Abstract
West Nile virus has evolved in concert with its expansion across North America, but little is known about the evolutionary dynamics of the virus on local scales. We analysed viral nucleotide sequences from mosquitoes collected in 2005, 2006, and 2007 from a known transmission ‘hot spot’ in suburban Chicago, USA. Within this approximately 11 × 14 km area, the viral envelope gene has increased approximately 0.1% yr−1 in nucleotide-level genetic diversity. In each year, viral diversity was higher in ‘residential’ sites characterized by dense housing than in more open ‘urban green space’ sites such as cemeteries and parks. Phylodynamic analyses showed an increase in incidence around 2005, consistent with a higher-than-average peak in mosquito and human infection rates that year. Analyses of times to most recent common ancestor suggest that WNV in 2005 and 2006 may have arisen predominantly from viruses present during 2004 and 2005, respectively, but that WNV in 2007 had an older common ancestor, perhaps indicating a predominantly mixed or exogenous origin. These results show that the population of WNV in suburban Chicago is an admixture of viruses that are both locally derived and introduced from elsewhere, containing evolutionary information aggregated across a breadth of spatial and temporal scales.
Keywords: West Nile virus, evolution, ecology, phylogenetics, phylodynamics
1. Introduction
West Nile virus (WNV) is an emerging mosquito-borne virus (family Flaviviridae, genus Flavivirus) maintained through an enzootic cycle of transmission involving birds and mosquitoes of the genus Culex (Weaver & Reisen 2010). The virus can cause febrile illness and neurologic disease in a range of avian and mammalian hosts, including humans. Since its introduction to North America in 1999 in the New York City area (Lanciotti et al. 1999), WNV spread rapidly throughout the United States, Canada (Gancz et al. 2004), Mexico (Deardorff et al. 2006) and Latin America and the Caribbean as far south as Argentina (Petersen & Hayes 2008). Now endemic in the Americas, WNV is transmitted in different geographical regions by local species of Culex. In the Northeastern and Midwestern USA, where transmission is primarily by Culex pipiens, WNV undergoes predictable seasonal ‘amplification,’ involving accelerated bird–mosquito transmission in late summer and ‘spillover’ to ‘dead-end’ hosts such as humans and horses (Theophilides et al. 2006).
WNV has evolved in concert with its geographical spread. Notably, a fitter ‘WN02’ variant replaced the original ‘NY99’ variant around 2002 (Snapinn et al. 2007), probably due to the WN02 variant's increased efficiency of replication in Culex vectors (Moudy et al. 2007), especially at elevated temperatures (Kilpatrick et al. 2008). Phylodynamic analyses of viral nucleotide sequences point to a logistic dynamic of epidemic growth for WNV across North America, reflecting the WN02 variant's initially rapid spread followed by a subsequent epidemic slowdown (Snapinn et al. 2007). WNV has as yet shown no evidence of phylogeographic subdivisioning within its North American range but rather appears to circulate panmictically, probably because of continuous long-distance transport by migrating birds and/or mosquitoes (Bertolotti et al. 2007; Venkatesan & Rasgon in press).
Although the evolution and population dynamics of WNV are well understood on continent-wide scales, spatio-temporally focused studies of WNV evolution have been rare. For example, how is viral diversity distributed across fine spatial scales, or how do local dynamics of seasonal WNV transmission characteristic of northern latitudes affect viral evolution? Previously, we analysed genetic diversity and phylogenetics of WNV within in a ‘hotspot’ of transmission in suburban Chicago, USA (Bertolotti et al. 2008). In this previous study, we demonstrated that viral genetic diversity was higher in vectors than in avian hosts, that viral diversity was higher in ‘residential’ areas than in nearby ‘urban green spaces,’ and that viral movement was apparently limited across relatively short distances of approximately 4 km. However, our previous study was cross-sectional, in that all viral sequences were from 2005, precluding examination of longitudinal trends in viral diversity and evolution.
Here, we analyse new viral nucleotide sequence data from the same suburban Chicago transmission ‘hot spot’ from two subsequent years, 2006 and 2007, yielding three years of data on viral genetic diversity and evolution within this important urban focus of transmission. By comparing our results with those obtained across coarser geographical scales, we provide a new perspective on the fine-scale molecular variation of WNV, including the extent to which WNV evolution in suburban Chicago reflects localized versus broader ecological processes.
