Abstract
Replication of HIV-1 under selective pressure frequently results in the evolution of virus variants that replicate more efficiently under the applied conditions. For example, in patients on antiretroviral therapy, such evolution can result in variants that are resistant to the HIV-1 inhibitors, thus frustrating the therapy. On the other hand, virus evolution can help us to understand the molecular mechanisms that underlie HIV-1 replication. For example, evolution of a defective virus mutant can result in variants that overcome the introduced defect by restoration of the original sequence or by the introduction of additional mutations in the viral genome. Analysis of the evolution pathway can reveal the requirements of the element under study and help to understand its function. Analysis of the escape routes may generate new insight in the viral life cycle and result in the identification of unexpected biological mechanisms. We have developed in vitro HIV-1 evolution into a systematic research tool that allows the study of different aspects of the viral replication cycle. We will briefly review this method of forced virus evolution and provide several examples that illustrate the power of this approach.
Keywords: HIV-1 evolution, antiviral therapy, drug-resistance, molecular mechanism, virus replication, RNA structure
1. Introduction to hiv-1 experimental evolution
There is abundant evidence for HIV-1 evolution in diverse settings. In fact, HIV-1 evolution follows the principles of Darwinian evolution, which is a two-step process. The first step consists of the generation of genetic variants. In the second step, this material is exposed to phenotypic selection. The error-prone reverse-transcription process is the major cause of the rapid generation of HIV-1 variants, resulting in a mixed viral population of related genomes, also termed quasispecies (Munoz et al. 1993; Domingo & Wain-Hobson 2009). When allowed to replicate, the HIV-1 variant with the best fitness will outcompete the other variants. It is very important to note that the mutation step of evolution is completely independent of the selection step. Even though the virus population is eventually shaped by phenotypic selection criteria, the nucleotide substitution rates will largely determine the set of mutants available within the viral quasispecies for this selection process. As we will present different virus evolution strategies, the importance of both steps will become apparent.
We prefer to start evolution experiments with a plasmid carrying a complete and infectious DNA copy of the HIV-1 genome. The use of such an infectious molecular clone as input material allows one to unequivocally score newly acquired mutations, which provides an important benefit over the use of a virus stock, which is likely to harbor diverse viral variants. The molecular clone also allows mutation of sequence or structured elements in the viral genome to study their function in replication. We usually start an evolution experiment by transfection of the molecular clone into T cells. Whereas the original virus (wild-type; wt) will replicate rapidly, replication of the mutant virus may be reduced or undetectable, in which case we maintain the cells in culture by splitting when needed. A low level of ongoing virus replication in combination with the error-prone viral replication machinery will drive evolution, which may result in the emergence of variants with improved replication potential and a concomitant increase in the viral load. Once signs of virus replication are observed, the virus is passaged onto fresh cells. Initially, a large inoculum is used that in addition to the virus also contains virus-producing cells. In most experiments, the replication capacity of the virus gradually increases and the size of the inoculum can be decreased in subsequent passages, till fast-replicating variants have evolved that can be passaged cell-free. At each passage, infected cells are isolated and stored for sequence analysis of the integrated proviral genome. The mere selection of a specific mutation does not necessarily mean that it is responsible for the restored replication capacity. It is necessary to experimentally demonstrate that the acquired mutation caused the increased replication capacity. To do so, the evolved sequence is re-cloned into the viral genome and the replication potential of this variant is compared with that of the original mutant virus. A similar protocol can be used to select virus variants that are resistant to specific virus inhibitors (e.g. reverse transcriptase or protease inhibitors) or that have adapted to specific conditions (e.g. replication in a specific cell type).
2. The hiv-1 mutation frequency as driver of evolution
The mutation frequency of HIV-1 varies considerably for different kinds of mutations. For example, the G-to-A mutation is observed most frequently and is considered to be relatively easy to generate. In general, transversions (purine to pyrimidine or pyrimidine to purine mutations) are seen less frequently than transitions (purine to purine or pyrimidine to pyrimidine mutations; Keulen et al. 1996). Moreover, double mutations are more difficult than single mutations and therefore much less frequently observed (Berkhout et al. 2001; Berkhout & de Ronde 2004). Given the considerable HIV-1 population size, combined with the rapid replication kinetics and the high error rate during viral replication, it can be calculated that all variant genotypes with single nucleotide substitutions will be available in an infected individual (Coffin 1995), but it is less likely that complex substitution patterns are available in the initial virus pool.
The virus populations that are handled in in vitro experiments will usually be much smaller, which will limit the available genotypic variation. The use of a molecular HIV-1 clone as starting material will reduce the initial genetic variation to almost zero. Since mutation is a chance process, one should not put too much value on the mutational pattern in an individual culture. One should ideally analyse virus variants that emerge in multiple independent cultures, which may provide different answers to the same problem. In most cases, multiple evolution routes are observed that provide a glimpse of the sequence space available to support virus replication. However, the repeated selection of the same or similar escape viruses is also very informative, as it indicates that the problem can only be solved by that frequently observed mutation.
