Abstract
Viral pathogens continue to emerge among humans, domesticated animals and cultivated crops. The existence of genetic variance for resistance in the host population is crucial to the spread of an emerging virus. Models predict that rapid spread decreases with the frequency and diversity of resistance alleles in the host population. However, empirical tests of this hypothesis are scarce. Arabiodpsis thaliana—tobacco etch potyvirus (TEV) provides an experimentally suitable pathosystem to explore the interplay between genetic variation in host's susceptibility and virus diversity. Systemic infection of A. thaliana with TEV is controlled by three dominant loci, with different ecotypes varying in susceptibility depending on the genetic constitution at these three loci. Here, we show that the TEV adaptation to a susceptible ecotype allowed the virus to successfully infect, replicate and induce symptoms in ecotypes that were fully resistant to the ancestral virus. The value of these results is twofold. First, we showed that the existence of partially susceptible individuals allows for the emerging virus to bypass resistance alleles that the virus has never encountered. Second, the concept of resistance genes may only be valid for a well-defined viral genotype but not for polymorphic viral populations.
Keywords: emergent viruses, experimental evolution, plant virus, resistance-breaking, resistance genes, virus evolution
1. Introduction
Emerging and re-emerging diseases, especially those induced by RNA viruses are gaining awareness not only among specialists but also among the general public because of their tremendous impact on human, animal and plant health (Woolhouse et al. 2005; Cleaveland et al. 2007; Parrish et al. 2008; Jones 2009). The first step in virus emergence (phase I) is the exposure of the new host species to the virus (Dennehy et al. 2006). The rate of exposure will be a complex function of the genetics, ecology and behaviour of the reservoir and naive hosts and the transmission biology of the virus (including any relevant vector that may be involved in the process). The second step (phase II) corresponds to the adaptation of the virus to its new host driven by changes in the viral genome. The final step of viral emergence (phase III) is the epidemiological spread of the new virus in the host population. In recent years, considerable experimental effort has been devoted to understanding the causes and consequences of RNA virus adaptation to new hosts (Elena & Sanjuán 2007).
The host represents the virus's ecosystem, therefore it should be the principal factor affecting virus evolution. The degree of viral adaptation to a particular host results from the balance between within-host selection and among-hosts gene flow (Turner & Elena 2000; Lajeunesse & Forbes 2001; Dennehy et al. 2006; Agudelo-Romero & Elena 2008). In ecological terms, viruses can evolve to become specialists, if their niche is restricted to one or few hosts (Fry 1996; Kaltz & Shykoff 1998), or generalists, if they are able to infect and transmit from a wide range of hosts (Woolhouse et al. 2001). In the case of specialist viruses, adaptation to a new host is often accompanied by the reduction of fitness in the reservoir host (Turner & Elena 2000; Agudelo-Romero et al. 2008b). This trade-off may result from two mutually non-exclusive causes: (i) antagonistic pleiotropy by which mutations that are beneficial in one host may be deleterious in the alternative one, or (ii) accumulation of neutral mutations in one host that are deleterious in the alternative unselected host. Given the compactness of RNA virus genomes with many cases of overlapping genes and multifunctional proteins, the existence of neutral loci is very unlikely and therefore, antagonistic pleiotropy is a more probable scenario (Remold et al. 2008; Agudelo-Romero et al. 2008b).
Plants can resist viruses by a number of mechanisms. Resistance pathways involving strain-specific recognition of a virus-encoded elicitor through direct or indirect interaction with a corresponding resistance gene (R gene) product can lead to a hypersensitive reaction involving localized cell death, activation of salicylic acid-mediated systemic acquired resistance (SAR) and limitation of the virus to initial infection foci (Jones & Dangl 2006). The genetic determinants of Arabidopsis thaliana's susceptibility to viral infection have been thoroughly studied during the last decade (Carr & Whitham 2007). One of such resistance systems described in A. thaliana is the Restricted Tobacco etch virus Movement (RTM) multigenic complex composed of three loci (RTM1, RTM2 and RTM3; Mahajan et al. 1998; Whitham et al. 1999, 2000; Chisholm et al. 2000, 2001). The presence of dominant alleles in all three loci is necessary for blocking the tobacco etch virus (TEV) systemic movement, while homozygous recessive mutations at any of the three loci result in systemic infection (Chisholm et al. 2000, 2001). RTM1 (At1g05760.1) encodes for a jacalin-like lectin protein with sequence similarities to several myrosinase-binding proteins involved in the defence response (Chisholm et al. 2000). RTM2 (At5g04890) encodes for a protein whose N-terminal region is similar to plant small heat shock proteins, whereas the C-terminal region has a transmembrane domain (Whitham et al. 2000). The RTM3 locus has not yet been characterized. RTM-mediated resistance differs from typical R gene-mediated resistance in that hypersensitive cell death does not occur around the infection and markers associated with SAR are not induced after infection with TEV (Whitham et al. 2000). The mechanism whereby, these proteins restrict long-distance movement of TEV is yet unclear, although they may cooperate in preventing TEV entry into, transport through or exit from the phloem; were RTM1 and RTM2 proteins are exclusively localized (Whitham et al. 2000; Chisholm et al. 2001). A. thaliana ecotypes vary in their susceptibility to TEV infection (Mahajan et al. 1998). While some ecotypes allow long-distance movement of the virus from inoculated rosette leaves to non-inoculated inflorescence tissue (e.g. Ler-0 and C24), many ecotypes (e.g. Col-0) support replication and cell-to-cell movement in inoculated leaves but do not allow systemic movement of the virus. The sequence for the susceptible rtm1 allele found in Ler-0 contains a stop codon at position 169, resulting in a truncated protein relative to the RTM1 allele of Col-0 (Chisholm et al. 2000). Similarly, rtm2 alleles are also associated with stop codons in the protein (Whitham et al. 2000).