2. Material and methods
(a). Study area and sample collection
The study was carried out in southwest suburban Chicago, IL (87°44′ W, 41°42′ N), a mosaic environment of residential and commercial/industrial areas interspersed with ‘urban green spaces’ such as parks and cemeteries. This area is a known ‘hot spot’ of arboviral transmission, as evidenced by a statistically significant cluster of human WNV cases in 2002, when the virus first reached the region, and an overlapping cluster in 1975 of human cases of St Louis encephalitis, caused by a related Flavivirus (Ruiz et al. 2004) and annually intensive episodes of transmission in the summer (Hamer et al. 2008). Mosquitoes were sampled from 11 residential sites and four ‘urban green space’ sites, equally spaced across the study area and chosen to represent a range of land cover types within our study area, as described previously (Bertolotti et al. 2008; Hamer et al. 2008).
Mosquitoes were collected from each of the 15 study sites in 2005, 2006 and in 2007 once every two weeks from mid-May trough mid-October. We used CO2-baited miniature light traps, gravid traps baited with rabbit pellet infusion (Lampman & Novak 1996), and battery-powered backpack aspirators (Meyer et al. 1983). Mosquitoes were identified to species (Andreadis et al. 2005) and pooled into groups of 25 or fewer, grouped by species, collection site and date. We processed mosquitoes in the laboratory and detected WNV RNA using real-time RT-PCR as previously described (Hamer et al. 2008).
(b). Molecular analyses
We generated full-length sequences of the WNV envelope (ENV) gene, the most variable in the WNV genome and the most commonly sequenced for phylogenetic studies. We conducted reverse transcription PCR on total RNA extracted from positive mosquito pools, and we sequenced amplicons directly following previously published protocols (Bertolotti et al. 2007). For mosquito pools with multiple positives that were identified by multiple peaks in chromatograms, we cloned amplicons into plasmid vectors for sequencing and resolution of individual haplotypes. Specifically, we amplified a long fragment of 1795 bp using forward primer WNV 871 (5′-CTGGTGGCAGCCGTCATTGGTTGG-3′) and reverse primer WNV 2666c (5′-AAATGTGGGAAGCAGTGAAGGACG-3′; primer set and PCR conditions described in Bertolotti et al. 2007). The cycling profile consisted of a denaturation step at 94°C, followed by 35 cycles of PCR (94°C for 30 s, annealing temperature of 55°C for 30 s, extension temperature of 72°C for 1.30 min), a terminal extension at 72°C for 7 min and an indefinite soak at 4°C. We then cloned amplicons using the EPICENTRE-CopyControl cDNA, Gene & PCR Cloning Kit with Chemically Competent TransforMax EPI300 Escherichia coli (EPICENTRE biotechnologies, Madison, WI), following the manufacturer's protocol. Clones were sequenced following previously published protocols (Bertolotti et al. 2007). Multiple haplotypes and all ambiguous bases were resolved by repeat sequencing, and all sequences were hand-edited and aligned with reference to published sequences prior to analysis.
(c). Evolutionary inferences
We quantified viral genetic diversity at the nucleotide level as Nei's (1987) nucleotide diversity, multiplied by 100 and thus expressed as per cent, using the computer programs MEGA4 (Tamura et al. 2007) and PAUP* 4.0b10 (Swofford 2003). We tested the statistical significance of differences in nucleotide diversity for different viral populations using 10 000 bootstrap resamplings of genetic distance matrices. We explored temporal trends in nucleotide diversity over the three-year sampling period using linear regression in the software R (R Development Core Team 2009). We tested the statistical significance of temporal trends using 1000 bootstrap re-samplings of the data.
To describe evolutionary relationships among WNV sequences from our study area, we constructed phylogenetic trees, selecting models of molecular evolution using a likelihood ratio test approach and the Akaike information criterion (Akaike 1973) implemented with the computer program Modeltest v. 3.7 (Posada & Crandall 1998), combined with Bayesian methods in the computer program MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003). We calculated tree statistics using PAUP* 4.0b10 (Swofford 2003).
To describe the demographic history of WNV in suburban Chicago, we applied Bayesian coalescent methods separately for each year (2005, 2006, 2007) and for all the three years together (2005–2007) using the computer programs BEAST v. 1.4.5 (Drummond & Rambaut 2007) and Tracer v. 1.4.1 (Rambaut & Drummond 2007). Specifically, we tested four parametric demographic models (constant population size, exponential growth, logistic growth and expansion population growth) and one non-parametric demographic model (Bayesian Skyline Plot, BSP) under both strict and relaxed (uncorrelated exponential and log-normal) clock models, all conducted under the HKY85 molecular substitution model (Hasegawa et al. 1985). We compared models by calculating the Bayes factor (BF), which is the ratio of the marginal likelihoods (marginal with respect to the prior) of the two models under consideration (Kass & Raftery 1995). Marginal likelihoods were estimated using the Newton & Raftery (1994) method with the modifications proposed by Suchard et al. (2001) and were compared as twice the natural logarithm of the BF, 2ln(BF) (Kass & Raftery 1995). Evidence against the null model (i.e. that the model under consideration had a lower marginal likelihood) was assessed as follows: 2ln(BF) = 2–6 indicates evidence against the null model; 2ln(BF) = 6–10 indicates strong evidence against the null model; 2ln(BF) more than 10 indicates very strong evidence against the null model.