We elaborated on the concept of easy and difficult mutations in the field of antiviral drug resistance. If relatively difficult drug-resistance mutations are consistently observed at certain positions in the viral genome, we hypothesized that it is likely that easier nucleotide substitutions at that codon did not pass the fitness selection criteria (Keulen et al. 1996). A notable example is provided by changes observed at codon 215 in the reverse transcriptase (RT) gene in HIV-1-infected individuals treated with the nucleotide inhibitor AZT (zidovudine). Variants were selected with either a Thr215Phe or a Thr215Tyr substitution. Both mutations require two nucleotide changes (ACC → TTC and ACC → TAC, respectively). There are five alternative amino acid substitutions (Ile, Ala, Ser, Pro, Asn) that are in fact easier to generate than Thr215Phe, and 10 substitutions (Ile, Ala, Ser, Pro, Asn, Val, Met, Leu, Asp, Gly) that are easier to generate than Thr215Tyr. Yet, none of these alternative substitutions have been detected in vivo and in vitro, arguing that they do not provide AZT-resistance or that they are incompatible with RT enzyme function and virus replication. These combined results suggest that only aromatic residues at position 215 can provide the AZT-resistance phenotype, which was confirmed by a mutational analysis (Lacey & Larder 1994).
3. Clonal evolution highlights the mutational step
Virus replication will result in variants that form the quasispecies. Subsequent competition will result in the outgrowth of the fittest variant. Thus, a single evolved variant will be selected over time and other less optimal variants will be missed. We described a limiting dilution evolution approach that prevents competition between different emerging replication-competent virus variants, which allows the outgrowth of sub-optimal virus variants. We applied this protocol for the selection of previously unidentified HIV-1 variants that are resistant to the nucleoside RT inhibitor 3TC (lamivudine) (Keulen et al. 1997). In this approach, infected cells were cultured in the presence of 3TC. Instead of maintaining a large cell-virus culture, the cells were serially diluted shortly after infection. The large-culture studies and 3TC-therapy of HIV-1-infected persons had yielded only two types of 3TC-resistant variants, with either a Met184Ile (ATG to ATA) or Met184Val (ATG to GTG) mutation in the catalytic domain of RT. The limiting dilution protocol did not only yield these Met184Ile and Met184Val variants (19 and 4 out of 32 clones, respectively), but also the novel Met184Thr variant (ATG to ACG; 9 out of 32 clones). Indeed, this 184Thr variant does replicate poorly and will easily be lost owing to competition in a larger population setting.
When the forces of competition are nullified, the likelihood of generating a particular mutation determines how the virus evolves. The results of the 3TC-selection experiment are fully consistent with this idea, as simple transitions are observed more frequently than more difficult transversions, and no double mutations were scored. One will only see the less likely events when a large number of independent evolution cultures are analysed. Among the 19 Met184Ile variants, we observed 18 times the expected Ile variant owing to a transition (ATG to ATA), but the alternative Ile codon owing to a single transversion (ATG to ATT) was scored only once. In larger viral populations, many possible variants may be present or within mutational reach, but selection will shape the quasispecies by the outgrowth of a limited number of top-fit virus variants.
In real life, virus evolution can depend on a complex interplay of the mutation and selection steps. For instance, 3TC-treated patients may initially show the 184Ile variant, which is made by the most frequently occurring G-to-A transition (Berkhout et al. 2001). This variant is subsequently outcompeted by the 184Val variant, which is more difficult to make via an A-to-G transition, but providing a subtly improved RT activity and viral fitness (Back et al. 1996). Thus, both mutation (Ile) and selection (Val) forces are visible in the evolution of 3TC-resistance.
4. Frustrating antiviral rnai approaches
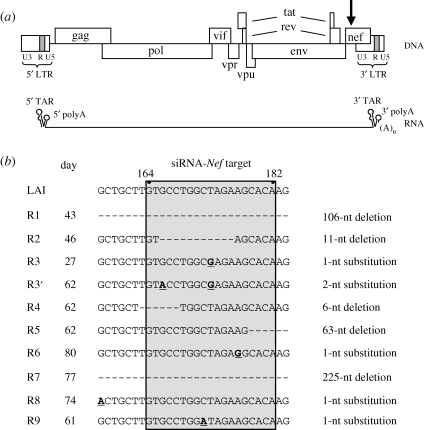
RNA interference (RNAi) is an evolutionarily conserved process that provides the cell with an additional tool to regulate gene expression and to control infecting viruses (Berkhout & Haasnoot 2006; Obbard et al. 2009). RNAi may also be a powerful method for intracellular immunization against HIV-1 (Haasnoot et al. 2007). We and others demonstrated long-term inhibition of virus replication in human T cells that stably express small-interfering RNAs (siRNAs) directed against the HIV-1 genome (Banerjea et al. 2003; Boden et al. 2003; Das et al. 2004a; Lee et al. 2005; ter Brake et al. 2006, 2008a; von Eije et al. 2008, 2009; von Eije & Berkhout 2009). However, viral escape variants can emerge that are no longer inhibited (Boden et al. 2003; Das et al. 2004a). For example, in a study in which the HIV-1 Nef gene was targeted, the virus escaped from the imposed RNAi pressure in multiple independent cultures (Das et al. 2004a). The escape routes demonstrate the exquisite sequence-specificity of RNAi, as all escape variants—except for the R8 variant—carried a mutation within the 19-nucleotide target sequence (figure 1). We observed point mutations at different positions, but also deletions that partially or completely delete the target sequence, thus highlighting the chance process of the mutational event that triggers escape. Further analysis of the exceptional R8 mutation demonstrated that this escape route affects the local RNA structure, such that the target sequence is masked and not accessible for the RNAi machinery (Westerhout et al. 2005). This result underscores why we usually perform so many evolution experiments in parallel. As evolution is a chance process, one may need to probe multiple evolution tracks before the more exotic escape paths will be disclosed. Other large-scale escape studies indicated that the sequence variation induced by RNAi pressure mimics the natural variation in HIV-1 isolates, arguing that viral fitness is a common and decisive determinant in both scenarios (ter Brake et al. 2008b; von Eije et al. 2008).