TEV is a member of the Potyviridae family belonging to the picornavirus supergroup of positive-stranded RNA viruses. Potyviruses form the largest family (approx. 30%) of plant viral pathogens and produce severe crop losses worldwide (Shukla et al. 1994). Potyviruses are transmitted by aphids and infect over nine plant families, mainly within the Solanaceae. In recent work, Agudelo-Romero et al. (2008a) carried out serial passages of TEV into A. thaliana ecotype Ler-0 (it has an rtm1 allele in homozygosis that encodes for a truncated protein). The resulting virus, hereafter referred to as TEV-At17, showed remarkable differences relative to the ancestral virus: approximately fivefold higher infectivity and 3-logs greater viral load and more severe symptoms that included stunting, vein clearing and leaf deformation. By adapting to its new host A. thaliana, TEV-At17 paid a fitness cost in the original one Nicotiana tabacum. Only 20 per cent of inoculated tobaccos were infected, in comparison with the 100 per cent infectivity with the ancestral genotype. Furthermore, infected plants only showed local chlorotic spots on the inoculated leaf instead of the severe systemic etching produced by the ancestral virus. The TEV-At17 genome differed from the ancestral virus in only five nucleotide changes, two of which were synonymous. A single amino acid replacement L2013F in the VPg domain of the NIa protein was sufficient to induce the severe syndrome, whereas the other two non-synonymous changes (A1047V in protein P3 and T1210M in the 6K1 small peptide) contributed epistatically to L2013F in enhancing the severity of symptoms. Mutations in VPg have been shown to affect the proper interaction with the eukaryotic translation initiation factor, eIF4E (Charron et al. 2008; Gallois et al. 2010). This suggests that L2013F facilitates translation of the viral genome, consequently leading to incremented TEV-At17 accumulation and, hence, increasing the likelihood of successful infection of new plants. P3 localizes within the cell nucleus and nucleolus in association with the NIa–VPg protein and interacts with CI protein during the process of amplification of viral genome (Urcuqui-Inchima et al. 2001). The 6K1 peptide is involved in pathogenicity (Urcuqui-Inchima et al. 2001). Interestingly, mutations in the same three cistrons were required for pea seedborne mosaic virus to overcome the resistance mediated by the sbm-1 gene (Hjulsager et al. 2002).
In this study, we sought to answer the question of whether adaptation of an emerging virus to a susceptible host genotype would facilitate its access to genotypes that were not accessible for the ancestral virus. The alternative hypothesis being that the virus is only capable of replicating in the host genotype to which it was locally adapted. To do so, we have analysed the infectivity and fitness of the ancestral TEV and evolved TEV-At17 isolates across a panel of 19 A. thaliana ecotypes, some of which carried the dominant alleles at the three RTM loci.
2. Material and methods
(a). Viruses and plants
Viral particles for both the ancestral TEV and the evolved TEV-At17 isolates were obtained as follows. Eight week-old N. tabacum var. Xhanti plants where inoculated by abrasion of the third true leaf with 5 µg of RNA obtained from in vitro transcription of infectious clone pTEV7DA (GenBank DQ986288) as described elsewhere (Carrasco et al. 2007a). Seven days post-inoculation (dpi), the infected plants were used to obtain a large stock of TEV particles following the protocol described in Carrasco et al. (2007a). TEV-At17 was purified from caulinar tissue of A. thaliana Ler-0 plants as described in Agudelo-Romero et al. (2008a). Viral extracts were prepared by adding 1 ml of 50 mM potassium phosphate buffer (pH 8.0) per gram of N2-frozen plant tissue and homogenized in a mortar. The homogenate was clarified by centrifugation at 4°C and 10 000g for 10 min. The concentration of infectious viral particles in the extracts was evaluated by the dilution–infectivity assay method on the local lesion host Chenopodium quinoa (Kleczkowski 1950).
Table 1 shows the 19 A. thaliana ecotypes used in this study. Allelic constitution for each locus and ecotype was either determined for this study (GenBank accessions GU396153 to GU396227) or kindly provided by Dr F. Revers (INRA Bordeaux, France). According to the allelic combinations, only Ei-2, Ler-0, Mrk-0 and St-0 should be susceptible to TEV infection.
Table 1.
List of A. thaliana ecotypes used in this study and the corresponding TEV-resistance phenotype according to the allelic combination at each RTM loci.
| ecotype | origin | expected phenotype |
|---|---|---|
| Akita-0a | Japan | resistant |
| Alc-0a | Spain | resistant |
| Bla-1b | Spain | resistant |
| Col-0a | USA | resistant |
| Cvi-0a | Cape Verde islands | resistant |
| Di-2b | France | resistant |
| Ei-2b | Germany | sensitive (rtm1/rtm1) |
| Ga-0b | Germany | resistant |
| Gy-0b | France | resistant |
| Ler-0a | Germany | sensitive (rtm1/rtm1) |
| Mrk-0b | Germany | sensitive (rtm1/rtm1) |
| Oy-0a | Norway | resistant |
| Ren-1b | France | resistant |
| Sorbo-0b | Tajikistan | resistant |
| St-0a,b | Sweden | sensitive (rtm3/rtm3) |
| Ta-0b | Czech Republic | resistant |
| Tsu-0a | Japan | resistant |
| Ws-0b | Russia | resistant |
| Wt-1b | Germany | resistant |
aRTM1, RTM2 and RTM3 loci genotyped by Dr F. Revers (INRA Bordeaux, France).
bGenotyped for this study. RTM3 alleles for Gy-0, Ga-0 and Di-2 have not been determined.
All plants were grown in a BSL-2 greenhouse at 25°C and 16 h light period. Plants were inoculated when the growth stages between 3.5 and 3.7 were attained following the scale of Boyes et al. (2001).
(b). Molecular confirmation of infections
Western blot analyses, including total protein extraction in non-denaturizing buffer, SDS–PAGE electrophoresis, electrophoretic transfer to Hybond ECL membranes (Amersham) and hybridization were performed with minor modifications of the procedures described in Lough et al. (1998) using commercial antibodies anti-coat protein conjugated with horseradish peroxidase (Agdia).
For RNA dot-blot hybridization, total RNA was extracted as follows. A tube containing a few caulinar leaves and one stainless steel ball (3 mm diameter) was submerged into liquid N2 and the plant tissue was powdered in a Tissuelyzer (MM300 Retsch Gmbh) for 30 s at 30 Hz. One millilitre of extraction buffer (100 mM Tris–HCl pH 8.0, 50 mM EDTA pH 7.0, 500 mM NaCl, 10 mM β-mercaptoetanol) was added to approximately 100 µg of plant tissue powder and briefly vortexed. After addition of 50 µl of 20 per cent SDS, the samples were incubated for 20 min at 65°C and mixed by inverting the tubes every 5 min. Prior to incubating 20 min on ice, 250 µl of 5 M potassium acetate was added. The tissue lysate was pelleted by centrifugation at 4°C and 13 000g for 15 min and 500 µl of supernatant was precipitated with 1 ml of 70 per cent ethanol. Nucleic acids were recovered by centrifugation (15 min at 13 000 rpm, 4°C). The pellet was air-dried and resuspended in 20 µl DEPC-treated water. The template for RNA probe synthesis was obtained by linearizing pTEV7DA with BglII (TaKaRa). A digoxigenin-labelled RNA probe was synthesized by in vitro transcription with T7 RNA polymerase following the manufacturer's instructions (DIG Northern Starter Kit Manual, Roche). The probe was homologous to 254 nt of the 3′ end of the CP cistron and part of the non-translated region. For detection with the DIG-labelled RNA probe, total RNA extracts were denatured at 95°C for 10 min and quickly cooled on ice. RNA samples of 3 µl were spotted onto a positively charged nylon membrane (Nytran SPC, Whatman, Sanford, USA) and fixed with UV light (180 mJ, 2.30 min). Hybridization with the probe was carried out overnight at 68°C with a standard hybridization solution. Two astringency washes, for 5 min each, with 2× SSC + 0.1 per cent SDS were performed at room temperature followed by two more washes, 15 min each in 0.1× SSC + 0.1 per cent SDS at 68°C. Chemoluminescent detection with anti-DIG alkaline phosphatase antibody conjugate was carried out according to manufacturer's recommendations (DIG Northern Starter Kit, Roche).