To evaluate the best-fit model describing WNV phylodynamics over the three-year period, we first carried out preliminary Bayesian coalescent analyses on 60 randomly selected WNV sequences from mosquitoes collected in our study area during the 2005, 2006 and 2007 transmission seasons separately (20 sequences per year). The best-fit model derived from these analyses was then applied to the full set of WNV sequences from the entire three-year period. The same best-fit models were also used to estimate viral evolutionary parameters (for each year separately and all three years together) and to evaluate WNV population dynamics. Evolutionary parameters were estimated using the Bayesian Markov chain Monte Carlo (MCMC) method implemented in the BEAST software package. Bayesian calculations consisted of several independent 50 000 000-generation MCMC runs for the single-year datasets separately, and four independent 100 000 000-generation MCMC runs for the multi-year dataset. For all MCMC runs, sampling occurred every 1000th generation. The independent runs were then combined with LogCombiner v. 1.4.7 (Drummond & Rambaut 2007). Convergence of the MCMC was assessed by calculating the Effective Sample Size (ESS) of the runs. All parameter estimates showed significant ESS (more than 200). Uncertainty in parameter estimates was represented as 95 per cent highest posterior-density (HPD) values. Results were visualized using the Tracer program. To test whether sequences contained sufficient signal to estimate evolutionary rates and divergence times, we randomized date–sequence relationships and repeated the BEAST analysis according to the methods of Ramsden et al. (2009).
To evaluate the strength and direction of selection in different viral populations, we calculated ratios (ω) of non-synonymous (Dn) to synonymous (Ds) substitutions per site for each year of sampling using the Datamonkey webserver (Pond & Frost 2005).
3. Results
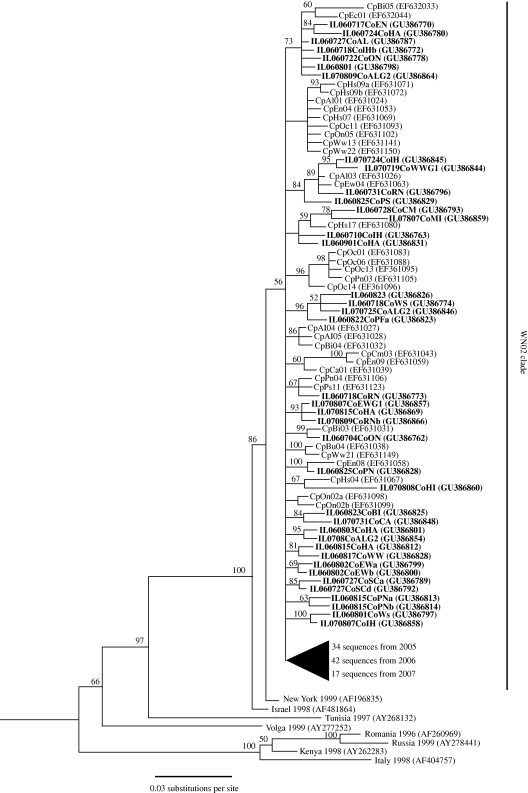
We analysed 254 WNV ENV sequences of 1575 nucleotides each from Culex mosquito pools, including 128 WNV sequences from 2005, 80 sequences from 2006 and 46 sequences from 2007. These samples came from all 15 study sites and contained 167 unique WNV ENV sequences (68 unique sequences out of 128 in 2005, 70 unique sequences out of 80 in 2006, 29 unique sequences out of 46 in 2007). Phylogenetic analysis yielded a tree similar to those described by (Davis et al. 2005) and (Bertolotti et al. 2008), in which all newly generated samples clustered within a single ‘WN02’ clade most closely related to the New York 1999 strain (figure 1). All newly generated sequences have been deposited into GenBank (accession numbers GU386 758-GU386 883).
Figure 1.