Figure 1.
HIV-1 escape variants that resist siRNA-Nef inhibition. (a) Schematic of the HIV-1 proviral genome with the LTR region subdivided into the U3, R and U5 domains. Transcription starts at the first nucleotide of the 5′ R region and the RNA transcripts are polyadenylated at the last nucleotide of the 3′ R. Both the 5′ and the 3′ end of the RNA molecule can fold a TAR and polyA hairpin. The position of the siRNA-Nef target sequence is indicated with an arrow. (b) HIV-1 variants resistant to siRNA-Nef were selected in nine independent cultures. The Nef target sequence (nucleotides 164–182 of the Nef gene) and flanking sequences are shown for the wt (HIV-1 LAI molecular clone) and the evolved RNAi-resistant viruses (R1–R9). The day at which the escape variants were sequenced is indicated. Deletions are shown as dashes, substitutions are underlined and in bold. In the R1 virus, nucleotides 125–230 of the Nef gene are deleted. In the R5 virus, nucleotides 179–241 are deleted. In the R7 virus, we observed deletion of nucleotides 44–268 and a T269A substitution. Adapted from Westerhout et al. (2005). © copyright 2005 Westerhout et al.
5. Evolution of hiv-1 vaccines
Virus evolution experiments can play an important role in the safety testing of candidate vaccines. For instance, whereas AIDS vaccines based on a live-attenuated virus have shown promise in the SIV-macaque model, virus evolution studies revealed that such vaccines will likely be unsafe (Johnson & Desrosiers 1998; Mills et al. 2000; Whitney & Ruprecht 2004; Koff et al. 2006). The major safety concern is that a chronic infection is established and low-level replication of the attenuated virus may eventually lead to the selection of fitter and pathogenic virus variants (Baba et al. 1995, 1999; Whatmore et al. 1995; Chakrabarti et al. 2003; Hofmann-Lehmann et al. 2003). The Δ3 HIV-1 variant, which is attenuated by deletions in three non-essential genome segments, was considered a vaccine candidate because it seemed impossible that the virus could evolve to a pathogenic variant through repair of the deletions. However, we observed that this virus can regain replication fitness in prolonged cell culture infections (Berkhout et al. 1999). As expected, the deletions were not repaired. However, an intriguing sequence duplication was selected in the LTR promoter, which doubled the number of Sp1 binding sites from three to six and greatly improved virus replication. Multiplication or deletion of repeat sequence motifs is more frequently observed and seems to be a popular virus evolution strategy (Berkhout et al. 1999; Marzio et al. 2001; Leonard et al. 2008; Berkhout 2009).
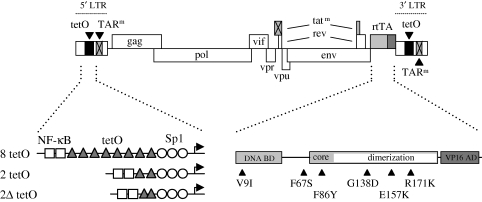
Evolution experiments may also help in the development of more safe HIV-1 vaccine candidates. For example, we successfully used virus evolution to improve a doxycycline-dependent HIV-1 variant, which was developed as a novel approach to increase the safety of a live-attenuated vaccine strain (Verhoef et al. 2001). In this conditionally live virus, named HIV-rtTA, the Tat–TAR transcription activation mechanism was functionally replaced by the Tet-ON system for inducible gene expression (Baron & Bujard 2000; Gossen & Bujard 2001). Both Tat and TAR were inactivated through mutations at essential positions in the protein and RNA hairpin, respectively (figure 2). The Tet-ON system was integrated by insertion of the gene encoding the doxycycline-inducible rtTA trans-activator protein at the site of the nef gene and insertion of eight rtTA-binding tetO sites in the LTR promoter region. The original HIV-rtTA construct replicated poorly, but stably maintained the introduced components of the Tet-ON system and the mutations in Tat and TAR. Long-term culturing of HIV-rtTA resulted in the selection of greatly improved variants. Analysis of these evolved viruses revealed that the components of the Tet-ON system had been optimized (figure 2). The number of tetO elements was reduced to two and the spacing between the remaining motifs was subsequently reduced from 41 to 26 or 27 nucleotides (Marzio et al. 2001). This new spacing remarkably resembles the 29-nt spacing of the tetO elements in the E. coli Tn10 operon from which the Tet-ON components are derived. Further analysis showed that the new tetO configuration improved viral replication by fine-tuning of the LTR promoter activity (Marzio et al. 2002). Upon long-term culturing of HIV-rtTA, mutations were also observed in the rtTA protein that significantly improve its activity and doxycycline-sensitivity (Das et al. 2004b; Zhou et al. 2006b, 2007). We demonstrated that these evolved rtTA variants not only improve replication of the conditionally live HIV-1 variant but also the performance of the Tet-ON system, which is widely used to regulate transgene expression. These new rtTA variants will be very useful in biological applications that require a more sensitive or active Tet-ON system. These studies demonstrate that the viral evolution strategy can be used to improve non-viral genes by making them an integral and essential part of the viral replication machinery.
Figure 2.
