus. Our results suggest that transmission via the respiratory route could be an important transmission route of AI virus in this species. Near-zero prevalence upon arrival on their wintering grounds, in combination with the epidemic nature of AI virus infections in white-fronted geese, suggests that white-fronted geese are not likely to disperse Asian AI viruses from their Siberian breeding grounds to their European wintering areas.
Keywords: disease dispersal, fitness consequences, migratory birds, virus epidemics, waterfowl
1. Introduction
Recent outbreaks of highly pathogenic avian influenza (HPAI) in poultry have initiated a range of studies examining the interplay between avian influenza (AI) A viruses and their wild hosts (Chen et al. 2005; Olsen et al. 2006; Dugan et al. 2008; Lang et al. 2008; Latorre-Margalef et al. 2009a; Rohani et al. 2009). Although it is considered unlikely that HPAI has repeatedly been introduced in poultry by migrating birds (Olsen et al. 2006), HPAI virus of the H5N1 subtype has been identified in apparently healthy birds (Chen et al. 2006; Gaidet et al. 2008; Kou et al. 2009), and wild birds can spread this virus to previously uninfected areas (Olsen et al. 2006). Furthermore, low pathogenic avian influenza (LPAI) virus of the H5 and H7 subtypes from wild birds can become highly pathogenic after spread in poultry (Röhm et al. 1996; Duan et al. 2007). Knowledge of the ecology and dynamics of LPAI may therefore help us better understand and anticipate the dynamics and consequences of HPAI outbreaks.
Waterbirds are considered to form the natural reservoir of AI viruses (Hinshaw et al. 1980; Webster et al. 1992). The habits of this species group seem particularly suitable for transmission, spread and persistence of AI viruses (Garamszegi & Møller 2007). Many waterbird species are long-distance migrants, breed in (mixed-species) colonies and congregate in large numbers in staging areas outside the breeding season. Here, viruses may readily be transmitted between individuals from the same or different species.
Despite the recent surge in papers on the topic, our current knowledge of the epidemiology of AI viruses, virus ecology in relation to host ecology, the temporal and spatial patterns of AI viruses in their natural hosts, and the role of potential new hosts in AI virus ecology is still very limited (Krauss et al. 2004; Munster et al. 2007). Many studies have focused on dabbling ducks of the genus Anas, the species group in which virus prevalence is generally highest (e.g. Olsen et al. 2006; Ip et al. 2008; Koehler et al. 2008; Latorre-Margalef et al. 2009a). AI virus prevalence in ducks peaks just after the breeding season, when a high percentage of the population consists of immunologically naive juveniles (Krauss et al. 2004; Wallensten et al. 2007). Just prior to and during the post-breeding migration to winter staging areas, AI virus prevalence may be as high as 60 per cent. Prevalence subsequently steadily declines to a seasonal low during spring migration (0.25–6.5%; Olsen et al. 2006). The intestinal tract is the main site of AI virus detection in ducks, while the respiratory tract plays a limited role (Munster et al. 2009). Ducks generally do not show any apparent disease signs after AI virus infection (e.g. Kida et al. 1980) and migration patterns are not affected by AI virus infection (Latorre-Margalef et al. 2009a).
It is unclear whether these patterns also apply to other species groups. For example, van Gils et al. (2007) found that the behaviour of two AI-virus-infected Bewick's swans (Cygnus columbianus bewickii) differed significantly from uninfected individuals. Infected swans experienced delayed migration, travelled shorter distances and fuelled at reduced rates. This suggests that temporal and spatial dynamics of AI may differ between species from different genera.
The Netherlands is the main winter staging area for geese in Europe with annual winter peak numbers of about two million geese (van Roomen et al. 2007). Since 1998, individuals of the most abundant species, the greater white-fronted goose, have been caught, measured and ringed with standard metal rings and a portion of them are additionally being equipped with uniquely identifiable neckbands before release. Since November 2005, most of these birds have been sampled consistently to test for AI virus infection. To improve our understanding of AI virus epidemiology, ecology and the dynamics of virus in waterbird hosts other than Anas species, we linked the capture-resighting data of individual white-fronted geese with their biometric data and AI virus infection test results. We asked how AI virus prevalence changes during the winter season, what factors are related to it in staging white-fronted geese, whether the intestinal tract or the respiratory tract is the main site of replication and whether there are ecological consequences associated with AI virus infection. Ultimately, we discuss what the consequences are of our findings for the spread of LPAI virus by white-fronted geese in northwestern Europe.
2. Material and methods
(a). Geese sampling
The greater white-fronted goose (henceforth white-fronted goose) breeds in the arctic tundra and has a circumpolar breeding range. The species is divided into five subspecies, of which the nominate subspecies Anser albifrons albifrons breeds between the Kanin Peninsula and the Khatanga and Popingay rivers east of the Taimyr Peninsula. This subspecies winters in Europe and southwestern Asia. In recent years, numbers of 700 000 and more wintering white-fronted geese have regularly been observed in The Netherlands, representing around 80 per cent of the European wintering population (van Roomen et al. 2007). During daytime, these birds graze in groups, mostly on agricultural grasslands and often together with other goose species. At night, they congregate in large numbers to roost on large water bodies, which are generally shared with a range of other waterbird species. White-fronted geese generally leave their breeding areas in September and start arriving in The Netherlands in November, where they stay until early March.