For one-step RT-PCR detection, approximately 100 mg of caulinar leaves tissue were homogenized in a 1.5 ml tube submerged into liquid N2 and mixed with 100 µl of extraction buffer (0.2 M Tris, 0.2 M NaCl, 50 mM EDTA, 2% SDS; pH 8). An equal volume of phenol–chloroform–isoamylic alcohol (25 : 25 : 1) was added, vortexed well and centrifuged at 12 000g for 5 min at 4°C. Eighty microlitres of upper aqueous phase together with 40 µl of 7.5 M LiCl + EDTA 50 mM solution was subjected to overnight precipitation at −20°C. The RNA precipitate was then centrifuged at 12 000g for 15 min at 4°C, washed twice with 70 per cent ice-cold ethanol, air-dried and resuspended in 10 µl of DEPC-treated ultrapure water. RNA concentration was measured spectrophotometrically. Total RNA was subjected to one-step RT-PCR in 10 µl reaction volume following manufacturer's instructions (TaKaRa). The reaction mix contained 1 U Taq polymerase (TaKaRa Ex Taq HS), 0.2 µl of PrimeScript RT enzyme Mix II, 0.8 µM of each primer and 100 ng of total RNA. The forward primer, PC90-95f 5′-GCTGTATTGAAAGTGCGAC and the reverse primer PC86-91r 5′-AGGCCCAACTCTCCGAAAG amplify 334 nucleotides of a conserved region from the NIb gene (Carrasco et al. 2007a). The amplification profile comprised 5 min at 42°C and 10 s at 95°C, followed by 40 cycles of 5 s at 95°C and 20 s at 60°C. PCR products were further visualized by 2 per cent agarose gel electrophoresis.
(c). Quantification of infectivity
TEV and TEV-At17 stocks were conveniently diluted to ensure that all infectivity experiments were always performed with the same inoculum concentration (Agudelo-Romero et al. 2008b). Prior to the inoculation, 10 per cent carborundum (100 mg ml−1) was added to each sample. Plants were rub-inoculated with 5 µl of viral preparation per leaf, two leaves per plant. Experiments were fully replicated five times; each experimental block contained all the ecotypes. A median of 26 plants per ecotype and per viral genotype were inoculated (most cases with greater than 16 but in few instances only five plants). As controls, one plant was always left non-inoculated and one mock-inoculated. Infections were confirmed 21 dpi by at least two of the three different molecular techniques described in §2b.
Infectivity (I) was estimated as the fraction of infected plants out of the total number of inoculated plants. LaPlace's point estimator for the Binomial frequency parameter was used instead of the commonly used maximum likelihood estimator because it provides more robust estimates for small sample sizes (Chew 1971). Binomial 95% confidence intervals (CI) were also computed. Point estimates and CI were computed using the www.measuringusability.org/wald.htm server. The full infectivity dataset was analysed using a general log-linear model in which ‘ecotype’ (Ei; i = 1,…,19) and ‘TEV genotype’ (Vj; j = 1, 2) were considered as orthogonal factors and the cell counts in the cross-tabulation (infected or not) were assumed to be Poisson-distributed: log Iijk = µ+ Ei + Vj + (E × V)ij + eijk, where µ represents the overall count and eijk the error term in the cell. A backward elimination procedure was used to obtain the simplest model that best explained the observed variation in infectivity. Finally, ecotypes were classified into homogeneous categories according to the infectivity of TEV-At17 by the k-means clustering method. The goodness-of-fit of nested models was assessed using the partial F-statistic. The model for which addition of one more cluster did not produce a significant reduction in the explained sum of squares was taken as the best one.
(d). Absolute real-time RT-PCR (RT-qPCR) determination of viral load
Viral RNA quantification was performed by using an in vitro transcript of TEV as external standard (Pfaffl 2004). The standard curve in RT-qPCR was drawn from six points corresponding to serial dilutions of TEV RNA. This method assured RT efficiency, as well as real-time PCR amplification efficiencies being identical for all samples, because RNA species subjected to reactions have the same replication kinetics. The TEV RNA transcripts were obtained from pTEV7DA as described in Carrasco et al. (2007a). To treat all samples equally in the quantification assay, this TEV RNA was cleaned up with RNeasy Plant Mini Kit (Qiagen), treated with Turbo DNA-free DNase (Ambion) and diluted into a preparation of total RNA extracted from non-inoculated plants in the same way. To minimize intra-assay variation, all transcripts used for the standard curve come from a single reaction and serial dilutions were aliquoted and preserved at −80°C.
Twenty-one dpi, infected whole plants were submerged into liquid N2 and ground in a mortar. Final RNA concentrations were spectrophotometrically measured three times, adjusted to 50 ng µl−1 and treated with Turbo DNA-free DNase (Ambion). Reaction standard dilutions, non-template control and 100 ng total RNA samples from infected plants were incubated with Multiscribe Reverse transcriptase (Applied Biosystems) for 10 min at 25°C, 30 min at 48°C and 5 min at 95°C in 20 µl reaction volume following manufacturer's instructions (SYBR Green PCR Master Mix and RT-PCR, Applied Biosystems). Primers for an absolute qPCR were designed using PrimerExpress v. 2 (Applied Biosystems). The primer pair TEV7DACP689F 5′-TTGGTCTTGATGGCAACGTG and TEV7DA739R 5′-TGTGCCGTTCAGTGTCTTCCT amplify a 30-nt fragment within the TEV CP cistron. Reactions were performed in 20 µl volume containing 10 µl 2× PCR Master Mix (SYBR Green PCR Master Mix and RT-PCR, Applied Biosystems), 300 nM of each primer and 2 µl of cDNA template. Reactions were set up in the dark. Amplifications were done using the ABI PRISM Sequence Analyzer 7000, according to the following profile: 10 min at 95°C, following 40 cycles of 15 s at 95°C and 1 min 60°C. Both RT and qPCR reactions were performed in triplicate, for each sample. Quantification results were further examined using SDS7000 software v. 1.2.3 (Applied Biosystems). Viral load was expressed as picograms of viral RNA per 100 ng of total RNA.