In vitro evolution of the doxycycline-dependent HIV-rtTA virus. In the HIV-rtTA virus, the Tat–TAR axis of transcription regulation has been inactivated by mutation of both Tat and TAR (tatm and TARm, crossed boxes). Transcription and replication of the virus were made doxycycline-dependent by introduction of eight tetO elements in the LTR promoter region and replacing the Nef gene by the rtTA gene. This 248-amino acid protein is a fusion of the E. coli Tet repressor (TetR, which can be subdivided into a DNA-binding domain (BD) and a regulatory core domain with a dimerization surface) and the VP16 activation domain (AD) of herpes simplex virus. Administration of the doxycycline effector induces a conformational switch in the rtTA protein, which subsequently can bind to the tetO-LTR promoter region and activate transcription of the proviral genome. Thus, transcription and replication of HIV-rtTA are critically dependent on doxycycline. Evolution of HIV-rtTA resulted in modifications in the tetO promoter region (lower left panel; the original eight tetO and evolved two tetO and 2Δ tetO configurations are shown, see text for details) and in the rtTA gene (lower right panel). The black triangles indicate amino acid substitutions in rtTA that were observed upon evolution of HIV-rtTA in multiple, independent long-term cultures, and that were found to improve the transcriptional activity and doxycycline sensitivity of rtTA.
In some extended cultures, we observed mutations at specific positions in rtTA that increased the background activity in the absence of doxycycline. These rtTA changes, which were always owing to one-nucleotide transitions, allowed virus replication in the absence of doxycycline. We therefore designed novel rtTA variants that require a more difficult nucleotide transversion or multiple simultaneous nucleotide substitutions to be converted to a doxycycline-independent variant (Zhou et al. 2006a,c). This higher genetic barrier did indeed block the undesired evolutionary routes of HIV-rtTA, which again illustrates the importance of the mutation step in viral evolution.
6. Virus evolution to study structured rna signals
We have successfully used evolutionary techniques to study important structured RNA motifs in the HIV-1 genome. Our initial studies addressed the TAR hairpin structure that is present at the 5′ and 3′ end of the viral RNA (figure 1a). The upper part of the 5′ TAR structure binds the viral transcriptional activator Tat protein and cellular co-factors, but no clear function for the lower stem region had been established. We reported that base substitutions in the lower stem region severely inhibited HIV-1 replication (Klaver & Berkhout 1994). Upon long-term culturing, better replicating viruses could be isolated with a great variety of genetic changes (point mutations, short deletions) that restored base-pairing of the lower stem region, which demonstrated that the stability of the TAR structure is essential for optimal HIV-1 replication.
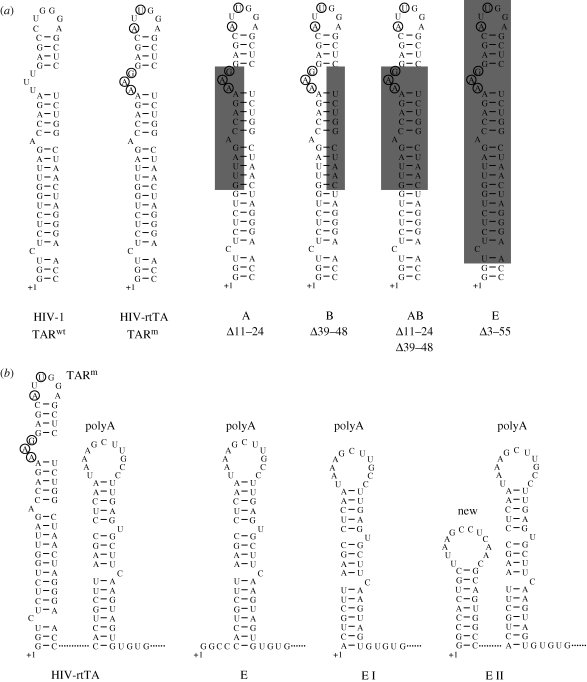
Several TAR mutagenesis studies suggested that the TAR hairpin has other functions in HIV-1 replication in addition to its role in transcription, such as translation, dimerization, packaging and reverse-transcription of the viral RNA (Das et al. 2007 and references therein). Most of these studies were complicated by the fact that mutations in TAR have a dominant negative effect on viral transcription, which obscures other effects in the viral life cycle. We therefore recently reinvestigated this issue in mutation–evolution studies using the HIV-rtTA variant that does not need TAR for the activation of transcription (Das et al. 2007). Surprisingly, deletions in either the left or right side of the TAR stem dramatically impaired HIV-rtTA replication (A and B mutants in figure 3a), while a double mutant that combines these deletions replicated efficiently (AB mutant). Evolution of the single-side mutants resulted in nucleotide substitutions in TAR that restored the stability of the stem and thereby improved viral replication. The nearly complete deletion of the TAR sequence also reduced HIV-rtTA replication (mutant E in figure 3a,b). In long-term cultures of this mutant, we observed two different evolution routes in which the remaining TAR nucleotides were either removed (variant E I in figure 3b) or replaced by an unrelated stable hairpin (variant E II). Both evolved TAR-deleted variants replicate efficiently, which demonstrates that TAR has no essential second function in HIV-1 replication, at least in vitro.
Figure 3.
HIV-1 requires a stable hairpin structure at the 5′ end of its RNA genome. (a) The HIV-1 molecular clone LAI contains a 57-nt wild-type TAR hairpin (TARwt). In HIV-rtTA, which is based on LAI, TAR was inactivated by nucleotide substitutions in both the bulge and loop motifs (encircled in TARm). The TARm sequence was partially or nearly completely deleted (mutants A–E). The deleted nucleotides are boxed in grey. (b) Secondary structure of the 5′ end of the viral RNA of the HIV-rtTA variant, the E mutant and the evolved E I and E II variants.
