Abstract
The vast majority of well-characterized eukaryotic viruses are those that cause acute or chronic infections in humans and domestic plants and animals. However, asymptomatic persistent viruses have been described in animals, and are thought to be sources for emerging acute viruses. Although not previously described in these terms, there are also many viruses of plants that maintain a persistent lifestyle. They have been largely ignored because they do not generally cause disease. The persistent viruses in plants belong to the family Partitiviridae or the genus Endornavirus. These groups also have members that infect fungi. Phylogenetic analysis of the partitivirus RNA-dependent RNA polymerase genes suggests that these viruses have been transmitted between plants and fungi. Additional families of viruses traditionally thought to be fungal viruses are also found frequently in plants, and may represent a similar scenario of persistent lifestyles, and some acute or chronic viruses of crop plants may maintain a persistent lifestyle in wild plants. Persistent, chronic and acute lifestyles of plant viruses are contrasted from both a functional and evolutionary perspective, and the potential role of these lifestyles in host evolution is discussed.
Keywords: persistent viruses, acute viruses, fungal viruses, virus ecology
1. Introduction
The study of viruses began in the 1890s with the discovery of an acute infectious agent in tobacco causing leaf spots (Beijerinck 1898). Tobacco mosaic virus (TMV) was the first virus characterized, and set a precedent for viruses as agents of disease. Today anyone you ask, from a scientist to a school child, will probably tell you that viruses make things sick. This is because the acute and chronic disease-causing viruses are the most well-studied, and not because there is any validity to the notion that most viruses cause disease.
Over the past 120 years, hundreds of viruses of plants have been described, almost all of which are pathogens of crop plants (Fauquet et al. 2005). The cryptic viruses were first noticed in plants in the 1960s and 1970s. These were described as viruses that cause no symptoms, are not horizontally transmissible and are at very low titre (Boccardo et al. 1987). These viruses are currently classified in the family Partitiviridae (Fauquet et al. 2005), and this family also includes viruses that infect fungi. Recently, other plant viruses have been described with similar characteristics, most notably the endornaviruses (reviewed in Fukuhara et al. 2006; Fukuhara & Moriyama 2008). These viruses remain the most poorly characterized viruses of plants. Here, we define these viruses as persistent plant viruses and compare and contrast them with acute and chronic plant viruses. Although often ignored, persistent viruses may have significant effects on the evolution of acute viruses, and on the evolution of their hosts.
2. Lifestyles of plant viruses
Plant viruses can have four different lifestyles: persistent, acute, chronic and endogenous. Some viruses may be able to change from one lifestyle to another, particularly between the acute and chronic lifestyles; other lifestyle changes probably occur but are rare. Persistent plant viruses have the following features: they are generally asymptomatic, although a lack of uninfected plants in most cases makes this difficult to fully assess; they are transmitted vertically via gametes (Blanc 2007), but not horizontally; they do not move between cells in plants, not even through grafts; rather they are found in every cell including the meristem. So far, all of the viruses in this classification have double-stranded (ds) RNA genomes, but this is not a part of the definition. All of the virus families containing persistent plant viruses also have members that infect fungi, and particularly endophytic fungi. In ecogenomic studies of wild plants, persistent viruses make up the majority of plant viruses, with incidence rates as high as 70 per cent in some plant families (M. J. Roossinck 2009, unpublished data). This definition of persistent plant viruses is distinct from the persistent mode of vector transmission (Gray & Banerjee 1999), which relates only to the transmission event and not to the lifestyle of the virus in the plant.
Acute plant viruses in contrast to persistent viruses, have a very different life cycle. They are most frequently transmitted horizontally, they may or may not be transmitted vertically in some hosts, and vertical transmission can be through embryos or gametes (Blanc 2007), they readily move from cell-to-cell and usually encode special proteins to facilitate this. They usually accumulate to high levels in plants, although accumulation is often cyclic. An important feature of acute viruses is that they flourish in monoculture. This is also true of the animal acute viruses, and it has been suggested that the acute virus lifestyle in human viruses is largely associated with large urban centres, and in other animals with domestication (Villarreal et al. 2000). However, animal acute viruses tend to be quite host-specific, whereas plant acute viruses can be either specialists, like barley stripe virus that only infects a few closely related plants (Timian 1974), or generalists, like cucumber mosaic virus with a host range of about 1200 species (Edwardson & Christie 1991). Specialists are more likely to spread rapidly in monoculture and very poorly in the mixed plant culture common in wildlands. However, plant viruses that often have a generalist host range are transmitted by specialist vectors, and this could have a similar effect to host specificity of the virus itself. The difference between acute and chronic plant viruses is subtle, and in general relates to the duration of infection. Acute virus infections are resolved in one of the three ways: death of the host, recovery of the host or conversion to chronic infection. Chronic plant viruses remain in the host plant for extended periods of time, and may or may not cause observable disease symptoms. The distinguishing features of persistent and acute or chronic plant viruses are summarized in table 1.