Viral loads achieved by the evolved virus (L) were log-transformed and analysed using Kruskal–Wallis non-parametric test. As above, ecotypes were classified into homogeneous categories according to the accumulation of TEV-At17 using the k-means clustering method. All statistical analyses were done using SPSS 16.0.1.
(e). Quantitative scale of symptoms
The symptoms produced by the two viral genotypes on each ecotype were classified according to the following semi-quantitative scale. Class 0: non-infected plants; class 1: infected plants but no visible symptoms (asymptomatic); class 2: infected symptomatic plants. Symptoms were also classified according to their intensity into three sub-categories: vein clearing and leaf curling were considered as mild symptoms (2.1), a moderate reduction in growth accompanied with vein clearing and stronger leaf deformation were considerate as intermediate symptoms (2.2); strong symptoms included lack of flower development, strong deformation of the rosette leaves and an overall chlorosis (2.3).
3. Results
(a). Ancestral and evolved TEV genotypes differ in infectivity across ecotypes
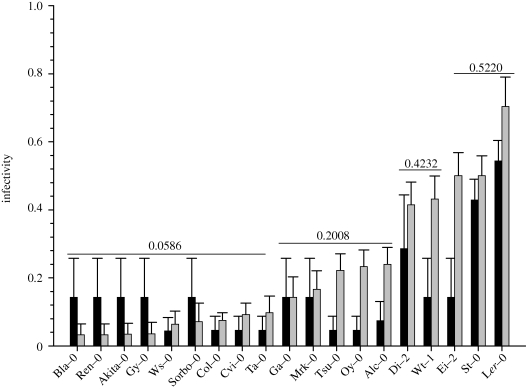
Figure 1 shows the results of the infectivity experiments for the ancestral and evolved TEV genotypes across all 19 ecotypes. Ecotypes have been ordered from the lowest to the highest infectivity values estimated for the evolved TEV-At17 genotype. The infectivity of the ancestral TEV genotype was significantly different from zero in only two ecotypes: Ler-0 and St-0. Two more ecotypes, Ei-2 and Mrk-0, were genotyped as homozygous for rtm1 alleles and should also be susceptible to the infection with the ancestral virus. However, we failed to successfully infect plants of these two ecotypes. This negative result can be explained by the very limited sample size used in these two cases (n = 5). In sharp contrast, the infectivity of the evolved TEV-At17 genotype was significantly different from zero in 13 of the tested ecotypes (figure 1), with the lowest significant value obtained for Col-0 (7.46%) and the highest value obtained for Ler-0 (54.41%). It is important to recall that TEV-At17 was evolved in the Ler-0 ecotype. Notice that the LaPlace estimator of infectivity produces non-zero values even though no plant is infected in a small sample size. It is important to highlight that in these cases, the 95% CI always contains zero.
Figure 1.
Estimates of infectivity obtained for the ancestral TEV (black bars) and evolved TEV-At19 (grey bars) genotypes across the 19 different A. thaliana ecotypes used in this study. Infectivity values were computed using the LaPlace's estimator for the Binomial parameter. Error bars represent the 95% CI. Horizontal lines represent groups of ecotypes for which TEV-At17 shows homogeneous infectivity according to the k-means clustering algorithm (average values shown above the line).
The log-linear model fitted to the data showed that the two viral genotypes differed in overall infectivity (χ2 = 438.9353, 1 d.f., p < 0.0001). Besides, differences in infectivity were also significant among A. thaliana ecotypes (χ2 = 496.6896, 18 d.f., p < 0.0001) and, more interestingly, a significant interaction existed between the viral genotype and the host ecotype (χ2 = 40.3606, 18 d.f., p = 0.0019). This significant interaction suggests that the magnitude of the difference in infectivity between the ancestral and evolved viruses depend on the particular A. thaliana ecotype employed in the experiments. We used the k-means clustering algorithm to detect which ecotypes were homogeneous in their response to the infection with the two viral genotypes. In the case of the ancestral TEV genotype, ecotypes were classified only into two significantly homogeneous groups (adding a third cluster produced a non-significant reduction in the residual sum of squares: partial F1,15 = 4.3488, p = 0.0545). The first group contained 17 ecotypes for which TEV infectivity was not significantly different from zero. The second group was formed by Ler-0 and St-0, with an average infectivity of 70.37 per cent. This grouping was clearly different from the one obtained with the infectivity data of the evolved TEV-At17 genotype. In this case, data were classified into one of the four significant groups, whose average infectivities are indicated in figure 1. The first group was constituted of the nine ecotypes for which TEV-At17 had a very low average infectivity (5.86%); including three ecotypes (Col-0, Cvi-0 and Ta-0) for which the point estimator of infectivity was significantly different from zero. The second group was formed by the five ecotypes in which the evolved virus had medium to low infectivity (average 20.08%). The third group was constituted of two ecotypes in which the TEV-At17 infectivity can be classified as medium to high (average 42.32%). Finally, the last group was formed by the three ecotypes in which the evolved virus has an average high infectivity (52.20%).
(b). Not all ecotypes support the same level of TEV-At17 replication
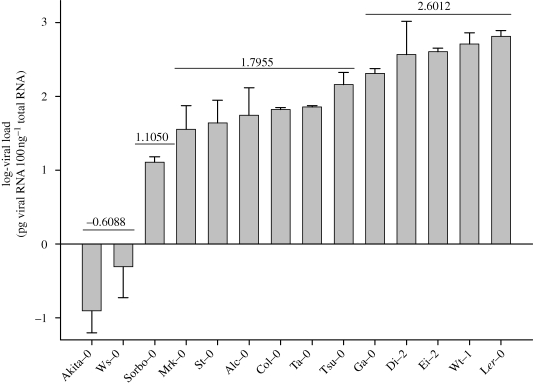
Next, we sought to estimate the fitness of TEV-At17 across a subset of A. thaliana ecotypes included in our study. As a proxy to within-host fitness, we used the decimal logarithm of the viral load estimated as the number of picograms of TEV-At17 genomic RNA accumulated per 100 ng of total RNA in the infected plant. Ecotypes Bla-0, Cvi-0, Gy-0, Oy-0 and Ren-0 were excluded from the study because no reliable quantifications were obtained after several trials. Figure 2 shows the distribution of fitness values. Since the data violated the assumptions of normality (Kolmogorov–Smirnov test: Z = 3.7085, p < 0.0001) and homocedasticity of variances among groups (Levene's test: F13,238 = 19.0746, p < 0.0001), a non-parametric Kruskal–Wallis test was used to test for TEV-At17 fitness differences among ecotypes. The test found a highly significant effect of ecotype when looking at within-host accumulation of the evolved virus (H = 98.3448, 13 d.f., p < 0.0001). Most of these differences were owing to differences between ecotypes (74.60%; REML method) rather than to error of measurements.