Using mechanical clap-nets and live decoys, white-fronted geese were caught and sampled for AI virus infection in four areas in The Netherlands: near Leeuwarden (53°12′06″ N, 5°52′56″ E), around Bunschoten (52°14′51″ N, 5°20′50″ E), near Lith (51°46′51″ N, 5°23′55″ E) and—only in the winters of 2005–2006 and 2006–2007—near Middelburg (51°31′23″ N, 3°39′16″ E). Sampling sizes and periods differed between locations and years (electronic supplementary material, table S1). In the winter of 2005–2006, 1754 individuals were caught in 158 successful attempts; in the 2006–2007 season, 580 individuals (53 attempts); in the 2007–2008 season, 1071 individuals (91 attempts); and in the 2008–2009 season, 1510 individuals (108 attempts). In the first two seasons, the cloaca of each bird was swabbed to test for AI virus infection. In the 2007–2008 season, samples were taken from the throat (55%), the cloaca (16%) or the throat and the cloaca (29%). In the final year of the study, all birds were sampled in both cloaca and throat except for 34 birds that were swabbed in the throat only. Each bird was subsequently weighed, sexed and aged, and head and wing length were determined. Before release, all birds were ringed and a random selection of 1787 white-fronted geese received neckbands with unique character combinations. Resightings of these individually identifiable birds by volunteers throughout much of northwestern Europe were entered directly into a central database through the website www.geese.org.
(b). RNA isolation and virus detection
Samples were collected using sterile cotton swabs and stored in transport medium (Hank's balanced salt solution containing 0.5% lactalbumin, 10% glycerol, 200 U ml−1 penicillin, 200 lg ml−1 streptomycin, 100 U ml−1 polymyxin B sulphate, 250 lg ml−1 gentamycin and 50 U ml−1 nystatin; ICN Pharmaceuticals, Zoetermeer, The Netherlands) and transported to the laboratory where they were stored at −80°C until analysis. Before transport, the samples were stored at 4°C for a maximum of two weeks. These storage conditions may affect the probability of isolating viable viruses, but not the probability of virus detection using PCR (Munster et al. 2009).
RNA isolation and virus detection were performed as described previously (Munster et al. 2009). In short, RNA was isolated using a MagnaPure LC system with the MagnaPure LC total nucleic acid isolation kit (Roche Diagnostics, Almere, The Netherlands) and influenza A virus was detected using a real-time reverse transcriptase-PCR (RRT-PCR) assay targeting the matrix gene (M). The specificity of M RRT-PCR does not differ between throat and cloacal swabs (Keawcharoen et al. 2008). Amplification and detection was performed on an ABI7700 machine with the TaqMan EZ RT-PCR Core Reagents kit (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). M RRT-PCR-positive samples were subsequently used for the detection of H5 influenza A viruses by using an RRT-PCR targeting the H5 gene (H5 RRT-PCR). H5 gene-positive samples were subsequently characterized further by nucleotide sequencing of the haemagglutinin (HA) region spanning the cleavage site (Munster et al. 2009).
(c). Virus isolation and characterization
All M RRT-PCR-positive samples were used for virus isolation. M RRT-PCR-positive samples were briefly centrifuged and 100 µl was inoculated into the allantoic cavity of 11-day-old embryonated chicken eggs. The allantoic fluid was harvested after 2 days and influenza A virus was detected using a haemagglutination assay with turkey erythrocytes. The HA subtype of virus isolates was characterized using a haemagglutination inhibition assay with turkey erythrocytes and subtype-specific hyperimmune rabbit antisera raised against all 16 HA subtypes (Fouchier et al. 2005). The neuraminidase (NA) subtype of virus isolates was determined by RT-PCR and sequencing, using primers specific for the non-coding regions of NA, as described previously (Munster et al. 2007). More methodological details can be found in Keawcharoen et al. (2008) and Munster et al. (2009).
(d). Analysis
Geese were sampled simultaneously in different sample sites, but sample sizes differed between locations and seasons (electronic supplementary material, table S1). Because white-fronted geese are very mobile in their wintering areas and display little site fidelity (Ebbinge 2009), we decided to pool the data from different sampling sites. Virus prevalence in ducks does not change linearly in the course of the winter season (Munster et al. 2007; Latorre-Margalef et al. 2009a). To analyse what factors explain the presence of AI virus in white-fronted geese, we therefore used general additive models (GAMs; Hastie & Tibshirani 1990) with a logistic link function and assuming a binomial error distribution, followed by a likelihood ratio test (or G-test). The flexibility of a GAM can be expressed by the degrees of freedom (d.f.) of the spline modelling the relationship between response and explanatory variable, with flexible curves having more d.f. The models included the factors sex, age (juvenile, adult) and season (2005–2006, 2006–2007, 2007–2008, 2008–2009), and the variables wing length (in centimetres), weight (in grams) and sampling date (days since 1 October). We included the interaction season × date because we were interested in whether prevalence patterns differed between years. Forward significance testing at the 5 per cent level was used to examine whether adding a d.f. improved the fit of the GAM. Data of geese belonging to the same family group were not entirely independent (they were less variable than those of geese from different family groups). Nevertheless, in the analyses, they were considered as independent because we had no accurate information on the family relations of the caught geese.