Phylogenetic tree constructed using Bayesian analysis of 254 WNV envelope gene sequences (length 1575 nucleotides) from suburban Chicago, plus eight out-group sequences. Taxon names generated from the Chicago study area indicate collection site (described in Bertolotti et al. 2008), identification number (only for WNV sequences from mosquitoes collected in 2005), dates of collection (YearMonthDay for sequences from mosquitoes collected in 2006 and 2007), and GenBank accession numbers (in parentheses). Posterior probabilities of clades are indicated above branches.
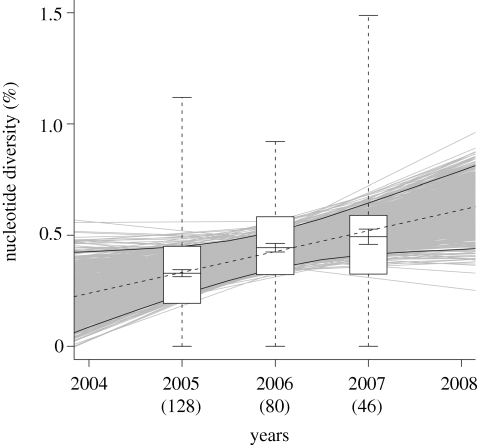
Mean nucleotide diversity for WNV sequences, summarized by year and by site, are shown in table 1. In all years, viral genetic diversity was lower among viruses from mosquitoes captured in urban green space sites than from mosquitoes captured in residential sites, a trend that we previously reported for 2005 (Bertolotti et al. 2008); however, this difference was statistically significant only for WNV sequences generated from mosquito pools collected in 2005 and 2006, but not in 2007, perhaps because of low sample sizes in the third study year (table 1). Analyses of nucleotide diversity by year showed a significant trend of increasing genetic diversity over time in our study area (R2 = 0.32, t = 36.8, p < 0.01; figure 2). The slope of this relationship, 0.94 × 10−3, indicates that the WNV ENV gene in our study area has diversified at a rate of 0.094% yr−1 between 2005 and 2007.
Table 1.
Nucleotide diversity of West Nile virus in Culex mosquitoes collected during the 2005, 2006 and 2007 transmission seasons from the suburban Chicago study area.
| year | location | N | nucleotide diversity (%) | standard error (%) | differencea |
|---|---|---|---|---|---|
| 2005 | residential sites | 76 | 0.37 | 0.022 | p < 0.001 |
| natural sites | 52 | 0.26 | 0.050 | ||
| all sites | 128 | 0.33 | 0.016 | ||
| 2006 | residential sites | 59 | 0.45 | 0.023 | p = 0.003 |
| natural sites | 21 | 0.42 | 0.037 | ||
| all sites | 80 | 0.44 | 0.019 | ||
| 2007 | residential sites | 38 | 0.50 | 0.039 | p = 0.107 |
| natural sites | 8 | 0.46 | 0.063 | ||
| all sites | 46 | 0.50 | 0.035 |
aStatistical difference between nucleotide diversity for viral sequences collected in residential versus natural sites within the same year, estimated from 10 000 bootstrap resamplings of the genetic distance matrices.
Figure 2.
WNV genetic diversity in Culex mosquitoes from suburban Chicago, 2005–2007. Genetic diversity is expressed as nucleotide diversity (Nei 1987), or mean proportion of nucleotide differences among sequences within each year, corrected for multiple substitutions using a model of molecular evolution derived from a hierarchical likelihood ratio test approach implemented using the computer program Modeltest, v. 3.7 (Posada & Crandall 2001). Lines and small error bars indicate means and standard errors, respectively. Boxes indicate third quartiles, and large error bars indicate ranges. The dashed line is a least squares regression line indicating the rate of increase of viral genetic diversity over time (0.94 × 10−3 yr−1, R2 = 0.32, t = 36.8, p < 0.01). Grey lines are least-squares regression lines derived from 1000 bootstrap replicates of the data; solid lines indicate 95% CI around the nucleotide diversity estimation. Sample sizes for each year are indicated in parentheses.
To infer the population dynamics of WNV, we analysed viral sequences using Bayesian coalescent methods. Because we knew exact dates of collection for all viral samples from the study area, we were able to estimate evolutionary parameters of the WNV ENV gene specifying days as units of time. In our study, the BF comparison among parametric and non-parametric demographic models selected the Bayesian Skyline Plot uncorrelated exponential relaxed clock as the best-fit model for each year separately and for all the three years together. Therefore, WNV evolutionary parameters and population dynamics were estimated under the Bayesian Skyline Plot uncorrelated exponential relaxed clock model.