Figure 2.
Distribution of TEV-At17 fitness across the panoply of A. thaliana ecotypes used in this study. Error bars represent ±1 s.e.m. Horizontal lines represent groups of ecotypes for which TEV-At17 shows homogeneous within-host fitness (average values shown above the line).
As we did with infectivity, now we classified A. thaliana ecotypes into groups for which the accumulation of TEV-At17 RNA genomes is homogeneous. The k-means classification method found that the minimum number of clusters necessary to explain the observed heterogeneity in the viral load was four (adding a fifth cluster did not improve the model prediction: partial F1,9 = 3.3847, p = 0.0989). Figure 2 indicates the average log-viral loads for each homogeneous cluster. The first cluster was formed by those ecotypes (Akita-0 and Ws-0) for which the log-viral load was low (average of −0.6088). Sorbo-0 was classified apart from all other ecotypes, characterized by a medium to low log-viral load (1.1050). The third cluster, the largest one, contained six ecotypes and was characterized by a medium to high viral accumulation (average of 1.7955). Finally, the fourth cluster was formed by five ecotypes for which viral load was high (average of 2.6012). The highest within-host fitness of TEV-At17 corresponded, not surprisingly, to the ecotype in which it was evolved, Ler-0.
(c). Association between TEV-At17 infectivity and within-host replicative fitness
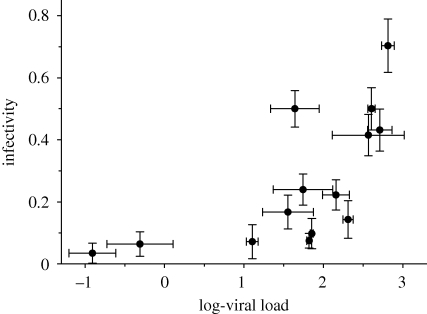
The results shown in the two previous sections indicated that both traits were affected by the plant ecotype in a remarkably parallel fashion. Therefore, we checked whether there was a significant statistical association between these two fitness traits. Figure 3 illustrates the relationship between these two traits, where each point represents the trait values estimated for each plant ecotype. A Spearman's correlation coefficient confirmed that, indeed, the two traits were not independent but positively correlated (ρS = 0.7481, 12 d.f., p = 0.0021). Therefore, it was possible to conclude that the higher infectivity of TEV-At17 in an A. thaliana ecotype, the more viral genomic RNA will be produced during infection.
Figure 3.
Correlation between the infectivity and within-host fitness values obtained for TEV-At17. Each dot represents the estimates obtained in a given A. thaliana ecotype. Error bars correspond to 95% CI in the case of infectivity and to ±1 s.e.m. for log-viral load.
It may be argued that this correlation could be a spurious consequence of the fact that differences in infectivity should also reflect differences in the amount of TEV-At17 particles that enter the plant during inoculation and successfully initiate replication and movement. If few particles were able to initiate a productive infection, the number of replication events during the time frame of our experiments should also be low. Conversely, the more infection foci that existed initially, the more replication rounds that could have occurred during the time frame of our experiments and, therefore, the more RNA genomes that could have accumulated. This being true, the differences in viral load observed between ecotypes would simply reflect differences in effective dosage at inoculation rather than differences in the TEV-At17 adaptation to each ecotype. A statistical way of ruling out this possibility was to fit an ANOVA model to the log-viral load data by using the four homogeneous clusters defined in the previous section as the only factor and treating infectivity as a covariable. If differences in log-viral load were spurious, then we expected to observe a significant interaction between the factor and the covariable. Ruling out this expectation, the interaction term of the model was not significant (F2,7 = 4.3036, p = 0.0604). Therefore, we should conclude that the observed differences in log-viral load among ecotypes do not depend on the degree of infectivity, although both fitness traits are positively correlated.
(d). Differences in symptomatology
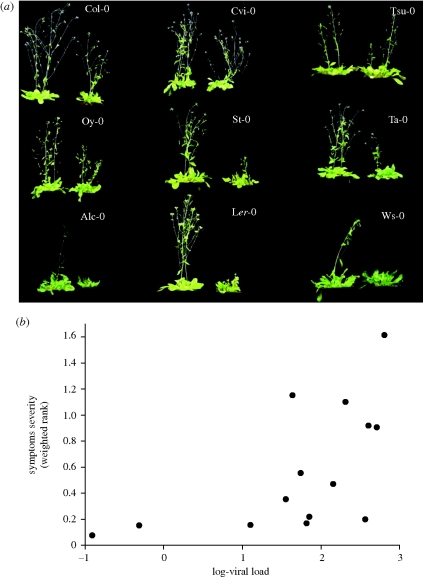
As illustrated in figure 4a, for a set of representative ecotypes, the symptoms induced by TEV-At17 strongly differed among ecotypes. Table 2 shows the semi-quantitative values describing symptoms' severity on each ecotype according to the scale defined in §2f. The severity of symptoms induced by TEV-At17 ranged between mild (Col-0, Cvi-0 and Tsu-0) to a very severe syndrome (Ler-0, Ws-0 or Alc-0).
Figure 4.
(a) Representative examples of the symptoms induced by TEV-At17 on different ecotypes. In all panels, the plant at the left is a healthy, mock-inoculated plant. (b) Correlation between the severity of symptoms and the accumulation of viral particles.
Table 2.