We analysed whether AI virus infection affected resighting probability using generalized linear models (GLMs) with a logistic link function and assuming a binomial error distribution. Models included the factors season, sex and age, and the variables wing length, weight and sampling date. Because resighting probability is proportional to the number of days between sampling date and the start of spring migration, only linear effects of sampling date were investigated. Distance between sampling and resighting after various periods of time was analysed using models including season and sampling date, but with an identity link and logarithmic error distribution. In this analysis, birds that were sampled and resighted on the same dates and locations were considered to belong to family groups. To avoid pseudoreplication, only one randomly selected individual of each family group was included in the analyses. All models were fitted using standard facilities in GenStat (Payne et al. 2002).
3. Results
(a). Factors related to AI virus prevalence
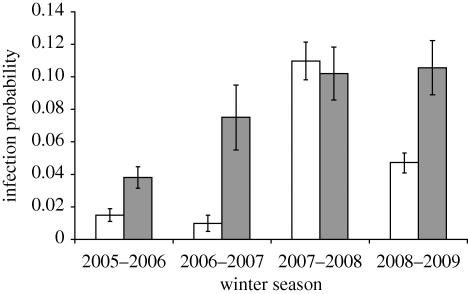
AI virus was detected in 5.5 per cent of the 4915 examined white-fronted goose samples. Initial analyses showed that infection rates differed significantly between seasons (2005–2006, 2.6%; 2006–2007, 2.9%; 2007–2008, 10.7%; 2008–2009, 6%; G-test, G = 77.7, 3 d.f., p < 0.001). Furthermore, the temporal patterns in prevalence differed between seasons (significant interaction between the curvilinear effects of sampling date and season; G = 54.7, 3 d.f., p < 0.001). Also, the model fitting the relationship between sampling date and AI virus infection independently for each season performed better than the model in which the same relationship was fitted in all four seasons (G = 140.06, 18 d.f., p < 0.001). We therefore examined the factors related to AI virus prevalence for each season separately. In all winters except that of 2007–2008, prevalence was significantly higher in juvenile compared with adult geese (table 1 and figure 1). Differences in AI virus prevalence were not related to the percentage of juveniles; in the four consecutive seasons, this was 46.5, 29.8, 32.2 and 22.6 per cent, respectively. Sex and wing length were not significantly related to AI virus infection in any of the years, but in 2008–2009 geese with a lower body weight had a higher AI infection probability (table 1; electronic supplementary material, fig. S1).
Table 1.
GAM test results of the relationship between LPAI virus infection of white-fronted geese and sampling date and bird characteristics.
| 2005–2006 |
2006–2007 |
2007–2008 |
2008–2009 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| d.f. | G | p | G | p | G | p | G | p | |
| sex | 1 | 2.70 | 0.100 | 0.70 | 0.396 | 1.00 | 0.317 | 0.58 | 0.448 |
| age | 1 | 7.95 | 0.005 | 16.19 | <0.001 | 0.11 | 0.737 | 13.94 | <0.001 |
| wing length | 1 | 0.03 | 0.872 | 2.42 | 0.128 | 2.95 | 0.086 | 1.41 | 0.235 |
| body mass | 1 | 0.03 | 0.868 | 0.10 | 0.747 | 0.05 | 0.829 | 10.32 | 0.001 |
| spline (date)a | 26.30 | <0.001 | 6.70 | 0.010 | 12.44 | <0.001 | 6.94 | 0.008 | |
aSpline (date) indicates the test statistics (G) of adding the final degrees of freedom (d.f.) that improved the fit of the model significantly (2005–2006, 1; 2006–2007, 2; 2007–2008, 8; 2008–2009, 8).
Figure 1.
AI virus infection probability of adult and juvenile white-fronted geese staging in The Netherlands during four consecutive winters. Bars indicate mean ± s.e. Unshaded bar, adult; shaded bar, juvenile.
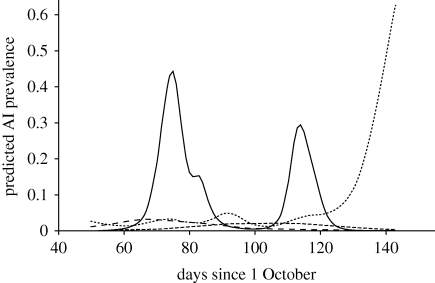
AI virus infection was significantly related to sampling date (table 1) in all winters, but the type of relationship differed between seasons (figure 2). In 2005–2006 and 2006–2007, AI virus prevalence followed approximate unimodal distributions with peaks around 5 and 29 December, respectively. Maximum infection probability in both seasons remained below 0.033 at all times. In the subsequent two seasons, AI virus prevalence showed pronounced peaks around 13 December 2007, 11 January 2008 and 21 February 2009, with in-between peaks, periods with zero or close to zero prevalence (figure 2). The 2007–2008 peaks were detected in geese sampled near Bunschoten and Lith; the 2008–2009 peak was detected primarily in geese sampled near Leeuwarden, with a small number of AI-virus-positive geese near Lith.
Figure 2.