Mean rates of evolution estimated under the best-fit model varied among years, with evolution being most rapid in 2006, albeit with overlapping HPD estimates (table 2). The rate of evolution across all years combined was 4.17 × 10−6 nucleotide substitutions per site per day, or 1.52 × 10−3 nucleotide substitutions per site per year (95% HPD = 1.22 × 10−3—1.84 × 10−3 substitutions per site per year). The mean ages of the most recent common ancestors for WNV from 2005 and 2006 were 1.5 and 1.1 years, respectively, whereas the mean age of the most recent common ancestor for WNV from 2007 was 5.6 years, again albeit with overlapping HPD estimates (table 2). Repeat analyses conducted on the date-randomized dataset showed largely different and non-overlapping 95 per cent HPD intervals, confirming that data contain adequate temporal signal and supporting estimation results.
Table 2.
Bayesian estimates of evolutionary parameters for WNV in suburban Chicago over timea.
| year | number of samples | range of dates of sample collection | substitution rateb (95% HPD) | TMRCAc (95% HPD) |
|---|---|---|---|---|
| 2005 | 128 | 21 July–4 October | 6.87 × 10−6 (1.11 × 10−6–1.50 × 10−5) | 532 (143–1243) |
| 2006 | 80 | 13 June–7 September | 13.11 × 10−6 (4.82 × 10−7–2.74 × 10−5) | 408 (92–1253) |
| 2007 | 46 | 5 July–9 September | 4.48 × 10−6 (6.73 × 10−8–1.38 × 10−5) | 2057 (122–7584) |
| all years | 254 | 21 July 2005–9 September 2007 | 4.17 × 10−6 (3.34 × 10−6–5.04 × 10−6) | 1206 (984–1531) |
aAll evolutionary parameters were estimated under the Bayesian skyline plot uncorrelated exponential relaxed clock model.
bMean number of nucleotide substitutions per site per day. HPD: high posterior density.
cMean time to the most recent common ancestor (TMRCA), in days.
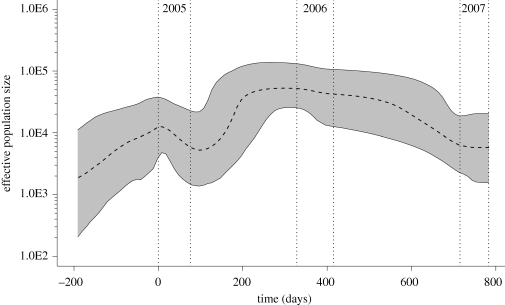
Figure 2 shows the non-parametric trajectory of WNV effective population size over time in suburban Chicago estimated using the Bayesian Skyline Plot model. The shape of the curve shows some evidence of increasing effective population size prior to 2005 and continuing until 2006, followed by a decrease in effective population size in 2007. WNV sequences from the Chicago study area collected in the different years (2005, 2006, 2007) showed overall ratios of non-synonymous substitutions per site that were comparable and well below 1 (ω = 0.100042 in 2005, 0.0714128 in 2006, 0.0701077 in 2007). Analyses at the codon level using the Datamonkey.org webserver showed no positively selected sites, indicating a pattern of strong stabilizing selection, as has previously been reported for WNV (Bertolotti et al. 2007, 2008; Jerzak et al. 2008).
4. Discussion
We have estimated patterns of genetic diversity and evolution of the West Nile virus circulating in Culex mosquitoes in a small geographic area in suburban Chicago (USA) over a three-year period (2005–2007). Our phylogenetic analyses indicate that WNV in this area has remained relatively stable, consisting of viral lineages within the WN02 clade that has spread across North America. The topology of our tree (figure 1) shows some resolution of sub-clades within the larger WN02 clade, but no evidence of phylogenetic sorting by year or sampling location. WNV in suburban Chicago therefore appears to be a relatively panmictic viral population that is roughly representative of WNV circulating in North America.
Our analyses of viral genetic diversity by year demonstrate a significant trend of increasing WNV genetic diversity in Culex mosquitoes from 2005 to 2007. Previously, we demonstrated a similar trend across the Midwestern USA of approximately 0.07 per cent diversification per year from 2002 to 2005, with similar overall levels of diversity (Bertolotti et al. 2008). Documenting a continuation of this trend on a much finer spatial scale than has previously been reported indicates that WNV in our suburban Chicago ‘hot spot’ is displaying similar population genetic dynamics as WNV across the Midwest. The increasing viral diversity we have documented has probably arisen by neutral evolution, as evidenced by our analyses of non-synonymous to synonymous substitution rates that show evidence of strong stabilizing selection, and by the fact that WNV evolutionary rates in our study area were similar across years.