Severity of symptoms induced by both viral genotypes on each ecotype. nd, not determined.
| ecotype | ancestral TEV | TEV-At17 |
|---|---|---|
| Akita-0 | 0 | 2.1 |
| Alc-0 | 0 | 2.3 |
| Bla-1 | 0 | nd |
| Col-0 | 0 | 2.2 |
| Cvi-0 | 0 | nd |
| Di-2 | 0 | 2.1 |
| Ei-2 | 0 | 2.2 |
| Ga-0 | 0 | 2.2 |
| Gy-0 | 0 | nd |
| Ler-0 | 1 | 2.3 |
| Mrk-0 | 0 | 2.1 |
| Oy-0 | 0 | 2.2 |
| Ren-1 | 0 | nd |
| Sorbo-0 | 0 | 2.1 |
| St-0 | 2.1 | 2.3 |
| Ta-0 | 0 | 2.2 |
| Tsu-0 | 0 | 2.1 |
| Ws-0 | 0 | 2.3 |
| Wt-1 | 0 | 2.1 |
Next, we sought to associate the severity of symptoms and the level of accumulation of TEV-At17. To do so, the severity values shown in table 2 were transformed into ranks and weighted by the frequency of plants of the corresponding ecotype that resulted infected. This weighted value represents a population average measure of the severity of symptoms. The strength of symptoms and the level of accumulation were positively correlated (figure 4b: ρS = 0.6791, 12 d.f., p = 0.0076), suggesting that, on an average, the more virus accumulated in a plant ecotype, the worse the symptoms developed by the plant.
4. Discussion
In this study, we have addressed the question of whether a virus adapted to a partially susceptible genotype of the new host would spill over into non-susceptible genotypes of the new host, or, in contrast, the adaptation of the emerging virus was local. To do so, we have explored the ability of two TEV genotypes that differ in their degree of adaptation to the susceptible ecotype Ler-0, to systemically infect a panel of 18 other ecotypes, most of which carry allelic combinations that confer the resistance to the ancestral TEV genotype. We found that as a correlated response to adaptation to the permissive host Ler-0, TEV-At17 also increased infectivity, within-host fitness and the severity of symptoms across the whole collection of A. thaliana ecotypes genetically heterogeneous for the RTM loci.
In some cases, the fitness of TEV-At17 and the symptoms induced in alternative ecotypes were undistinguishable from the values quantified in Ler-0, suggesting that expanding the host range to genetically different genotypes imposed no fitness burden. This suggests that the mutations responsible for adaptation to Ler-0 have a pleiotropic positive effect in other hosts' genetic backgrounds. Or in other words, no fitness trade-off exists that may favour the evolution of specialist viruses. This observation has two important implications. First, very few experiments have tested whether the effects of adaptive mutations remain beneficial across a set of different environments or are environment-specific. In a pioneering study, Ostrowski et al. (2005) found that mutations fixed in Escherichia coli lineages adapted to glucose as the only carbon source were also beneficial in five other carbon sources, concluding that positive pleiotropism was a norm for the bacterium. Second, our observation is highly relevant for the study of emerging viral diseases as it suggests that mutations conferring selective advantage for the emerging virus in a given genotype of the new host would open the possibility for a virus to successfully replicate and induce disease in other genotypes of the host, including those carrying resistance genes. In other words, local adaptation does not influence the dynamics of the actual infection process (Ben-Ami et al. 2008).
The evolved TEV-At17 genotype was produced in a previous study after a process of experimental evolution consistent in 17 serial undiluted passages of the viral population in the Ler-0 ecotype (Agudelo-Romero et al. 2008a). TEV-At17 accumulation was three orders of magnitude higher than that of the ancestral virus and induced a severe syndrome in the plants, while infection of Ler-0 with the ancestral virus was asymptomatic. The evolved virus also increased its infectivity in Ler-0. The molecular basis of this adaptive process was studied and three amino acid replacements in NIa-VPg, P3 and 6K1, respectively, were necessary for phenocopying the symptoms produced by TEV-At17 (Agudelo-Romero et al. 2008a). The NIa–VPg covalent attachment to the 5′ end of the RNA is essential for RNA replication and correct establishment of the CAP complex that recruits the host's eIF4G necessary for translation (Nicaise et al. 2007). This interaction has been described as a key determinant of host genotype-specificity and systemic movement (Charron et al. 2008; Gallois et al. 2010). With this information in hand, we postulated that mutation L2013F improved the interaction between NIa–VPg and the eIF4G allele of Ler-0 in such a way that it optimized RNA transcription, protein synthesis and increased virus accumulation. The mutations in P3 and 6K1 further enhanced the severity of symptoms. Previous work has described a causal relationship between changes in viral structural proteins, including RNA-binding ones, and host-range expansions, tissue tropisms and immune escapes. For instance, work with bacteriophage φX174 showed that a single mutation in a DNA-binding viral protein played a stepping stone role in the process of fixation of other successive mutations that allow infection of different E. coli genotypes previously inaccessible to the virus (Pepin & Wichman 2007; Pepin et al. 2008). Similarly, studies with animal viruses such as vesicular stomatitis virus (Remold et al. 2008), SARS coronavirus (Poon et al. 2005), influenza A virus (Parrish & Kawaoka 2005) and canine parvovirus (Shackelton et al. 2005) have also confirmed that host range can be expanded by one or a few changes in structural genes.
Regoes et al. (2000) have analysed theoretically possible effects of genetic variability for susceptibility in the host population on the virulence of emerging pathogens. Models predict that such heterogeneity would impose a limit in the virulence and that pathogens should evolve as generalists, being capable of infecting different host genotypes. Although our results do not allow directly addressing these predictions, they give some support to the second one. Our experiments provide evidence that TEV-At17 has evolved as a within-species generalist because of its capacity to infect with different efficiencies A. thaliana genotypes that were inaccessible to the ancestral virus. Day et al. (2006) also suggested from a theoretical point of view that host heterogeneity for susceptibility would limit the rate of epidemic expansion. Our current results do not allow for testing this interesting prediction, but ongoing experiments in which TEV-At17 is evolving on polymorphic RTM populations would enable a direct test. Someone may argue that our host system lacks enough heterogeneity because all ecotypes belong to the same species. However, there is strong evidence showing that A. thaliana population structure is highly polymorphic and phenotypically diverse (Nördborg et al. 2005; Schmid et al. 2006; Platt et al. 2010).
Virulence does not represent an obvious advantage to parasites and most models seeking to explain the evolution of pathogen virulence assume that it is a side effect of within-host replication and accumulation (Brown et al. 2006). Despite the obvious interest of this question, data directly testing the existence of the predicted correlation are, at least, scarce, and the molecular basis of virulence is poorly understood. The few available results are somehow contradictory. Carrasco et al. (2007b) found no association between fitness effects and virulence in a collection of single-nucleotide substitution mutants tested for these two traits in the natural host N. tabacum. Two main reasons can be argued to explain the difference between this result and the positive correlation reported here. First, the host species used in both experiments were different. Second, Carrasco et al. (2007a,b) studied single-nucleotide mutants in a single host genotype, whereas here we study the virulence of a single genotype across a panel of closely related but still genetically heterogeneous hosts. In a recent set of studies, Pagán et al. (2007, 2008) found that the virulence of three different isolates of CMV varied across a panel of 21 A. thaliana ecotypes, but that not in all combinations, was the level of accumulation associated with the severity of symptoms. The association was only significant for ecotypes with a short life cycle and investing most of their resources in reproduction rather than in vegetative growth. These results suggest that a particular combination of life-history trait values of the host may increase tolerance to virus infection.