Table 1.
Distinguishing characteristics of persistent versus acute plant viruses.
| persistent lifestyle | acute/chronic lifestylesa |
|---|---|
| does not induce obvious symptoms | often symptomatic; may be lethal |
| maintains infection for lifetime of host | infection may be resolved by recovery |
| does not move from cell to cell; no movement protein | moves systemically through host, usually with the aid of a virus-encoded protein |
| seed transmission nearly 100% via gametes | may or may not be seed transmitted via gametes or embryo invasion; rarely to high frequency |
| not transmitted horizontally | horizontally transmitted, often by insect vectors |
| generally low titre | may establish very high titre |
aThe distinction between acute and chronic viruses is discussed in the text, and refers to whether or not the infection is resolved by recovery or death (acute) or remains for prolonged periods (chronic).
Animal viruses have been characterized as persistent or acute based largely on the duration of infection. In recent years, these lifestyles have been discussed in depth by Luis Villarreal (Villarreal et al. 2000; Villarreal 2007, 2009), and persistent viruses are described in terms of chronic infections and of asymptomatic infections. Asymptomatic persistence is common in wild animals, and probably contributes to resistance to acute viral infections, in a scenario where viruses switch from persistent to acute lifestyles. In the persistent infections, acquired early in life, the virus establishes a low level of infection that induces an immune state and prevents later acute infections (reviewed in Villarreal 2009). This is similar to the well-documented effects of cross-protection in plants, where infection by a mild variant of a virus protects against a more severe variant (Fraser 1998). In fact, similar to plants, animals in the wild seem to be mostly healthy, even though, when examined, they are full of viruses and other parasites. This may be because there are positive effects of parasites under some conditions (Thomas et al. 2000; Brown et al. 2006). Farmed animals however, like farmed plants, are frequently plagued with disease-causing or lethal virus infections. Animal persistent viruses are believed to be responsible for emerging acute viral diseases (Villarreal et al. 2000). These viruses can switch between persistent and acute lifestyles. It is not clear if this is true with plant viruses, as it has not been demonstrated. It seems unlikely, or at least quite rare, because persistent plant viruses would have to acquire the abilities to move horizontally and systemically in plants in order to become acute. This would involve a major leap that would be more likely to occur through recombination with another virus (discussed further below).
An additional group of plant viruses are the endogenous viruses. These are viruses that are integrated into the genome of the plant. A majority of these are defective remnants of ancient virus infections, but some can be activated under certain conditions. The endogenous viruses have been recently reviewed (Hohn et al. 2008), and will not be discussed further here.
3. Plant persistent viruses: origins and evolution
The most well studied of the plant persistent viruses are those previously called cryptic viruses (Boccardo et al. 1987). These are now classified as members of the Partitiviridae. The taxonomy of this family is changing as more and more viruses are being discovered and sequenced. The family includes viruses with fungal hosts, plant hosts and recently protozoan hosts (Nibert et al. 2009). They are widespread in plants, and have been found in gymnosperms (Veliceasa et al. 2006) as well as algae (Koga et al. 2003). In algae, they are associated with chloroplasts leading to the proposal that they are of prokaryotic origin (Ishihara et al. 1992). In plants, they appear to persist indefinitely. For example, years of continuous tissue culture, thermotherapy and meristem tip culture were unable to cure plants of partitiviruses (Szegö et al. 2005). Pepper cryptic virus, isolated from Jalapeño peppers (Arancibia et al. 1995), is found in all cultivars tested, indicating its presence at least as far back as the origin of the Jalapeño cultivar.