Our analyses confirm our previous observation from 2005 data that WNV sequences from mosquitoes captured in residential sites are more genetically diverse than are those from mosquitoes captured in urban green space sites (Bertolotti et al. 2008). The direction of this trend was consistent across all years, but did not reach statistical significance in 2007, probably due to a small sample size (2007 was characterized by relatively low transmission, and consequently only eight sequences from natural sites were available from that year). This trend may reflect altered transmission dynamics in the two types of habitats, perhaps due to fine-scale variation in the distribution of avian hosts or microclimate. Although the ecological reasons for this trend remain obscure, it nevertheless confirms that fine-scale features of the urban landscape can influence the genetic diversity and evolution of arboviral pathogens such as WNV.
Previously, Snapinn et al. (2007) performed phylodynamic analyses of WNV sequences from across North America, arguing that WNV on a continent-wide scale displayed a logistic pattern of epidemic growth between 2001 and 2005, interpreted as reflecting an initial rapid expansion of the WN02 genotype followed by saturation of available hosts. In our study, we applied similar methods at a finer spatial scale (the suburban Chicago ‘hot spot’) and temporal scale (days versus years). As shown in figure 3, the effective number of WNV infections increased prior to 2005 and until approximately 2006, after which it remained fairly constant before declining moderately in 2007. Importantly, we found no evidence for continued overall population expansion, consistent with the Snapinn et al. (2007) results, and no evidence of regular, annual population expansions and contractions, as might have been expected based on the well-known ‘amplification’ of WNV in our study area and other northern latitude sites in late summer and early autumn (Theophilides et al. 2006). WNV phylodynamics in our study area are therefore apparently ‘decoupled’ from the local epidemic pattern of highly seasonal transmission and amplification. We note, however, that the Chicago region registered a very hot and dry summer in 2005, with temperatures that were the third highest since 1958 and precipitation that was approximately 30 cm below recent averages, and that the resulting combination of high temperatures and low rainfall provided favourable conditions for the reproduction of Culex mosquitoes, leading to a higher-than-average peak in mosquito infection rate that year and a correspondingly high incidence of human cases (Ruiz et al. in press). By contrast, WNV transmission in 2007 was atypically low, as reflected in the numbers of human cases in Cook and DuPaige Counties (the counties containing Chicago and its major suburbs) recorded by the Illinois Department of Public Health, which for 2005, 2006, and 2007 were 182, 159 and 53, respectively (Ruiz et al. in press). The expansion of the WNV effective population size through 2005 and a subsequent decline in 2007 as shown in figure 3 is consistent with these epidemiological observations.
Figure 3.
Bayesian skyline plot of WNV in suburban Chicago, USA. The black bold dashed line represents the median estimate of effective number of infections through time in suburban Chicago, derived from ENV gene sequences using a Bayesian coalescent approach. Genetic distances were transformed into a time scale of days taking into account the different sampling times of WNV in our study area and enforcing a relaxed molecular clock (see §2). Ninety-five per cent HPD values shaded. Vertical dashed grey lines indicate the sampling periods (21 July–4 October 2005, 13 June–7 Septmber 2006, 5 July–9 September 2007) for the WNV sequences analysed in this study.
Using the same Bayesian coalescent approach, we were able to estimate the mean age of WNV (most recent common ancestor) in our study area for each year (table 2). Our best estimates for the ages of the most recent common ancestors of WNV sampled during the 2005 and 2006 transmission seasons (1.4 years and 1.1 years, respectively) suggest that WNV lineages in these years derived predominantly from viruses present the previous year, which implies a dominant evolutionary signal of overwintering of WNV in our study area in these years. However, the age of the most recent common ancestors for WNV sampled in 2007 was 5.6 years, suggesting that our 2007 sample of WNV contained lineages that had diverged much earlier than the previous year. This observation implies a stronger signal of re-introduction of exogenous and divergent WNV into our study area in 2007 than in the two previous years. In this light, we note that the 2007 winter was unusually cold in Chicago, which might have reduced the rate of overwintering and consequently increased the proportion of exogenous viruses sampled during the subsequent 2007 transmission season. These preliminary analyses therefore suggest that WNV in our study area may derive from both local and distant sources and that the relevant proportions of viral sequences originating from these two sources may vary among years in response to winter weather conditions. We note, however, that our high posterior density estimates are wide for all years, such that we cannot at this point definitively differentiate a predominant pattern of overwintering from one of reintroduction for any given year. More intensive sampling and more extensive sequencing should help clarify the relative importance of these phenomena.