AI virus infection in staging white-fronted geese during four consecutive winters. Lines indicate the predicted infection probability for male adult geese with average wing length and weight, based on the models given in table 1. In the 2005–2006 and 2006–2007 seasons, AI virus was sampled using cloacal swabs; in the 2007–2008 and 2008–2009 seasons, cloacal and throat swabs were used. An illustration of the observed infection probability is given in the electronic supplementary material, fig. S2. Long-dashed line, 2005–2006; short-dashed line, 2006–2007; solid line, 2007–2008; dotted line, 2008–2009.
Of the 231 MA RRT-PCR-positive samples, seven (3%) were of the H5 subtype as determined by the H5 RRT-PCR. All H5 RRT-PCR-positive samples could be characterized by sequencing of the region of HA spanning the cleavage site. All samples positive for the H5 subtype were of the low-pathogenic genotype. AI virus isolates were obtained from 19 (7.1%) M RRT-PCR positive samples. AI virus isolates were of the H1N1 (5.3%), H5N2 (15.8%), H5N3 (5.3%) and H6N2 (5.3%) subtype combinations. The majority (63.2%) of AI viruses were, however, of the H6N8 subtype combination that was detected in all sampling years except 2006–2007, in which only one isolate (H1N1) was obtained.
(b). Prevalence in intestinal versus respiratory tracts
Previous studies have shown that AI viruses in migratory mallards Anas platyrrhynchos are predominantly shed via the intestinal tract (Webster et al. 1992; Ellstrom et al. 2008; Munster et al. 2009). In total, 1705 individuals had been sampled concurrently in the cloaca and in the throat; 21 geese were found positive in both swab types, 19 only in the cloacal swabs and 75 only in the throat swabs. The difference in detection rate (2.4 times higher in throat compared with cloacal swabs) was significant (G = 24.73, 1 d.f., p < 0.001) and did not differ between the two seasons (2007–2008 and 2008–2009) for which both cloaca and throat (or oropharyngeal) samples were available (season × swab type interaction: G = 2.22, 1 d.f., p = 0.136). In these years, the rate of AI virus detection in cloacal samples was 2.3 per cent, and therefore similar to the rate of virus detection in the years 2005–2006 and 2006–2007, when only cloacal swabs were used.
The threshold cycle (CT) value is the first real-time amplification cycle in which target gene amplification is detectable. A low CT value indicates a high number of virus genome copies, and thus virus particles in the sample, whereas a high CT value indicates a small amount of virus (Munster et al. 2009). CT values of the 21 geese that tested positive for AI virus in both cloacal and throat samples did not differ significantly between the two sample types (paired t-test: t20 = −0.67, p = 0.511; mean cloacal swab Ct value: 32.56; mean throat swab Ct value: 33.07).
(c). Ecological consequences of AI virus infection?
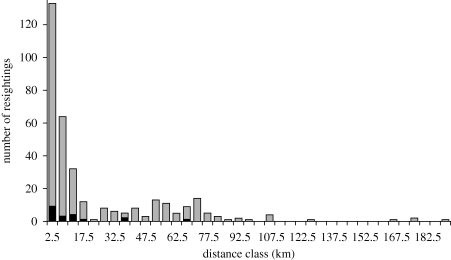
Ninety per cent of the 1787 white-fronted geese that received neckbands were resighted at least once after release. The percentage was somewhat higher for the 67 geese that tested positive, but this was not significantly different from AI-negative geese (93.7 versus 89.3; G = 0.17, 1 d.f., p = 0.676). Sex, age, wing length or body weight did not significantly affect the resighting probability (G < 1.07, 1 d.f., p > 0.30). The duration of AI virus infection in ducks is approximately 3–8 days (Latorre-Margalef et al. 2009a). We therefore examined the maximum distance covered by geese 1, 2, 4, 8 and 12 days after sampling. AI virus infection did not affect the mean maximum distance geese covered between 1 and 12 days after testing (p > 0.12). Note that the separate tests are not independent as they are to a considerable extent based on the same observations: the 503 geese that were resighted within 12 days of sampling were observed on average 1.8 times and 51 per cent were observed only once. The day after sampling, resighted geese were observed at a mean maximum distance of about 5 km from the ringing site. Between 1 and 12 days after ringing, the mean maximum distance increased by 2.2 km per day. There was a lot of variation between geese in the absolute maximum distance covered, however. The frequency distribution of the maximum distance between ringing site and resighting after 8 days followed an exponentially declining distribution (figure 3). The absolute maximum distance covered by a goose after 2, 4 and 8 days amounted to 93, 178 and 191 km, respectively.
Figure 3.
The frequency distribution of the maximum distance of resightings of white-fronted geese 8 days after they were sampled. Each label indicates the centre of a 5 km distance class. Grey bar, AI negative; black bar, AI positive.
4. Discussion
(a). AI virus prevalence in white-fronted geese in winter staging areas
Mean virus prevalence in the first two seasons was low and in the range of findings for other goose species (Bonner et al. 2004; Jonassen & Handeland 2007; Munster et al. 2007). The average prevalence of 10.7 per cent in the 2007–2008 season is among the highest AI virus prevalence reported thus far for a species outside the genus Anas. In waterfowl, high AI virus prevalence is often associated with the presence of a high proportion of immunologically naive juvenile birds in the population (Krauss et al. 2004). This did not explain the variation in AI virus prevalence in our study, however, as in the 2007–2008 season the percentage of juveniles was similar or lower than in the previous seasons, but AI virus prevalence was at least 3.7 times higher. Furthermore, in this season, prevalence in adult white-fronted geese was similar to that in juveniles.