Generally, we can conclude that the capacity of expansion of an emergent virus is not necessarily restricted by genetic heterogeneity of potential hosts. In terms of epidemiology and emergence, our results strongly suggest that partially susceptible plants could constitute a springboard for the invasion of a new host species. Even if part of this genetic variability consists of virus resistance, as it may be in the case of RTM loci, it is possible that the emergent virus gained capacity to overcome the resistance in quantitative mode, rather than qualitative. It must be remembered that TEV and A. thaliana do not share a past coevolutionary history and, hence the RTM system has not been tuned by natural selection to specifically resist TEV in an efficient way. In this sense, we suggest that RTM represents a sort of circumstantial resistance likely due to an inefficient interaction between viral and cell factors. Our experimental evolution protocol selected for a virus with increased replication ability in A. thaliana. By doing so, interactions between viral and cell proteins may have been optimized, overcoming any unspecific resistance. Following this interpretation, concerns must be raised about the notion of resistance genes, deeply rooted in the community of plant pathologists. Our results clearly indicate that RTM ‘resistance genes’ only have sense in the combination of Col-0 and the ancestral TEV genotype pathosystem used in the original experiments by Chisholm et al. (2000, 2001) and others, and as a starting point for our evolution experiments. Genetic variation in viral proteins quickly and dramatically affects the delicate interplay between these proteins and cellular factors, pushing the equilibrium towards a situation which cannot be predicted simply by looking at the alleles present at the RTM loci.
Acknowledgements
We thank F. de la Iglesia for fantastic technical support and S. Bedhomme for comments and fruitful discussion. We are immensely grateful to Dr F. Revers (INRA Bordeaux, France) for sharing with us his unpublished RTM genotypes of many of the ecotypes. This work was supported by grant BFU2009-06993 from the Spanish Ministry of Science and Innovation to S. F. E. J. L. and P. C. are supported by CSIC's JAE programme.
Footnotes
One contribution of 14 to a Theme Issue ‘New experimental and theoretical approaches towards the understanding of the emergence of viral infections’.
References
- Agudelo-Romero P., Elena S. F.2008The degree of plant resilience to infection correlates with virus virulence and host-range. Span. J. Agric. Res 6, 160–169 [Google Scholar]
- Agudelo-Romero P., Carbonell P., Pérez-Amador M. A., Elena S. F.2008aVirus adaptation by manipulation of host's gene expression. PLoS ONE 3, e2397 (doi:10.1371/journal.pone.0002397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudelo-Romero P., De la Iglesia F., Elena S. F.2008bThe pleiotropic cost of host-specialization in tobacco etch potyvirus. Infect. Genet. Evol 8, 806–814 [DOI] [PubMed] [Google Scholar]
- Ben-Ami F., Regoes R. R., Ebert D.2008A quantitative test of th relationship between parasite dose and infection probability across different host–parasite combinations. Proc. R. Soc. B 275, 853–859 (doi:10.1098/rspb.2007.1544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes C. D., Zayed A. M., Ascenzi R., McCaskill A. J., Hoffman N. E., Davis K. R., Görlach J.2001Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510 (doi:10.1105/tpc.13.7.1499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. F., Wickham M. E., Coombes B. K., Finaly B. B.2006Crossing the line: selection and evolution of virulence traits. PLoS Pathog. 2, e42 (doi:10.1371/journal.ppat.0020042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr T., Whitham S. A.2007An emerging model system: Arabidopsis as a viral host plant. Plant Cell Monogr. 7, 159–183 (doi:10.1007/7089_2006_104) [Google Scholar]
- Carrasco P., Daròs J. A., Agudelo-Romero P., Elena S. F.2007aA real-time RT-PCR assay for quantifying the fitness of tobacco etch virus in competition experiments. J. Virol. Methods 139, 181–188 (doi:10.1016/j.jviromet.2006.09.020) [DOI] [PubMed] [Google Scholar]
- Carrasco P., De la Iglesia F., Elena S. F.2007bDistribution of fitness and virulence effects caused by single-nucleotide substitutions in tobacco etch virus. J. Virol. 81, 12 979–12 984 (doi:10.1128/JVI.00524-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron C., Nicolaï M., Gallois J.-L., Robaglia C., Moury B., Palloix A., Carauta C.2008Natural variation and functional analyses provide evidence for evolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68 (doi:10.1111/j.1365-313X.2008.03407.x) [DOI] [PubMed] [Google Scholar]
- Chew V.1971Point estimation of the parameter of the Binomial distribution. Am. Stat. 25, 47–50 (doi:10.2307/2686085) [Google Scholar]
- Chisholm S. T., Mahajan S. K., Whitham S. A., Yamamoto M. L., Carrington J. C.2000Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement in tobacco etch virus. Proc. Natl Acad. Sci. USA 97, 489–494 (doi:10.1073/pnas.97.1.489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S. T., Parra M. A., Anderberg R. J., Carrington J. C.2001Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 127, 1667–1675 (doi:10.1104/pp.010479) [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S., Haydon D. T., Taylor L.2007Overivews of pathogen emergence: wich pathogens emerge, when and why? Curr. Opin. Microbiol. Immunol. 315, 85–111 (doi:10.1007/978-3-540-70962-6_5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy J. J., Friedenberg N. A., Holt R. D., Turner P. E.2006Viral ecology and the maintenance of novel host use. Am. Nat. 167, 429–439 (doi:10.1086/499381) [DOI] [PubMed] [Google Scholar]
- Elena S. F., Sanjuán R.2007Virus evolution: insights from an experimental approach. Annu. Rev. Ecol. Evol. Syst. 38, 27–52 (doi:10.1146/annurev.ecolsys.38.091206.095637) [Google Scholar]
- Fry J. D.1996The evolution of host specialization: are trade-offs overrated? Am. Nat. 148, S84–S107 (doi:10.1086/285904) [Google Scholar]
- Gallois J. L., et al. 2010Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso)4E and eIF(iso)4G. J. Gen. Virol. 91, 288–293 (doi:10.1099/vir.0.015321-0) [DOI] [PubMed] [Google Scholar]