We also caution that, despite our systematic and prospective sampling effort across three consecutive years, our data nevertheless suffer some limitations due to lack of phylogenetic resolution. Although we were able to resolve some sub-clades within the larger WN02 clade shown in figure 1, relationships among many of our sequences remained unresolved. Greater phylogenetic resolution might have allowed us to focus our phylodynamic analyses on sub-clades representing clusters of locally transmitted viruses, as has proved useful for examining co-circulating and epidemiologically interacting lineages in the case of influenza virus, for example (Holmes et al. 2005). Because we could not resolve such sub-clades, however, it is probable that our phylodynamic analyses offer a spatially aggregated picture of WNV population dynamics that includes both locally evolved WNV lineages and lineages introduced from elsewhere. Indeed, our analyses of times to most recent common ancestor showing TMRCAs of approximately one year for 2005 and 2006 but greater than one year for 2007 tentatively support this notion and should therefore be viewed as preliminary.
Overall, our results suggest that the population of WNV in suburban Chicago is broadly representative of that on coarser spatial scales, but also that it contains population genetic and phylogenetic signatures of local ecological and epidemiological processes, such as urban landscape type and climate. Moreover, WNV in suburban Chicago appears to show substantial temporal variability, both in its propensity to increase in diversity across years and in its variable evolutionary dynamics from year to year. WNV in our small ‘hot spot’ thus appears to represent an admixture of viruses that are both locally derived and introduced from elsewhere and that reflect transmission dynamics aggregated across a breadth of spatial and temporal scales. We suggest that further studies of WNV on fine spatial and temporal scales in other regions might help clarify the importance of micro-scale processes to the transmission and evolution of this and other emerging arboviral pathogens.
Acknowledgements
We are grateful for the assistance of S. Loss, T. Thompson, D. Gohde, M. Goshorn, B. Pultorak, M. Neville, S. Dallmann and J. McClain for assistance in the field, T. Tranby, B. Bullard and L. Abernathy for assistance in the laboratory, E. Holmes for assistance with phylogenetic and phylodynamic analyses, and S. Frost and one anonymous reviewer for constructive comments on the manuscript. We also thank J. Fahey and the Archdiocese of Chicago and the municipalities of Evergreen Park, Palos Hills, Burbank, Alsip, Blue Island, the City of Chicago, and private landowners in these municipalities for allowing us to conduct this research, as well as to the Village of Oak Lawn for providing field laboratory facilities and logistical support. This material is based upon work supported by the National Science Foundation/National Institutes of Health Ecology of Infectious Diseases programme under Award no. 0840403.
Footnotes
One contribution of 14 to a Theme Issue ‘New experimental and theoretical approaches towards the understanding of the emergence of viral infections’.
References
- Akaike H.1973Maximum likelihood identification of Gaussian autoregressive moving average models. Biometrica 60, 255–265 (doi:10.1093/biomet/60.2.255) [Google Scholar]
- Andreadis T. G., Thomas M. C., Shepard J. J.2005Identification guide to the mosquitoes of Connecticut. New Haven, CT: The Connecticut Agricultural Experiment Station [Google Scholar]
- Bertolotti L., Kitron U., Goldberg T. L.2007Diversity and evolution of West Nile virus in Illinois and The United States, 2002–2005. Virology 360, 143–149 (doi:10.1016/j.virol.2006.10.030) [DOI] [PubMed] [Google Scholar]
- Bertolotti L., Kitron U. D., Walker E. D., Ruiz M. O., Brawn J. D., Loss S. R., Hamer G. L., Goldberg T. L.2008Fine-scale genetic variation and evolution of West Nile virus in a transmission ‘hot spot’ in suburban Chicago, USA. Virology 374, 381–389 (doi:10.1016/j.virol.2007.12.040) [DOI] [PubMed] [Google Scholar]
- Davis C. T., et al. 2005Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: evidence for the emergence of a dominant genotype. Virology 342, 252–265 (doi:10.1016/j.virol.2005.07.022) [DOI] [PubMed] [Google Scholar]
- Deardorff E., et al. 2006Introductions of West Nile virus strains to Mexico. Emerg. Infect. Dis. 12, 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A.2007BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancz A. Y., Barker I. K., Lindsay R., Dibernardo A., McKeever K., Hunter B.2004West Nile virus outbreak in North American owls, Ontario, 2002. Emerg. Infect. Dis. 10, 2135–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G. L., et al. 2008Rapid amplification of West Nile virus: the role of hatch-year birds. Vector Borne Zoonotic Dis. 8, 57–67 (doi:10.1089/vbz.2007.0123) [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T.1985Dating of the human–ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174 (doi:10.1007/BF02101694) [DOI] [PubMed] [Google Scholar]