The high AI virus prevalence in the 2007–2008 and 2008–2009 seasons was largely due to distinct peaks with maximum AI virus prevalence of 30 to 60 per cent. The narrowness of the peaks is in agreement with the most recent insights into AI virus infection duration in mallards, lasting 3–8 days (Latorre-Margalef et al. 2009a) and suggests multiple distinct outbreaks of AI virus. It is unclear whether the prevalence peaks in the last two seasons were localized events or occurred simultaneously throughout the entire wintering population. Some peaks, but not others, were detected simultaneously in more than one sample site, but between-site differences in size and temporal distribution of samples (electronic supplementary material, table S1) makes the interpretation of these findings difficult. However, white-fronted geese are very mobile on their wintering grounds and the core wintering area is relatively small. Twenty-one per cent of the geese sampled in this study were resighted within 8 days beyond 50 km, and 2.6 per cent beyond 100 km (e.g. figure 3). The distance between our sampling sites varied between 50 and 220 km, and this range covers most of the wintering area of the white-fronted geese (electronic supplementary material, fig. S3). Assuming that our sampling was representative, the peaks in AI virus infection were therefore probably present in a large proportion of the wintering population of white-fronted geese.
Although within-season multi-modal distributions of AI prevalence have been reported before for mallards (Halvorson et al. 1983; Hinshaw et al. 1985), they are at odds with the AI virus dynamics in surface-feeding ducks that have been reported in most recent studies. Most recent studies on ducks report a peak in prevalence just after the breeding season, a steady decline during southward autumn migration and a seasonal low during spring migration to the breeding grounds (Krauss et al. 2004; Bin Muzaffar et al. 2006; Munster et al. 2007; Wallensten et al. 2007; Ip et al. 2008). Because it is present in the population throughout the year, AI is considered to be endemic in ducks (Webster et al. 1992; Krauss et al. 2004). The multi-modal AI prevalence distribution suggests that AI virus infections behave epidemically in white-fronted geese. The near absence of AI virus prevalence upon arrival of white-fronted geese on their wintering grounds could be explained by white-fronted geese picking up AI virus only after their arrival on their wintering grounds. This would make white-fronted geese secondary hosts of AI virus at best. This hypothesis needs further testing and could be supported by the absence of AI virus infection on the Arctic breeding grounds of white-fronted geese.
Interestingly, the predominant AI virus detected within our study was of the H6N8 subtype (63%), which was detected in all sampling years except 2006–2007. The relative abundance of the detection of the H6 subtype within the white-fronted goose population does not correlate with the predominant subtypes detected within mallards, which are regarded as one of the major AI virus hosts and are the most frequently sampled bird in The Netherlands. In mallards, subtypes H1–H12 are continuously detected. Predominant HA subtypes detected in mallards in The Netherlands during the same sampling periods were: H3 (13.1%) and H4 (28.3%) in 2005–2006; H3 (23.8%) and H4 (27.0%) in 2006–2007; H4 (38.9%) and H6 (27.8%) in 2007–2008; and H3 (35.2%) and H4 (16.9%) in 2008–2009. Compared with other subtypes, the H6 AI viruses appear to have a broader host range, as they have been obtained frequently from dabbling ducks, gulls, auks, swans and geese (Munster et al. 2007).
(b). AI virus prevalence in intestinal versus respiratory tracts
The high prevalence of AI virus in the last two seasons of the study was largely due to the oropharyngeal (throat) samples that were taken in these but not the other seasons of the study. AI virus prevalence in cloacal samples was similar in all 4 years. The fact that M RRT-PCR-positive cloacal and oropharyngeal samples were obtained from white-fronted geese suggests that AI viruses replicate in both respiratory and intestinal tracts. However, white-fronted geese displayed a 2.4 times lower detection frequency in cloacal than in oropharyngeal samples. This contrasts with mallards, in which AI virus detection in cloacal samples was 2.2 times higher than in oropharyngeal samples (Munster et al. 2009). The similar virus loads in cloacal and oropharyngeal samples (indicated by their similar CT values) suggest that differences in transmission or ecology rather than in replication of AI viruses are causing the higher detection frequency in oropharyngeal samples. Dabbling ducks defecate in surface waters where they also forage. This probably makes faecal–oral transmission an effective route for AI virus persistence in populations of these species. Geese, on the other hand, forage mainly on land where they defecate in compact droppings, making it less probable that viruses are transmitted by uptake of faecal matter. We therefore speculate that direct infection from respiratory tract to respiratory tract (as in humans) is more effective in these species, which would be in agreement with the higher AI virus prevalence in oropharyngeal samples. In any case, the implementation of oropharyngeal sample collection from geese species in AI virus surveillance studies is essential to study the circulation of AI viruses in these host populations.