Subsequent analyses revealed that the single-side deletions that opened TAR caused an unforeseen interaction between the remaining TAR nucleotides and nucleotides downstream of the polyA hairpin (Vrolijk et al. 2008). This interaction resulted in an extension of the polyA hairpin that—like TAR—is present at both the 5′ and 3′ end of the viral RNA (figure 1a). This interaction altered the conformation of the untranslated leader at the 5′ end of the viral RNA, which affected the processes of RNA dimerization and packaging (Vrolijk et al. 2008 and unpublished results). Moreover, stabilization of the polyA hairpin at the 3′ end of the viral RNA affected the process of polyadenylation, which further inhibited viral replication (Vrolijk et al. 2009). These studies demonstrate that mutational analysis of TAR is risky because it can induce unwanted side effects that affect non-related replication steps. These effects can easily be misinterpreted as providing evidence for secondary TAR functions.
We used similar virus evolution experiments to study other structured RNA signals in HIV-1 (e.g. polyA and SD hairpin; Das et al. 1997, 1999a; Abbink & Berkhout 2007), which eventually led to the description of an alternative HIV-1 RNA leader structure in which the polyA hairpin is opened to allow base-pairing with the DIS domain (Huthoff & Berkhout 2001; Abbink & Berkhout 2003; Ooms et al. 2004). The equilibrium between the two leader conformations may function as a riboswitch that is important in controlling RNA dimerization and packaging. Virus evolution approaches have also successfully been used by others to study RNA signals in the RNA bacteriophage MS2 (Olsthoorn & van Duin 1996; Klovins et al. 1997; Licis et al. 2000).
7. Helping the mutation step by sequence randomization
The Systematic Evolution of Ligands by EXponential enrichment (SELEX) technique is a powerful method for the selection of molecules with unique properties (Breaker 2004). Whereas Darwinian evolution usually applies to living organisms over long periods of time, SELEX allows for the rapid in vitro evolution of functionally active nucleic acids from a pool of randomized sequences. We described an in vivo version of this nucleic acid evolution protocol in which selection and amplification take place inside living cells by means of HIV-1 replication (Berkhout & Klaver 1993). In brief, we generated a library of HIV-1 DNA genomes with random sequences in a particular genetic domain. This mixture of HIV-1 genomes was transfected in human T cells and the outgrowth of the fittest viruses was observed within two weeks of viral replication. Compared with the laborious in vitro selection and amplification steps, much time and effort is saved in this in vivo approach. Moreover, adaptive changes may arise outside the randomized genome segment during virus replication due to the error-prone viral replication machinery. We used this approach to probe the sequence requirements for the HIV-1 TAR hairpin structure (Berkhout & Klaver 1993). We randomized the 3-nt bulge region to which the viral Tat transcriptional activator protein binds. Replication of this mixture of viruses resulted in the rapid selection of the wild-type bulge sequence UCU, which is also present in 80 per cent of the natural HIV-1 isolates. The method has also been successfully applied to study the replication of other retroviruses (Doria-Rose & Vogt 1998; Morris et al. 2002), hepatitis B virus (Rieger & Nassal 1995), plant viruses (Sun et al. 2005; Zhang & Simon 2005) and herpes simplex virus type 1 (Yoon & Spear 2004). Although many RNA viruses exist as a quasispecies of closely related but genetically distinct genotypes, their evolutionary potential is restricted because they probe only into a limited area of sequence space around the quasispecies. In vivo SELEX provides the opportunity to sample a larger part of the sequence space by randomization of multiple nucleotides, yielding valuable information and new molecules with interesting properties.
8. Second-site mutations
In most mutation–evolution studies, HIV-1 will restore the function of the mutated element through sequence changes in this domain. The mutated virus can also acquire additional mutations outside the mutated region that improve viral replication. For example, in evolution studies with a Tat-inactivated HIV-1 variant, the virus acquired an additional mutation in another domain of the viral Tat protein. This second-site mutation partially restored Tat activity and viral replication, although a structural explanation is currently lacking (Verhoef & Berkhout 1999). Second-site mutations can also occur in other genome segments to impose a general replication improvement that is not related to the initial defect. For instance, we selected an up-mutation in the Envelope protein in studies with an HIV-1 variant that was translationally crippled through modification of the 5′ untranslated leader region (Das et al. 1998). Although it is attractive to think about a functional coupling of these two issues, further analysis revealed that the second-site mutation presented a general improvement of viral replication fitness that is not directly linked to the leader modifications (Das et al. 1999b). Indeed, this same Envelope mutation appeared later in a completely unrelated evolution study (Baldwin & Berkhout 2006). Some recent examples indicate that the misinterpretation of such a second-site mutation as a direct effect can become a major problem in the interpretation of HIV-1 evolution studies (Hache et al. 2008, 2009; Leonard et al. 2008; Berkhout 2009).
9. Discussion
Several basic and applied research lines on HIV-1 were presented in which we used diverse virus evolution methods. These studies ranged from the selection of drug-resistant HIV-1 variants by virus evolution under selective pressure to the study of critical RNA hairpin motifs in the HIV-1 genome by means of forced evolution of virus mutants. These examples underscore the enormous potential of incorporating evolution methods in the design of virus experiments. It may therefore be surprising that only a few laboratories have used the virus evolution method in a systematic manner. One should obviously keep in mind the limitations of such in vitro HIV-1 studies, which will—among other things—miss the major selection pressure imposed by the adaptive immune system. A major advantage of virus evolution approaches over regular hypothesis-driven research methods is that one will frequently stumble on new phenomena that are disclosed in the physiologically relevant setting of replicating virus. For those researchers that are willing to listen to the virus, there will be many unforeseen discoveries that the virus is trying to teach us about. We have trained ourselves and our students to foster unexpected viral evolution routes because they can form the basis for truly new discoveries. This sometimes means that the scientist has to abandon the traditional way of doing science, in which a pre-existing model is tested by means of specific experiments. To be able to carefully listen to what the virus is telling us, it is preferable that the researcher starts with an unbiased view. This paradigm shift in how to perform scientific experiments may be the most difficult part of performing pioneering virus evolution studies.