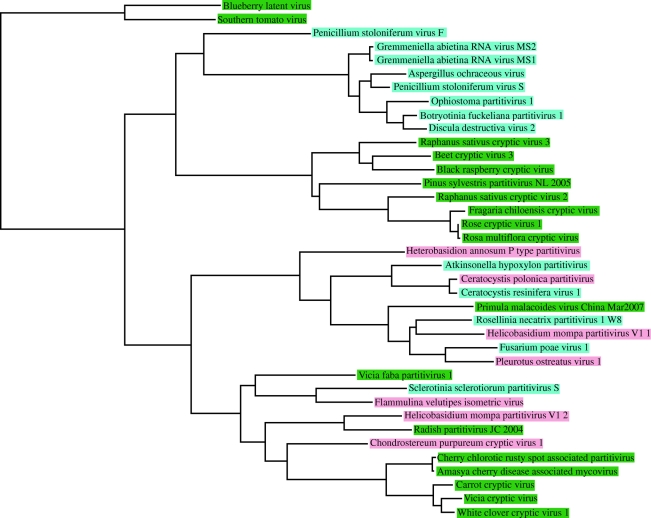
Recent publications have suggested that some plant partitiviruses are more similar to fungal partitiviruses than other plant partitiviruses (Veliceasa et al. 2006; Sabanadzovic & Ghanem-Sabanadzovic 2008; Li et al. 2009). A phylogenetic analysis using two related plant viruses, southern tomato virus (Sabanadzovic et al. 2009) and blueberry latent virus as outgroups to the Partitiviridae indicates that while some clades contain only plant or ascomycete partitiviruses, others have mixtures of viruses from all three hosts (figure 1, table 2). This is strongly suggestive of transmission of these viruses, both among fungi and between plants and fungi. Previously, I proposed that plant cryptic viruses could have originated as fungal viruses that were trapped in plants during an endophytic association, and that these could have acquired the necessary movement proteins from their plant hosts to evolve into acute plant viruses (Roossinck 1997). Given the complexity of the relationships (and it seems likely that these relationships will become even more interesting as more of these viruses are discovered and characterized), another scenario is that these are fungal viruses that use plants as their vectors. Transmission is likely a rare event because it would require entry into the germline, but it is certainly plausible, and a more reasonable explanation than that these viruses are so ancient that they have co-diverged with hosts, as was suggested for the totiviruses (Bruenn 1993). There is no evidence in the phylogeny for co-divergence, i.e. there is no congruence between the viral phylogeny and the host phylogeny.
Figure 1.
Phylogenetic relationships of partitvirus RdRps. Amino acid sequences for partitivirus RdRps were downloaded from GenBank, aligned with MAFFT and alignments were thoroughly edited in Mesquite (Maddison & Maddison 2009). Aligned sequences were analysed using MrBayes (Ronquist & Huelsenbeck 2003) under a fixed rate model, with 10 000 generations. The consensus tree after burn-in was displayed in Mesquite. Viruses names are coloured by the host group: green, plant host; pink, basidiomycete host; turquoise, ascomycete host. A nearly identical tree was obtained with maximum parsimony (not shown).
Table 2.
Names and accession numbers of viruses used in figure 1.
| virus name | host | accession number |
|---|---|---|
| Amasya cherry disease-associated mycovirus | (plant) | YP_138537.1 |
| Aspergillus ochraceous virus | ascomycete | ABV30675.1 |
| Atkinsonella hypoxylon partitivirus | ascomycete | NP_604475.1 |
| Beet cryptic virus 3 | plant | AAB27624.1 |
| Black raspberry cryptic virus | plant | ABU55400.1 |
| Blueberry latent virus | plant | ABO36236.2 |
| Botryotinia fuckeliana partitivirus 1 | ascomycete | YP_001686789.1 |
| Carrot cryptic virus | plant | ACL93278.1 |
| Ceratocystis polonica partitivirus | ascomycete | AAP86639.1 |
| Ceratocystis resinifera virus 1 | basidiomycete | AAU26069.1 |
| Cherry chlorotic rusty spot associated partitivirus | plant | CAH03668.1 |
| Chondrostereum purpureum cryptic virus 1 | basidiomycete | CAQ53729.1 |
| Discula destructiva virus 2 | ascomycete | NP_620301.1 |
| Flammulina velutipes isometric virus | basidiomycete | BAH08700.1 |
| Fragaria chiloensis cryptic virus | plant | YP_001274391.1 |
| Fusarium poae virus 1 | ascomycete | NP_624349.1 |
| Gremmeniella abietina RNA virus MS1 | ascomycete | AAM12240.1 |
| Gremmeniella abietina RNA virus MS2 | ascomycete | YP_138540.1 |
| Helicobasidium mompa partitivirus V1-1 | basidiomycete | BAD32677.1 |
| Helicobasidium mompa partitivirus V1-2 | basidiomycete | BAD32678.1 |
| Heterobasidion annosum P-type partitivirus | basidiomycete | AAL79540.1 |