(c). Ecological consequences of AI virus infection for white-fronted geese
This is the first study that quantitatively examines the consequences of AI virus infection for dispersal of waterbirds (and thus spread of the virus) at the short time scale relevant to the virus infection cycle. Previously, van Gils et al. (2007) addressed this issue in Bewick's swans using GPS collars but could only examine dispersal behaviour of two AI-virus-infected birds. It is unclear how representative the observed reduced travelling distances are for AI-virus-infected Bewick's swans in general. Latorre-Margalef et al. (2009a) examined dispersal of mallards during the course of the wintering season and found no differences between infected and uninfected birds. Because of the extended period of time between AI virus sampling and recovery of the mallards in the study by Latorre-Margalef et al. (2009a), any effects on dispersal behaviour during the infection period could have been masked by dispersal behaviour when birds were virus-free.
In three of four seasons, infected and uninfected birds did not differ in body mass, which is in line with previous studies (e.g. Webster et al. 1978; Kida et al. 1980; van Gils et al. 2007). In the fourth season, however, AI-virus-infected birds had significantly lower body mass than uninfected birds. Similarly, Latorre-Margalef et al. (2009a) found significant year × sex interactions for effects of LPAI virus infection on body weight of mallards, suggesting that effects of LPAI may differ between years. It remains unclear whether AI virus infection results in lower body mass or birds with lower body mass are more likely to become infected with AI virus (Flint & Franson 2009; Latorre-Margalef et al. 2009b). It does suggest that AI epidemics may have different characteristics in different years. This is also indicated by the fact that juveniles had higher AI virus prevalence in the first two seasons and the last season (in agreement with findings of Ip et al. 2008), but not in the third season. Whether these differences are the result of the dominance of different virus subtypes in different years or of the same subtype combinations under different environmental conditions remains to be examined.
Dispersal of infected white-fronted geese during the first 12 days after sampling did not differ in any way from that of uninfected birds (note that the statistical power of analyses on dispersal up to 4 days was low due to small numbers of observed geese). The potential spread of AI virus by white-fronted geese in the wintering season can therefore be explored using all birds caught and resighted. The results highlight the unpredictable nature of the dynamics of AI virus spread in the wintering white-fronted goose population, even without considering the potential effects on other waterbird species. AI virus prevalence varies considerably in the course of the wintering season (figure 2), and maximum dispersal distance and direction varies markedly between individuals (figure 3). This makes dispersal of the AI virus a stochastic process, dependent on the number of geese that are infected, as well as the distance and direction in which they travel during the infectious lifespan of the virus in its host.
Nevertheless, the results do indicate that AI viruses can easily and rapidly spread through the entire western European wintering white-fronted goose population. More than 700 000 individuals of this species winter between November and March in The Netherlands (van Roomen et al. 2007). Additionally, a significant number of white-fronted geese winter in neighbouring parts of Belgium and Germany. The furthest distance covered by any of the 345 geese resighted after 8 days was 191 km. Using the four-season average AI virus prevalence of 5.5 per cent yields an estimate of a daily average of 112 white-fronted geese dispersing AI virus at least 191 km within 8 days of infection (700 000 × 1/345 × 0.055). If anything, this is a conservative estimate because the maximum distance at which geese were resighted underestimates the real maximum dispersal distance. Birds with shorter dispersal distances that were not resighted do not bias the results, whereas missed birds with longer dispersal distances do.
(d). Implications for spread of AI virus by white-fronted geese
Migration connects bird populations in time and space. Populations wintering in geographically widely separated areas may share breeding areas or, conversely, birds breeding in markedly different locations may use the same staging areas outside the breeding season (Bairlein 2001). This raises concern about the possibility of migratory waterbirds spreading HPAI H5N1 virus across continents (Chen et al. 2005; Liu et al. 2005; Olsen et al. 2006). The epidemic nature of AI virus infections in white-fronted geese, in combination with the near-zero prevalence upon arrival on their wintering grounds, suggests that white-fronted geese are not likely to disperse Asian AI viruses from their Siberian breeding grounds to their European wintering areas. An AI-virus-free departure of white-fronted geese from their breeding grounds followed by repeated infections on their different staging areas could explain the patterns observed in this study.
Our findings illustrate the large potential of waterbirds to disperse AI virus. The wintering population of white-fronted geese in western Europe forms a single reservoir in which AI viruses can be introduced and spread. AI virus picked up from other species at common roosting sites may thus spread rapidly through the entire population, as well as being passed on to other species. It is unknown what this implies for the potential role of white-fronted geese in the spread of HPAI because knowledge of LPAI viruses in wild birds cannot simply be extrapolated to HPAI viruses (Olsen et al. 2006). Experimental infection with H5N1 virus in Chinese geese Anser cygnoides killed all infected birds within 7 days (Chen et al. 2006). Prior infection with LPAI virus may protect waterfowl against a lethal H5N1 subtype challenge (Pasick et al. 2007; Kalthoff et al. 2008). Nevertheless, no HPAI has been isolated so far from apparently healthy goose species and the bar-headed goose Anser indicus was the main victim of one of the largest outbreaks of HPAI in wild birds worldwide (Chen et al. 2006). These species are closely related to white-fronted geese, suggesting that HPAI may not be readily spread by this species because of its pathogenic effects.