Footnotes
One contribution of 14 to a Theme Issue ‘New experimental and theoretical approaches towards the understanding of the emergence of viral infections’.
References
- Abbink T. E. M., Berkhout B.2003A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the gag start codon. J. Biol. Chem. 278, 11601–11611 (doi:10.1074/jbc.M210291200) [DOI] [PubMed] [Google Scholar]
- Abbink T. E., Berkhout B.2007RNA structure modulates splicing efficiency at the HIV-1 major splice donor. J. Virol. 82, 3090–3098 (doi:10.1128/JVI.01479-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T. W., Jeong Y. S., Penninck D., Bronson R., Greene M. F., Ruprecht R. M.1995Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science 267, 1820–1825 (doi:10.1126/science.7892606) [DOI] [PubMed] [Google Scholar]
- Baba T. W., et al. 1999Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 5, 194–203 (doi:10.1038/8859) [DOI] [PubMed] [Google Scholar]
- Back N. K. T., Nijhuis M., Keulen W., Boucher C. A. B., Oude Essink B. B., van Kuilenburg A. B. P., Van Gennip A. H., Berkhout B.1996Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15, 4040–4049 [PMC free article] [PubMed] [Google Scholar]
- Baldwin C. E., Berkhout B.2006Second site escape of a T20-dependent HIV-1 variant by a single amino acid change in the CD4 binding region of the envelope glycoprotein. Retrovirology 3, 84 (doi:10.1186/1742-4690-3-84) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjea A., Li M. J., Bauer G., Remling L., Lee N. S., Rossi J., Akkina R.2003Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol. Ther. 8, 62–71 (doi:10.1016/S1525-0016(03)00140-0) [DOI] [PubMed] [Google Scholar]
- Baron U., Bujard H.2000Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 327, 401–421 (doi:10.1016/S0076-6879(00)27292-3) [DOI] [PubMed] [Google Scholar]
- Berkhout B.2009A new Houdini act: multiple routes for HIV-1 escape from RNAi-mediated inhibition. Future Microbiol. 4, 151–154 (doi:10.2217/17460913.4.2.151) [DOI] [PubMed] [Google Scholar]
- Berkhout B., de Ronde A.2004APOBEC3G versus reverse transcriptase in the generation of HIV-1 drug-resistance mutations. AIDS 18, 1861–1863 (doi:10.1097/00002030-200409030-00022) [DOI] [PubMed] [Google Scholar]
- Berkhout B., Haasnoot J.2006The interplay between virus infection and the cellular RNA interference machinery. FEBS Lett. 580, 2896–2902 (doi:10.1016/j.febslet.2006.02.070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Klaver B.1993In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res. 21, 5020–5024 (doi:10.1093/nar/21.22.5020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Verhoef K., van Wamel J. L., Back N. K.1999Genetic instability of live, attenuated human immunodeficiency virus type 1 vaccine strains. J. Virol. 73, 1138–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Das A. T., Beerens N.2001HIV-1 RNA editing, hypermutation and error-prone reverse transcription. Science 292, 7 (doi:10.1126/science.292.5514.7a) [DOI] [PubMed] [Google Scholar]
- Boden D., Pusch O., Lee F., Tucker L., Ramratnam B.2003Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 77, 11531–11535 (doi:10.1128/JVI.77.21.11531-11535.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R. R.2004Natural and engineered nucleic acids as tools to explore biology. Nature 432, 838–845 (doi:10.1038/nature03195) [DOI] [PubMed] [Google Scholar]
- Chakrabarti L. A., Metzner K. J., Ivanovic T., Cheng H., Louis-Virelizier J., Connor R. I., Cheng-Mayer C.2003A truncated form of Nef selected during pathogenic reversion of simian immunodeficiency virus SIVmac239Deltanef increases viral replication. J. Virol. 77, 1245–1256 (doi:10.1128/JVI.77.2.1245-1256.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J. M.1995HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267, 483–489 (doi:10.1126/science.7824947) [DOI] [PubMed] [Google Scholar]
- Das A. T., Klaver B., Klasens B. I. F., van Wamel J. L. B., Berkhout B.1997A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 71, 2346–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. T., van Dam A. P., Klaver B., Berkhout B.1998Improved envelope function selected by long-term cultivation of a translation-impaired HIV-1 mutant. Virology 244, 552–562 [DOI] [PubMed] [Google Scholar]
- Das A. T., Klaver B., Berkhout B.1999aA hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 73, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. T., Land A., Braakman I., Klaver B., Berkhout B.1999bHIV-1 evolves into a non-syncytium-inducing virus upon prolonged culture in vitro. Virology 263, 55–69 (doi:10.1006/viro.1999.9898) [DOI] [PubMed] [Google Scholar]
- Das A. T., Brummelkamp T. R., Westerhout E. M., Vink M., Madiredjo M., Bernards R., Berkhout B.2004aHuman immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 78, 2601–2605 (doi:10.1128/JVI.78.5.2601-2605.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. T., Zhou X., Vink M., Klaver B., Verhoef K., Marzio G., Berkhout B.2004bViral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 279, 18776–18782 (doi:10.1074/jbc.M313895200) [DOI] [PubMed] [Google Scholar]