| Ophiostoma partitivirus 1 | ascomycete | CAJ31886.1 |
| Penicillium stoloniferum virus F | ascomycete | YP_271922.1 |
| Penicillium stoloniferum virus S | ascomycete | AAN86834.2 |
| Pinus sylvestris partitivirus NL-2005 | plant | AAY51483.1 |
| Pleurotus ostreatus virus 1 | basidiomycete | YP_227355.1 |
| Primula malacoides virus China/Mar2007 | plant | YP_003104768.1 |
| Radish partitivirus JC-2004 | plant | AAU88207.1 |
| Raphanus sativus cryptic virus 2 | plant | YP_001686783.1 |
| Raphanus sativus cryptic virus 3 | plant | YP_002364401.1 |
| Rosa multiflora cryptic virus | plant | ABV89762.1 |
| Rose cryptic virus 1 | plant | YP_001686786.1 |
| Rosellinia necatrix partitivirus 1-W8 | ascomycete | YP_392480.1 |
| Sclerotinia sclerotiorum partitivirus S | ascomycete | YP_003082248.1 |
| Southern tomato virus | plant | ABO36238.1 |
| Vicia cryptic virus | plant | YP_272124.1 |
| Vicia faba partitivirus 1 | plant | ABJ99996.1 |
| White clover cryptic virus 1 | plant | YP_086754.1 |
A second group of persistent viruses in plants are the endornaviruses. These viruses can also be found in fungi and protists and are quite highly conserved among their different hosts (Kozlakidis et al. 2010). The endornavirus genome is also dsRNA, although there is a nick in the coding strand (Pfeiffer 1998). The genomes contain an unusual single large open reading frame (ORF) of around 5000 amino acids although this is almost certainly either a polyprotein, or it is expressed as separate ORFs from subgenomic RNAs (Fukuhara & Moriyama 2008).
Endornaviruses do not appear to package their RNA, but are associated with cytoplasmic membrane vesicles that also contain their RNA-dependent RNA polymerase (RdRp; Lefebvre et al. 1990). There are very few sequences available for the endornaviruses, so it is not possible to compare their phylogenetic relationships with those of their hosts with any reliability. However, the endornaviruses of domestic and wild rice are quite closely related, whereas those of broad bean and kidney bean are not, suggesting that transmission may have occurred with these viruses as well (Fukuhara & Moriyama 2008). Endornaviruses appear to be quite common in plants, but it is likely that they often go unnoticed because they can be easily mistaken for contaminating DNA in a standard dsRNA analysis (M. J. Roossinck 2006, unpublished data). Similar to partitiviruses, endornavirus persistence seems to be very stable. All cultivars of bell pepper contain pepper endornavirus that can be vertically transmitted to other cultivars of pepper through crosses (Valverde & Gutierrez 2007).
The RdRp of the endornaviruses falls into the pFAM00978 group (NCBI) that includes the alphaviruses of plants and animals (Gibbs et al. 2000), single-stranded plus-sense viruses. Hence, these viruses may have evolved from acute, infectious viruses, although an alternate hypothesis is that the acute alphaviruses evolved from an ancestral endornavirus, consistent with the idea that emerging acute viruses can evolve from persistent viruses. Elements related to known endornaviruses are also quite common in wild plants, although not as prevalent as partitiviruses (M. J. Roossinck 2009, unpublished data).
The titres of persistent viruses in plants are generally low, but can vary considerably. The level of endornavirus in rice cells is very low, estimated at about 100 copies per cell. In rice suspension cultures, the levels rise about 10-fold, but return to normal levels in regenerated plants, indicating some type of developmental control of virus levels (Moriyama et al. 1996). Interestingly, an indirect measure of white clover cryptic virus levels suggested that virus titres are reduced during nodulation in white clover (Nakatsukasa-Akune et al. 2005).