Acknowledgements
Thanks are due to the ringers, E. van Oort, F. Leurs and S. Bakker, and all geese catchers, notably K. Polderdijk, H. van Kessel, A. van Schoten, M. van Schoten, A. Vegter and B. Sjonger for geese sampling. The help of P. Lexmond, O. Vuong, M. Schutten and J. Guldenmeester in the laboratory and P. Goedhart with the statistical analyses is gratefully acknowledged. Comments by P. Flint improved this manuscript. This work was carried out in the framework of the projects ‘Flyway conservation of migratory waterbirds’, BO-10-003-002, and ‘Voortzetting monitoring ganzen en Smienten ivm het voorkomen van vogelgriepvirussen in 2008/2009’, Verpl. Nr. 2001126, funded by the Dutch ministry of Agriculture, Nature Conservation and Food Quality, and projects by Erasmus Medical Center (EU DG-SANCO, EU FP6 project New Flubird, SSP/8.1 no 044490, and NIAID-NIH contract HHSN266200700010C).
References
- Bairlein F.2001Results of bird ringing in the study of migration routes. Ardea 89, 7–19 [Google Scholar]
- Bin Muzaffar S., Ydenberg R. C., Jones I. L.2006Avian influenza: an ecological and evolutionary perspective for waterbird scientists. Waterbirds 29, 243–257 [Google Scholar]
- Bonner B. M., et al. 2004Do Canada geese (Branta canadensis Linnaeus, 1758) carry infectious agents for birds and man? Europ. J. Wildlife Res. 50, 78–84 (doi:10.1007/s10344-004-0044-1) [Google Scholar]
- Chen H., Smith G. J. D., Zhang S. Y., Qin K., Wang J., Li K. S., Webster R. G., Peiris J. S. M., Guan Y.2005H5N1 virus outbreak in migratory waterfowl. Nature 436, 191–192 (doi:10.1038/nature03974) [DOI] [PubMed] [Google Scholar]
- Chen H., et al. 2006Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl Acad. Sci. USA 103, 2845–2850 (doi:10.1073/pnas.0511120103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., et al. 2007Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 81, 7529–7539 (doi:10.1128/JVI.00327-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan V. G., et al. 2008The evolutionary genetics and emergence of avian influenza viruses in wild birds. PloS Pathog. 4, e1000076 (doi:10.1371/journal.ppat.1000076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbinge B. S.2009Evaluatie Opvangbeleid 2005–2008 overwinterende ganzen en smienten. Deelrapport 4. Invloed opvangbeleid op de internationale verspreiding van overwinterende ganzen in NW-Europa Alterra-rapport 1842 Wageningen, The Netherlands: Alterra [Google Scholar]
- Ellstrom P., Latorre-Margalef N., Griekspoor P., Waldenstrom J., Olofsson J., Wahlgren J., Olsen B.2008Sampling for low-pathogenic avian influenza A virus in wild mallard ducks: oropharyngeal versus cloacal swabbing. Vaccine 26, 4414–4416 (doi:10.1016/j.vaccine.2008.06.027) [DOI] [PubMed] [Google Scholar]
- Flint P. L., Franson J. C.2009Does influenza A affect body condition of wild mallard ducks, or vice versa? Proc. R. Soc. B 276, 2345–2346 (doi:10.1098/rspb.2008.1962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R. A., Munster V., Wallensten A., Bestebroer T. M., Herfst S., Smith D., Rimmelzwaan G. F., Olsen B., Osterhaus A. D. M. E.2005Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79, 2814–2822 (doi:10.1128/JVI.79.5.2814-2822.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidet N., et al. 2008Evidence of infection by H5N2 highly pathogenic avian influenza viruses in healthy wild waterfowl. PloS Pathog. 4, e1000127 (doi:10.1371/journal.ppat.1000127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garamszegi L. Z., Møller A. P.2007Prevalence of avian influenza and host ecology. Proc. R. Soc. B 274, 2003–2012 (doi:10.1098/rspb.2007.0124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorson D., Karunakaran D., Senne D., Kelleher C., Bailey C., Abraham A., Hinshaw V., Newman J.1983Epizootiology of avian influenza—simultaneous monitoring of sentinel ducks and turkeys in Minnesota. Avian Dis. 27, 77–85 (doi:10.2307/1590374) [PubMed] [Google Scholar]
- Hastie T. J., Tibshirani R. J.1990Generalized additive models London, UK: Chapman and Hall; [DOI] [PubMed] [Google Scholar]
- Hinshaw V. S., Webster R. G., Turner B.1980The perpetuation of orthomyxoviruses and paramyxoviruses in Canadian waterfowl. Can. J. Microbiol. 26, 622–629 [DOI] [PubMed] [Google Scholar]
- Hinshaw V. S., Wood J. M., Webster R. G., Deibel R., Turner B.1985Circulation of influenza viruses and paramyxoviruses in waterfowl originating from 2 different areas of North America. Bull. WHO 63, 711–719 [PMC free article] [PubMed] [Google Scholar]
- Ip H. S., et al. 2008Prevalence of influenza A viruses in wild migratory birds in Alaska: patterns of variation in detection at a crossroads of intercontinental flyways. Virol. J. 5 (doi:10.1186/1743-422X-5-71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen C. M., Handeland K.2007Avian influenza virus screening in wild waterfowl in Norway, 2005. Avian Dis. 51, 425–428 (doi:10.1637/7555-033106R1.1) [DOI] [PubMed] [Google Scholar]