- Das A. T., Harwig A., Vrolijk M. M., Berkhout B.2007The TAR hairpin of human immunodeficiency virus type-1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 81, 7742–7748 (doi:10.1128/JVI.00392-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Wain-Hobson S.2009The 30th anniversary of quasispecies. Meeting on ‘Quasispecies: past, present and future’. EMBO Rep. 10, 444–448 (doi:10.1038/embor.2009.61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N. A., Vogt V. M.1998In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J. Virol. 72, 8073–8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Bujard H.2001Tetracyclines in the control of gene expression in eukaryotes. In Tetracyclines in biology, chemistry and medicine (eds Nelson M., Hillen W., Greenwald R. A.), pp. 139–157 Basel, Switzerland: Birkhäuser Verlag [Google Scholar]
- Haasnoot J., Westerhout E. M., Berkhout B.2007RNA interference against viruses: strike and counterstrike. Nat. Biotechnol. 25, 1435–1443 (doi:10.1038/nbt1369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G., Shindo K., Albin J. S., Harris R. S.2008Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 18, 819–824 (doi:10.1016/j.cub.2008.04.073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G., Abbink T. E., Berkhout B., Harris R. S.2009Optimal translation initiation enables Vif-deficient human immunodeficiency virus type 1 to escape restriction by APOBEC3G. J. Virol. 83, 5956–5960 (doi:10.1128/JVI.00045-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann-Lehmann R., Vlasak J., Williams A. L., Chenine A. L., McClure H. M., Anderson D. C., O'Neil S., Ruprecht R. M.2003Live attenuated, nef-deleted SIV is pathogenic in most adult macaques after prolonged observation. AIDS 17, 157–166 (doi:10.1097/00002030-200301240-00004) [DOI] [PubMed] [Google Scholar]
- Huthoff H., Berkhout B.2001Two alternating structures for the HIV-1 leader RNA. RNA 7, 143–157 (doi:10.1017/S1355838201001881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. P., Desrosiers R. C.1998Protective immunity induced by live attenuated simian immunodeficiency virus. Curr. Opin. Immunol. 10, 436–443 (doi:10.1016/S0952-7915(98)80118-0) [DOI] [PubMed] [Google Scholar]
- Keulen W., Boucher C., Berkhout B.1996Nucleotide substitution patterns can predict the requirements for drug-resistance of HIV-1 proteins. Antiviral Res. 31, 45–57 (doi:10.1016/0166-3542(96)00944-8) [DOI] [PubMed] [Google Scholar]
- Keulen W., Back N. K. T., van Wijk A., Boucher C. A. B., Berkhout B.1997Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of the human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 71, 3346–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaver B., Berkhout B.1994Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 13, 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovins J., Tsareva N. A., de Smit M. H., Berzins V., van Duin J.1997Rapid evolution of translational control mechanisms in RNA genomes. J. Mol. Biol. 265, 372–384 (doi:10.1006/jmbi.1996.0745) [DOI] [PubMed] [Google Scholar]
- Koff W. C., et al. 2006HIV vaccine design: insights from live attenuated SIV vaccines. Nat. Immunol. 7, 19–23 (doi:10.1038/ni1296) [DOI] [PubMed] [Google Scholar]
- Lacey S. F., Larder B. A.1994Mutagenic study of codons 74 and 215 of the human immunodeficiency virus type 1 reverse transcriptase, which are significant in nucleoside analog resistance. J. Virol. 5, 3421–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., et al. 2005Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood 106, 818–826 (doi:10.1182/blood-2004-10-3959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J. N., Shah P. S., Burnett J. C., Schaffer D. V.2008HIV evades RNA interference directed at TAR by an indirect compensatory mechanism. Cell Host Microbe 4, 484–494 (doi:10.1016/j.chom.2008.09.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licis N., Balklava Z., van Duin J.2000Forced retroevolution of an RNA bacteriophage. Virology 271, 298–306 (doi:10.1006/viro.2000.0327) [DOI] [PubMed] [Google Scholar]
- Marzio G., Verhoef K., Vink M., Berkhout B.2001In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc. Natl Acad. Sci. USA 98, 6342–6347 (doi:10.1073/pnas.111031498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G., Vink M., Verhoef K., de Ronde A., Berkhout B.2002Efficient human immunodeficiency virus replication requires a fine-tuned level of transcription. J. Virol. 76, 3084–3088 (doi:10.1128/JVI.76.6.3084-3088.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills J., Desrosiers R., Rud E., Almond N.2000Live attenuated HIV vaccines: proposal for further research and development. AIDS Res. Hum. Retroviruses 16, 1453–1461 (doi:10.1089/088922200750005976) [DOI] [PubMed] [Google Scholar]
- Morris S., Johnson M., Stavnezer E., Leis J.2002Replication of avian sarcoma virus in vivo requires an interaction between the viral RNA and the TpsiC loop of the tRNA(Trp) primer. J. Virol. 76, 7571–7577 (doi:10.1128/JVI.76.15.7571-7577.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J. L., Parks W. P., Wolinsky S. M., Korber B. T., Hutto C.1993HIV-1 reverse transcriptase. A diversity generator and quasispecies regulator. Ann. NY Acad. Sci. 693, 65–70 (doi:10.1111/j.1749-6632.1993.tb26257.x) [DOI] [PubMed] [Google Scholar]