(a). Suspected/potential persistent viruses of plants
Recent ecogenomic studies of plant viruses in wild plants (Roossinck et al. 2010) have found other families of viruses that are also similar, i.e. they do not express any apparent symptoms, they do not encode obvious movement proteins and they seem not to be readily transmitted horizontally. These include viruses in the families of Totiviridae and Chrysoviridae. The totiviruses are found in fungi and protists. Several recent reports have also described totiviruses in plants (Covelli et al. 2004; Kozlakidis et al. 2006; Marais et al. 2009; Roossinck et al. 2010; NCBI Taxonomy ID: 463392 and 508679; M. J. Roossinck 2009, unpublished data). In some cases, these have been attributed to endophytic fungi. In cherry chlorotic rusty spot and Amasya cherry diseases no fungus could be cultured from the plants, but fungal mycelia were visible by microscopic examination (Alioto et al. 2003). Interestingly, these plants also harboured partitiviruses and chrysoviruses. Chrysovirus-like sequences are also found frequently in plant virus biodiversity studies (Roossinck et al. 2010; M. J. Roossinck 2009, unpublished data). While it remains to be finally determined whether or not these viruses are actually fungal or are replicating in the plant cells, the latter seems more plausible given the very small amount of fungal tissue that is found in plants harbouring endophytes.
(b). Persistent viruses and quasispecies
The quasispecies nature of RNA viruses is well known, and is related to the error prone nature of replication (Bull et al. 2005). Quasispecies refers to a high level of genetic variation in a single replicating virus population, and has numerous biological implications that have been thoroughly reviewed elsewhere (Domingo 2006). Whether or not persistent viruses maintain a quasispecies nature is still largely unknown. Very little work has been done on this topic, but in one study with mouse hepatitis virus there was no detectable variation in virus populations in persistent infections, while acute infections are known to have a quasispecies nature (Stühler et al. 1997). There have been no quasispecies studies to date on persistent viruses in plants. However, Curvularia thermal tolerance virus, a novel, persistent, bipartite fungal virus with a dsRNA genome (Márquez et al. 2007) did not show any sequence variation in 10-fold coverage of its genome (M. J. Roossinck 2004, unpublished observation). Low levels of variation would provide an additional hurdle for the evolution of persistent viruses to acute emerging viruses.
(c). Effects of persistent viruses on their hosts
Only a few reports have appeared in the literature concerning any effects of persistent plant viruses on their hosts. However, in a few cases some effects have been seen. For example, the presence of Vicia faba endornavirus is correlated with male sterility, although the mechanism is not known (Pfeiffer 1998). In beets, the presence of beet cryptic virus reduced the yields in some fields, but not others. The fields where yields were not affected were also subjected to drought (Xie et al. 1994). Can persistent viruses affect drought tolerance in plants, as acute viruses can (Xu et al. 2008)? In a transcriptome analysis of Rhizobium colonization in white clover, a transcript that was significantly decreased was later found to be the CP for white clover cryptic virus. When this gene was expressed in lotus, another legume species, it suppressed nodulation through the plant hormone abscisic acid (Nakatsukasa-Akune et al. 2005). It seems likely that similar examples exist, that plants have co-opted the genes of persistent viruses for their own purposes. Viral sequences may be common in the EST databases, and plants may use these mRNAs as epigenetic material for many functions. These would be most important during rapid changes in the plant's environment, when the slow process of Darwinian evolution could not allow for the required flexibility. Maintaining the persistent state of the virus would then be of prime importance to the plant, establishing a mutualistic symbiosis. This has been observed in a plant–fungus–virus interaction, where a persistent virus in an endophytic fungus is required for thermal tolerance of plants growing in geothermal soils (Márquez et al. 2007).
In most studies of persistent plant viruses no effect has been found on the host. However, it is not always possible to find an uninfected isogenic plant for comparison, so subtle effects are probably not detected.
Persistent viruses may affect their hosts in more subtle ways and certainly could be involved in the evolution of new acute viruses by reassortment/recombination. The evolution of many plant viruses has been modular (Roossinck 2005). It seems likely that persistent viruses that are so common in plants could be a source for the generation of novel viruses by recombining/reassorting with infecting acute viruses. A recent analysis of the plant virus genus Ourmiavirus suggests that its members contain elements from viruses of fungi and plants supporting the potential role of persistent viruses in the evolution of acute viruses (Rastgou et al. 2009).
4. Conclusions
The plant persistent viruses are a very common, but largely unstudied group of viruses. Most of the focus of plant virology has been centred on acute or chronic viruses. However, the persistent viruses have the potential for a profound effect on their plant hosts, as epigenetic elements providing novel genes, or as sources for newly emerging viruses. There is still a lot to learn: can acute viruses switch to a persistent lifestyle, or vice versa? Are persistent viruses of plants actually trapped fungal viruses? Are fungal viruses and plant persistent viruses transmitted across kingdoms? For the most part, this field is at the discovery phase. With more and more metagenomic and ecogenomic data becoming available, some of these questions may be answered, and more will certainly be posed. Placing plant viruses into categories based on lifestyle is useful in thinking about their potential roles as mutualists rather than agents of disease. However, as in all of biology, strict adherence to definitions is not useful, and it is likely that viruses exist on a continuum from persistent to acute lifestyles. Virus biodiversity studies will undoubtedly reveal viruses in all parts of the continuum.