- Hjulsager C. K., Lund O. S., Johansen I. E.2002A new pathotype of Pea seedborne mosaic virus explained by properties of the P3-6K1-and viral genome-linked protein (VPg)-coding regions. Mol. Plant-Microb. Interact. 15, 169–171 (doi:10.1094/MPMI.2002.15.2.169) [DOI] [PubMed] [Google Scholar]
- Jones R. A. C.2009Plant virus emergence and evolution: origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 141, 113–130 (doi:10.1016/j.virusres.2008.07.028) [DOI] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L.2006The plant immune system. Nature 444, 323–329 (doi:10.1038/nature05286) [DOI] [PubMed] [Google Scholar]
- Kaltz O., Shykoff J.1998Local adaptation in host–parasite systems. Heredity 81, 361–370 (doi:10.1046/j.1365-2540.1998.00435.x) [Google Scholar]
- Kleczkowski A.1950Interpreting relationships between the concentration of plant viruses and number of local lesions. J. Gen. Microbiol. 4, 53–69 [DOI] [PubMed] [Google Scholar]
- Lajeunesse M. J., Forbes M. R.2001Host range and local parasite adaptation. Proc. R. Soc. Lond. B 269, 703–710 (doi:10.1098/rspb.2001.1943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough T. J., Balmori E., Beck D. L., Forster L. S.1998Western analysis of transgenic plants. In Plant virology protocols (eds Foster G. D., Taylor S. C.), pp. 447–451 Totowa, NJ: Humana Press Inc; [DOI] [PubMed] [Google Scholar]
- Mahajan S. K., Chisholm S. T., Whitham S. A., Carrington J. C.1998Identification and charaterization of a locus (RTM1) that restrics long-distance movement of tobacco etch virus in Arabidopsis thaliana. Plant J. 14, 177–186 (doi:10.1046/j.1365-313X.1998.00105.x) [DOI] [PubMed] [Google Scholar]
- Nicaise V., et al. 2007Coordinated and selective recruitment of eIF4E and eIF4G factors by potyvirus infection in Arabidopsis thaliana. FEBS Lett. 581, 1041–1046 (doi:10.1016/j.febslet.2007.02.007) [DOI] [PubMed] [Google Scholar]
- Nördborg M., et al. 2005The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3, e196 (doi:10.1371/journal.pbio.0030196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski E. A., Rozen D. E., Lenski R. E.2005Pleiotropic effects of beneficial mutations in Escherichia coli. Evolution 59, 2343–2352 [PubMed] [Google Scholar]
- Pagán I., Alonso-Blanco C., García-Arenal F.2007The relationship of within-host multiplication and virulence in a plant–virus system. PLoS ONE 8, e786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán I., Alonso-Blanco C., García-Arenal F.2008Host responses in life-history traits and tolerance to virus infection in Arabidopsis thaliana. PLoS Pathog. 4, e1000124 (doi:10.1371/journal.ppat.1000124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish C. R., Kawaoka Y.2005The origins of new pandemic viruses: the acquisition of new ost ranges by canine parovirus and influenza A viruses. Annu. Rev. Microbiol. 59, 553–586 (doi:10.1146/annurev.micro.59.030804.121059) [DOI] [PubMed] [Google Scholar]
- Parrish C. R., Holmes E. C., Moreus D. M., Park E.-C., Burke D. S., Calisher C. H., Laughlin C. A., Saif L. J., Daszak P.2008Cross-species transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72, 457–470 (doi:10.1128/MMBR.00004-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin K. M., Wichman H. A.2007Variable epistatic effects between mutations at host recognition sites in ϕX174 bacteriophage. Evolution 67, 1710–1724 [DOI] [PubMed] [Google Scholar]
- Pepin K. M., Domsic J., McKenna R.2008Genomic evolution in a virus under specific selection for host recognition. Infect. Genet. Evol. 8, 825–834 (doi:10.1016/j.meegid.2008.08.008) [DOI] [PubMed] [Google Scholar]
- Pfaffl M. V.2004Quantification strategies in real-time PCR. In A-Z of quantitative PCR (ed. Bustin S. A.), pp. 87–112 La Joya, CA: International University Line [Google Scholar]
- Platt A., et al. 2010The scale of population structure in Arabidopsis thaliana. PLoS Genet. 6, e1000843 (doi:10.1371/journal.pgen.1000843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L. L., Leung C. S., Chan K. H., Yuen K. Y., Guan Y., Peiris J. S.2005Recurrent mutations associated with isolation and passages of SARS coronavirus in cells from non-human primates. J. Med. Virol. 76, 435–440 (doi:10.1002/jmv.20379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes R. R., Nowak M. A., Bonhoeffer S.2000Evolution of virulence in a heterogeneous host population. Evolution 54, 64–71 [DOI] [PubMed] [Google Scholar]
- Remold S. K., Rambaut A., Turner P. E.2008Evolutionary genomics of host adaptation in Vesicular stomatitis virus. Mol. Biol. Evol. 25, 1138–1147 (doi:10.1093/molbev/msn059) [DOI] [PubMed] [Google Scholar]
- Schmid K. J., Törjék O., Meyer R., Schmuths H., Hoffmann M. H., Altmann T.2006Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor. Appl. Genet. 112, 1104–1114 (doi:10.1007/s00122-006-0212-7) [DOI] [PubMed] [Google Scholar]
- Shackelton L. A., Parrish C. R., Truyen U., Holmes E. C.2005High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl Acad. Sci. USA 102, 379–384 (doi:10.1073/pnas.0406765102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D. D., Ward C. W., Brunt A. A.1994The Potyviridae Wallingford, CT: CAB International [Google Scholar]
- Turner P. E., Elena S. F.2000Cost of host radiation in an RNA virus. Genetics 156, 1465–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcuqui-Inchima S., Haenni A. L., Bernardi F.2001Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175 (doi:10.1016/S0168-1702(01)00220-9) [DOI] [PubMed] [Google Scholar]
- Whitham S. A., Yamamoto M. L., Carrington J. C.1999Selectable viruses and altered susceptibility mutants in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 96, 772–777 (doi:10.1073/pnas.96.2.772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S. A., Anderberg R. J., Chisholm S. T., Carrington J. C.2000Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 12, 569–582 (doi:10.1105/tpc.12.4.569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M. E. J., Haydon D. T., Antia R.2005Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20, 238–244 (doi:10.1016/j.tree.2005.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M. E., Taylor L. H., Haydon D. T.2001Population biology of multihost pathogens. Science 292, 1109–1112 (doi:10.1126/science.1059026) [DOI] [PubMed] [Google Scholar]