- Kalthoff D., Breithaupt A., Teifke J. P., Globig A., Harder T., Mettenleiter T. C., Beer M.2008Highly pathogenic avian influenza virus (H5N1) in experimentally infected adult mute swans. Emerg. Inf. Dis. 14, 1267–1270 (doi:10.3201/eid1408.080078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keawcharoen J., van Riel D., van Amerongen G., Bestebroer T., Beyer W. E., van Lavieren R., Osterhaus A. D. M. E., Fouchier R. A. M., Kuiken T.2008Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg. Inf. Dis. 14, 600–607 (doi:10.3201/eid1404.071016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H., Yanagawa R., Matsuoka Y.1980Duck influenza lacking evidence of disease signs and immune response. Infect. Immun. 30, 547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A. V., Pearce J. M., Flint P. L., Franson J. C., Ip H. S.2008Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta). Mol. Ecol. 17, 4754–4762 (doi:10.1111/j.1365-294X.2008.03953.x) [DOI] [PubMed] [Google Scholar]
- Kou Z., et al. 2009The survey of H5N1 flu virus in wild birds in 14 provinces of China from 2004 to 2007. PLoS ONE 4, e6926 (doi:10.1371/journal.pone.0006926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S., Walker D., Pryor S. P., Niles L., Chenghong L., Hinshaw V. S., Webster R. G.2004Influenza A viruses of migrating wild aquatic birds in North America. Vector-Borne Zoonotic Dis. 4, 177–189 (doi:10.1089/vbz.2004.4.177) [DOI] [PubMed] [Google Scholar]
- Lang A. S., Kelly A., Runstadler J. A.2008Prevalence and diversity of avian influenza viruses in environmental reservoirs. J. Gen. Virol. 89, 509–519 (doi:10.1099/vir.0.83369-0) [DOI] [PubMed] [Google Scholar]
- Latorre-Margalef N., et al. 2009aEffects of influenza A virus infection on migrating mallard ducks. Proc. R. Soc. B 276, 1029–1036 (doi:10.1098/rspb.2008.1501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Margalef N., et al. 2009bDoes influenza A affect body condition of wild mallard ducks, or vice versa? A reply to Flint and Franson. Proc. R. Soc. B 276, 2347–2349 (doi:10.1098/rspb.2009.0275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., et al. 2005Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309, 1206–1206 (doi:10.1126/science.1115273) [DOI] [PubMed] [Google Scholar]
- Munster V. J., et al. 2007Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3, e61 (doi:10.1371/journal.ppat.0030061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V. J., et al. 2009Practical considerations for high-throughput influenza A virus studies of wild birds by use of molecular diagnostic tests. J. Clin. Microbiol. 47, 666–673 (doi:10.1128/JCM.01625-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B., Munster V. J., Wallensten A., Waldenström J., Osterhaus A. D. M. E., Fouchier R. A. M.2006Global patterns of influenza A virus in wild birds. Science 312, 384–388 (doi:10.1126/science.1122438) [DOI] [PubMed] [Google Scholar]
- Pasick J., et al. 2007Susceptibility of Canada geese (Branta canadensis) to highly pathogenic avian influenza virus (H5NI). Emerg. Inf. Dis. 13, 1821–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne R. W., et al. 2002Genstat for windows, 6th edn Oxford, UK: VSN International [Google Scholar]
- Rohani P., Breban R., Stallknecht D. E., Drake J. M.2009Environmental transmission of low pathogenicity avian influenza viruses and its implications for pathogen invasion. Proc. Natl Acad. Sci. USA 106, 10 365–10 369 (doi:10.1073/pnas.0809026106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhm C., Suss J., Pohle V., Webster R. G.1996Different hemagglutinin cleavage site variants of H7N7 in an influenza outbreak in chickens in Leipzig, Germany. Virology 218, 253–257 (doi:10.1006/viro.1996.0187) [DOI] [PubMed] [Google Scholar]
- van Gils J. A., Munster V. J., Radersma R., Liefhebber D., Fouchier R. A. M., Klaassen M.2007Hampered foraging and migratory performance in swans infected with low-pathogenic avian influenza A virus. PLoS ONE 2, e184 (doi:10.1371/journal.pone.0000184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roomen M., van Winden E., Koffijberg K., van den Bremer L., Ens B., Kleefstra R., Schoppers J., Vergeer J.-W., Soldaat L. & SOVON Ganzen- en Zwanenwerkgroep 2007Waterbirds in The Netherlands in 2005–2006. SOVON-monitoringrapport 2007/03 Beek-Ubbergen, The Netherlands: SOVON Vogelonderzoek Nederland; [In Dutch] [Google Scholar]
- Wallensten A., et al. 2007Surveillance of influenza A virus in migratory waterfowl in northern Europe. Emerg. Inf. Dis. 13, 404–411 (doi:10.3201/eid1303.061130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Yakhno M., Hinshaw V. S., Bean W. J., Murti K. G.1978Intestinal influenza—replication and characterization of influenza-viruses in ducks. Virology 84, 268–278 (doi:10.1016/0042-6822(78)90247-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y.1992Evolution and ecology of influenza-A viruses. Microbiol. Rev. 56, 152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]