Acknowledgements
This work was supported by the Samuel Roberts Noble Foundation; the National Science Foundation (grant number EF-0627108 and EPS-0447262); and the United States Department of Agriculture (grant no. OKLR-2007-01012). The author thanks the anonymous reviewers who provided numerous helpful suggestions, and the conference organizers Santiago Elena and Remy Froissart.
Footnotes
One contribution of 14 to a Theme Issue ‘New experimental and theoretical approaches towards the understanding of the emergence of viral infections’.
References
- Alioto D., Zaccaria F., Covelli L., Serio F. D., Ragozzino A., Milne R. G.2003Light and electron microscope observations on chlorotic rusty spot, a disorder of cherry in Italy. J. Plant Pathol. 85, 215–218 [Google Scholar]
- Arancibia R. A., Valverde R. A., Can F.1995Properties of a cryptic virus from pepper (Capsicum annuum). Plant Pathol. 44, 164–168 [Google Scholar]
- Beijerinck M. W.1898Concerning a contagium vivum fluidum as cause of the spot disease of tobacco leaves. In Phylopathological classics, No. 7 (ed. Johnson J.). St Paul, MN: American Phytopathological Society [Google Scholar]
- Blanc S.2007Virus transmission–getting in and out. In Viral transport in plants (eds Waigman E., Heinlein M.), pp. 1–28 Berlin Heidelberg, Germany: Springer [Google Scholar]
- Boccardo G., Lisa V., Luisoni E., Milne R. G.1987Cryptic plant viruses. Adv. Virus Res. 33, 171–214 (doi:10.1016/S0065-3527(08)60477-7) [DOI] [PubMed] [Google Scholar]
- Brown S. P., Lechat L., Depaepe M., Taddei F.2006Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr. Biol. 16, 2048–2052 (doi:10.1016/j.cub.2006.08.089) [DOI] [PubMed] [Google Scholar]
- Bruenn J. A.1993A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res. 21, 5667–5669 (doi:10.1093/nar/21.24.5667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J. J., Ancel M. L., Lachmann M.2005Quasispecies made simple. PLoS Biol. 1, 0450–0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covelli L., Coutts R. H. A., Serio F. D., Citir A., Acikoz S., Hernadez C., Ragozzino A., Flores R.2004Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral-like double-stranded RNAs. I. Characterization of a new species in the genus Chrysovirus. J. Gen. Virol. 85, 3389–3397 (doi:10.1099/vir.0.80181-0) [DOI] [PubMed] [Google Scholar]
- Domingo E. (ed.)2006Quasispecies: concepts and implications for virology Berlin, Germany: Springer [Google Scholar]
- Edwardson J. R., Christie R. G. Cucumoviruses. CRC handbook of viruses infecting legumes. 1991. Boca Raton, FL: CRC Press. [Google Scholar]
- Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (eds) 2005Virus taxonomy Eighth report of the International Committee on taxonomy of viruses San Diego, CA: Elsevier Academic Press [Google Scholar]
- Fraser R. S. S.1998Introduction to classical cross protection. In Plant virology protocols (eds Foster G. D., Taylor S. C.), pp. 13–24 Totowa, NJ: Humana Press [Google Scholar]
- Fukuhara T., Moriyama H.2008Endornaviruses. In Encyclopedia of virology (eds Mahy B. W. J., Regenmortel M. H. V. V.). Amsterdam, The Netherlands: Elsevier [Google Scholar]
- Fukuhara T., et al. 2006The wide distribution of endornaviruses, large double-stranded RNA replicons with plasmid-like properties. Arch. Virol. 151, 995–1002 (doi:10.1007/s00705-005-0688-5) [DOI] [PubMed] [Google Scholar]
- Gibbs M. J., Koga R., Moriyama H., Pfeiffer P., Fukuhara T.2000Phylogenetic analysis of some large double-stranded RNA replicons from plants suggests they evolved from a defective single-stranded RNA virus. J. Gen. Virol. 81, 227–233 [DOI] [PubMed] [Google Scholar]