- Obbard D. J., Gordon K. H., Buck A. H., Jiggins F. M.2009The evolution of RNAi as a defence against viruses and transposable elements. Phil. Trans. R. Soc. B 364, 99–115 (doi:10.1098/rstb.2008.0168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn R. C. L., van Duin J.1996Evolutionary reconstruction of a hairpin deleted from the genome of an RNA virus. Proc. Natl Acad. Sci. USA 93, 12 256–12 261 (doi:10.1073/pnas.93.22.12256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M., Huthoff H., Russell R., Liang C., Berkhout B.2004A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 78, 10 814–10 819 (doi:10.1128/JVI.78.19.10814-10819.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger A., Nassal M.1995Distinct requirements for primary sequence in the 5′- and 3′-part of a bulge in the hepatitis B virus RNA encapsidation signal revealed by a combination in vivo selection/in vitro amplification system. Nucleic Acids Res. 23, 3909–3915 (doi:10.1093/nar/23.19.3909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhang G., Simon A. E.2005Short internal sequences involved in replication and virion accumulation in a subviral RNA of turnip crinkle virus. J. Virol. 79, 512–524 (doi:10.1128/JVI.79.1.512-524.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Brake O., Konstantinova P., Ceylan M., Berkhout B.2006Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol. Ther. 14, 883–892 (doi:10.1016/j.ymthe.2006.07.007) [DOI] [PubMed] [Google Scholar]
- ter Brake O., 't Hooft K., Liu Y. P., Centlivre M., von Eije K. J., Berkhout B.2008aLentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 16, 557–564 (doi:10.1038/sj.mt.6300382) [DOI] [PubMed] [Google Scholar]
- ter Brake O., von Eije K. J., Berkhout B.2008bProbing the sequence space available for HIV-1 evolution. AIDS 22, 1875–1877 (doi:10.1097/QAD.0b013e328309efe3) [DOI] [PubMed] [Google Scholar]
- Verhoef K., Berkhout B.1999A second-site mutation that restores replication of a Tat-defective human immunodeficiency virus. J. Virol. 73, 2781–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef K., Marzio G., Hillen W., Bujard H., Berkhout B.2001Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 75, 979–987 (doi:10.1128/JVI.75.2.979-987.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eije K. J., Berkhout B.2009RNA-interference-based gene therapy approaches to HIV type-1 treatment: tackling the hurdles from bench to bedside. Antivir. Chem. Chemother. 19, 221–233 [DOI] [PubMed] [Google Scholar]
- von Eije K. J., ter Brake O., Berkhout B.2008Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J. Virol. 82, 2895–2903 (doi:10.1128/JVI.02035-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eije K. J., ter Brake O., Berkhout B.2009Stringent testing identifies highly potent and escape-proof anti-HIV short hairpin RNAs. J. Gene Med. 11, 459–467 (doi:10.1002/jgm.1329) [DOI] [PubMed] [Google Scholar]
- Vrolijk M. M., Ooms M., Harwig A., Das A. T., Berkhout B.2008Destabilization of the TAR hairpin affects the structure and function of the HIV-1 leader RNA. Nucleic Acids Res. 36, 4352–4363 (doi:10.1093/nar/gkn364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrolijk M. M., Harwig A., Berkhout B., Das A. T.2009Destabilization of the TAR hairpin leads to extension of the polyA hairpin and inhibition of HIV-1 polyadenylation. Retrovirology 6, 13 (doi:10.1186/1742-4690-6-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhout E. M., Ooms M., Vink M., Das A. T., Berkhout B.2005HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 33, 796–804 (doi:10.1093/nar/gki220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore A. M., Cook N., Hall G. A., Sharpe S., Rud E. W., Cranage M. P.1995Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J. Virol. 69, 5117–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney J. B., Ruprecht R. M.2004Live attenuated HIV vaccines: pitfalls and prospects. Curr. Opin. Infect. Dis. 17, 17–26 (doi:10.1097/00001432-200402000-00004) [DOI] [PubMed] [Google Scholar]
- Yoon M., Spear P. G.2004Random mutagenesis of the gene encoding a viral ligand for multiple cell entry receptors to obtain viral mutants altered for receptor usage. Proc. Natl Acad. Sci. USA 101, 17 252–17 257 (doi:10.1073/pnas.0407892101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Simon A. E.2005Importance of sequence and structural elements within a viral replication repressor. Virology 333, 301–315 [DOI] [PubMed] [Google Scholar]
- Zhou X., Vink M., Berkhout B., Das A. T.2006aModification of the Tet-On regulatory system prevents the conditional-live HIV-1 variant from losing doxycycline-control. Retrovirology 3, 82 (doi:10.1186/1742-4690-3-82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Vink M., Klaver B., Berkhout B., Das A. T.2006bOptimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther. 13, 1382–1390 (doi:10.1038/sj.gt.3302780) [DOI] [PubMed] [Google Scholar]
- Zhou X., Vink M., Klaver B., Verhoef K., Marzio G., Das A. T., Berkhout B.2006cThe genetic stability of a conditional-live HIV-1 variant can be improved by mutations in the Tet-On regulatory system that restrain evolution. J. Biol. Chem. 281, 17084–17091 (doi:10.1074/jbc.M513400200) [DOI] [PubMed] [Google Scholar]
- Zhou X., Symons J., Hoppes R., Krueger C., Berens C., Hillen W., Berkhout B., Das A. T.2007Improved single-chain transactivators of the Tet-On gene expression system. BMC Biotechnol. 7, 6 (doi:10.1186/1472-6750-7-6) [DOI] [PMC free article] [PubMed] [Google Scholar]