- Holmes E. C., et al. 2005Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 3, e300 (doi:10.1371/journal.pbio.0030300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzak G., Brown I., Shi P.-Y., Kramer L. D., Ebel G. D.2008Genetic diversity and purifying selection in West Nile virus populations are maintained during host switching. Virology 374, 256–260 (doi:10.1016/j.virol.2008.02.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E., Raftery A. E.1995Bayes factors. J. Am. Statist. Assoc. 90, 773–795 (doi:10.2307/2291091) [Google Scholar]
- Kilpatrick A. M., Meola M. A., Moudy R. M., Kramer L. D.2008Temperature, viral genetics, and the transmission of West Nile virus by Culex pipiens mosquitoes. PLoS Pathog. 4, e1000092 (doi:10.1371/journal.ppat.1000092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampman R. L., Novak R. J.1996Oviposition preferences of Culex pipiens and Culex restuans for infusion-baited traps. J. Am. Mosq. Control Assoc. 12, 23–32 [PubMed] [Google Scholar]
- Lanciotti R. S., et al. 1999Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286, 2333–2337 (doi:10.1126/science.286.5448.2333) [DOI] [PubMed] [Google Scholar]
- Meyer R. P., Reisen W. K., Hill B. R., Martinez V. M.1983The AFS sweeper, a battery-powered backpack mechanical aspirator for collecting adult mosquitoes. Mosquito News 43, 346–350 [Google Scholar]
- Moudy R. M., Meola M. A., Morin L. L., Ebel G. D., Kramer L. D.2007A newly emergent genotype of West Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. Am. J. Trop. Med. Hyg. 77, 365–370 [PubMed] [Google Scholar]
- Nei M.1987Molecular evolutionary genetics New York, NY: Columbia University Press [Google Scholar]
- Newton M. A., Raftery A. E.1994Approximate Bayesian inference with the weighted likelihood bootstrap. J. R. Statist. Soc. Ser. B 56, 3–48 [Google Scholar]
- Petersen L. R., Hayes E. B.2008West Nile virus in the Americas. Med. Clin. North Am. 92, 1307–1322 (doi:10.1016/j.mcna.2008.07.004) [DOI] [PubMed] [Google Scholar]
- Pond S. L., Frost S. D.2005Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 (doi:10.1093/bioinformatics/bti320) [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K. A.1998Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2009R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Rambaut A., Drummond A. J.2007. Tracer v.1.4. Available from http://beast.bio.ed.ac.uk/Tracer
- Ramsden C., Holmes E. C., Charleston M. A.2009Hantavirus evolution in relation to its rodent and insectivore hosts: no evidence for codivergence. Mol. Biol. Evol. 26, 143–153 (doi:10.1093/molbev/msn234) [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P.2003MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- Ruiz M. O., Tedesco C., McTighe T. J., Austin C., Kitron U.2004Environmental and social determinants of human risk during a West Nile virus outbreak in the greater Chicago area, 2002. Int. J. Health. Geogr. 3, 8 (doi:10.1186/1476-072X-3-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M. O., Chaves L. F., Hamer G. L., Sun T., Brown W. M., Walker E. D., Haramis L., Goldberg T. L., Kitron U. D.In press Local impact of temperature and precipitation on West Nile virus infection in Culex species mosquitoes in northeast Illinois, USA. Parasit. Vectors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapinn K. W., Holmes E. C., Young D. S., Bernard K. A., Kramer L. D., Ebel G. D.2007Declining growth rate of West Nile virus in North America. J. Virol. 81, 2531–2534 (doi:10.1128/JVI.02169-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M. A., Weiss R. E., Sinsheimer J. S.2001Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18, 1001–1013 [DOI] [PubMed] [Google Scholar]
- Swofford D. L.2003PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer Associates [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S.2007MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- Theophilides C. N., Ahearn S. C., Binkowski E. S., Paul W. S., Gibbs K.2006First evidence of West Nile virus amplification and relationship to human infections. Int. J. Geogr. Inf. Sci. 20, 103–115 (doi:10.1080/13658810500286968) [Google Scholar]
- Venkatesan M., Rasgon J. L.In press Population genetic data suggest a role for mosquito-mediated dispersal of West Nile virus across the western United States. Mol. Ecol. (doi:10.1111/j.1365-294X.2010.04577.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S. C., Reisen W. K.2010Present and future arboviral threats. Antiviral Res. 85, 328–345 (doi:10.1016/j.antiviral.2009.10.008) [DOI] [PMC free article] [PubMed] [Google Scholar]