- Gray S. M., Banerjee N.1999Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63, 128–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn T., Richert-Pöggeler K. R., Staginnus C., Harper G., Schwarzacher T., Teo C. H., Teycheney P.-Y., Iskra-Caruana M.-L., Hull R.2008Evolution of integrated plant viruses. In Plant virus evolution (ed. Roossinck M. J.), pp. 53–81 Heidelberg, Germany: Springer [Google Scholar]
- Ishihara J., Pak J. Y., Fukuhara T., Nitta T.1992Association of particles that contain double-stranded RNAs with algal chloroplasts and mitochondria. Planta 187, 475–482 (doi:10.1007/BF00199965) [DOI] [PubMed] [Google Scholar]
- Koga R., Horiuchi H., Fukuhara T.2003Double-stranded RNA replicons associated with chloroplasts of a green alga, Bryopsis cinicola. Plant Mol. Biol. 51, 991–999 (doi:10.1023/A:1023003412859) [DOI] [PubMed] [Google Scholar]
- Kozlakidis Z., Covelli L., Diserio F., Citir A., Acikgöz S., Hernández C., Ragozzino A., Flores R., Coutts R. H. A.2006Molecular characterization of the largest mycoviral-like double-stranded RNAs associated with Amasya cherry disease, a disease of presumed fungal aetiology. J. Gen. Virol. 87, 3113–3117 (doi:10.1099/vir.0.82121-0) [DOI] [PubMed] [Google Scholar]
- Kozlakidis Z., Brown N. A., Jamal A., Phoon X., Coutts R. H. A.2010Incidence of endornaviruses in Phytophthora taxon douglasfir and Phytophthora ramorum. Virus Genes 40, 130–134 (doi:10.1007/s11262-009-0421-7) [DOI] [PubMed] [Google Scholar]
- Lefebvre A., Scalla R., Pfeiffer P.1990The double-stranded RNA associated with the ‘447’ cytoplasmic male sterility in Vicia faba is packaged together with its replicase in cytoplasmic membranous vesicles. Plant Mol. Biol. 14, 477–490 (doi:10.1007/BF00027494) [DOI] [PubMed] [Google Scholar]
- Li L., Tiam Q., Du Z., Duns G. J., Chen J.2009A novel double stranded RNA virus detected in Primula malacoides is a plant-isolated partitivirus closely related to partivirus infecting fungal species. Arch. Virol. 154, 565–572 (doi:10.1007/s00705-009-0342-8) [DOI] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R.2009. Mesquite: a modular system for evolutionary analysis, 2.72 ed. See http://mesquiteproject.org
- Marais A., Faure C., Couture C., Svanella L., Hullé M., Leromancer M., Candresse T.2009Characterisation of plant virus populations by a metagenomic approach: survey in French sub-Antarctic islands. In Understanding emergence of infectious diseases: focus on new experimental and theoretical approaches to virus evolution Roscoff, France: Conférences Jacques-Monod [Google Scholar]
- Márquez L. M., Redman R. S., Rodriguez R. J., Roossinck M. J.2007A virus in a fungus in a plant—three way symbiosis required for thermal tolerance. Science 315, 513–515 (doi:10.1126/science.1136237) [DOI] [PubMed] [Google Scholar]
- Moriyama H., Kanaya K., Wang J. Z., Nitta T., Fukuhara T.1996Stringently and developmentally regulated levels of a cytoplasmic double-stranded RNA and its high-efficiency transmission via egg and pollen in rice. Plant Mol. Biol. 31, 713–719 (doi:10.1007/BF00019459) [DOI] [PubMed] [Google Scholar]
- Nakatsukasa-Akune M., et al. 2005Suppression of root nodule formation by artificial expression of the TrEnodDR1 (coat protein of White clover cryptic virus 2) gene in Lotus japonicus. Mol. Plant Microbe Interact. 18, 1069–1080 (doi:10.1094/MPMI-18-1069) [DOI] [PubMed] [Google Scholar]
- Nibert M. L., Woods K. M., Upton S. J., Ghabrial S. A.2009Cryspovirus: a new genus of protozoan viruses in the family Partitiviridae. Arch. Virol. 154, 1959–1965 (doi:10.1007/s00705-009-0513-7) [DOI] [PubMed] [Google Scholar]
- Pfeiffer P.1998Nucleotide sequence, genetic organization and expression strategy of the double-stranded RNA associated with the ‘447 cytoplasmic male sterility trait in Vicia faba. Virology 79, 2349–2358 [DOI] [PubMed] [Google Scholar]
- Rastgou M., Habibi M. K., Izdpanah K., Masenga V., Milne R. G., Wold Y. I., Koonin E. V., Turina M.2009Molecular characterization of the plant virus genus Ourmiavirus and evidence on inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 90, 2525–2535 (doi:10.1099/vir.0.013086-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P.2003MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- Roossinck M. J.1997Mechanisms of plant virus evolution. Ann. Rev. Phytopathol. 35, 191–209 (doi:10.1146/annurev.phyto.35.1.191) [DOI] [PubMed] [Google Scholar]
- Roossinck M. J.2005Symbiosis versus competition in the evolution of plant RNA viruses. Nat. Rev. Microbiol. 3, 917–924 (doi:10.1038/nrmicro1285) [DOI] [PubMed] [Google Scholar]
- Roossinck M. J., Saha P., Wiley G. B., Quan J., White J. D., Lai H., Chavarría F., Shen G., Roe B. A.2010Ecogenomics: using massively parallel pyrosequencing to understand virus ecology. Mol. Ecol. 19, 81–88 (doi:10.1111/j.1365-294X.2009.04470.x) [DOI] [PubMed] [Google Scholar]
- Sabanadzovic S., Ghanem-Sabanadzovic N. A.2008Molecular characterization and detection of a tripartite cryptic virus from rose. J. Plant Pathol. 90, 287–293 [Google Scholar]
- Sabanadzovic S., Valverde R., Brown J. K., Martin R. R., Tzanetakis I. E.2009Southern tomato virus: the link between the families Totiviridae and Partitiviridae. Virus Res. 140, 130–137 (doi:10.1016/j.virusres.2008.11.018) [DOI] [PubMed] [Google Scholar]
- Stühler A., Flory E., Wege H., Lassmann H., Wege H.1997No evidence for quasispecies populations during persistence of the coronavirus mouse hepatitis virus JHM: sequence conservation within the surface glycoprotein gene S in Lewis rats. J. Gen. Virol. 78, 747–756 [DOI] [PubMed] [Google Scholar]
- Szegö A., Tóth E. K., Potyondi L., Lukás N.2005Detection of high molecular weight dsRNA persisting in Dianthus species. Acta Biol. Szegediensis 49, 17–19 [Google Scholar]
- Thomas F., Poulin R., Guégan J.-F., Michalakis Y., Renaud F.2000Are there pros as well as cons to being parasitized? Parasitol. Today 16, 533–536 (doi:10.1016/S0169-4758(00)01790-7) [DOI] [PubMed] [Google Scholar]
- Timian R. G.1974The range of symbiosis of barley and barley stripe mosaic virus. Phytopathology 64, 342–345 [Google Scholar]
- Valverde R. A., Gutierrez D. L.2007Transmission of a dsRNA in bell pepper and evidence that it consists of the genome of an endornavirus. Virus Genes 35, 399–403 (doi:10.1007/s11262-007-0092-1) [DOI] [PubMed] [Google Scholar]
- Veliceasa D., Enünlü N., Kós P. B., Köster S., Beuther E., Morgun B., Deshmukh S. D., Lukács N.2006Searching for a new putative cryptic virus in Pinus sylvestris L. Virus Genes 32, 177–186 (doi:10.1007/s11262-005-6874-4) [DOI] [PubMed] [Google Scholar]
- Villarreal L. P.2007Virus-host symbiosis mediated by persistence. Symbiosis 44, 1–9 [Google Scholar]
- Villarreal L. P.2009Persistence pays: how viruses promote host group survival. Curr. Opin. Microbiol. 12, 467–472 (doi:10.1016/j.mib.2009.06.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal L. P., Defillippis V. R., Gottlieb K. A.2000Acute and persistent viral life strategies and their relationship to emerging diseases. Virology 272, 1–6 (doi:10.1006/viro.2000.0381) [DOI] [PubMed] [Google Scholar]
- Xie W. S., Antoniw J. F., White R. F., Jolliffe T. H.1994Effects of beet cryptic virus infection on sugar beet in field trials. Ann. Appl. Biol. 124, 451–459 (doi:10.1111/j.1744-7348.1994.tb04150.x) [Google Scholar]
- Xu P., Chen F., Mannas J. P., Feldman T., Sumner L. W., Roossinck M. J.2008Virus infection improves drought tolerance. New Phytol. 180, 911–921 (doi:10.1111/j.1469-8137.2008.02627.x) [DOI] [PubMed] [Google Scholar]