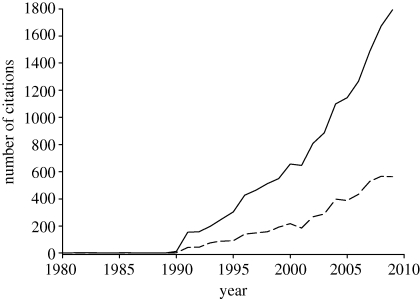
Emerging viral infectious diseases have been defined as either (i) a new disease provoked by a new virus (new in its nature, transmission mode, expression and/or adaptation to new host) or (ii) a known disease but expressing a new phenotype and associated with increase, in a relatively short time scale, of its incidence, impact or geographical range. Interestingly, the concept of ‘emerging infectious diseases’ was almost never used before the 1990s and its use has increased drastically until now (figure 1). During this period, approximately one-third of scientific articles dealing with emerging infectious diseases were concerned with viruses. In fact, new and potentially devastating viruses have been emerging during the last years as a consequence of climatic change and of the increasing introduction of human societies and its domestic animals and plants into the few remaining virgin areas of the planet (e.g. Jones et al. 2008; Anderson et al. 2004b). Socio-economic, environmental and ecological factors thus bring local hosts into contact with viruses which may jump the species barrier and become epidemic. Examples of emerging viral infectious diseases are Dengue, Junin, Lassa, Machupo, Hantavirus, Norwalk, SARS, yellow fever, West Nile encephalitis and Ebola, just to mention a few. As well as this, there have arisen new genotypes of classic viruses, such as the H5N1 avian and H1N1 swine influenza A viruses, or the new recombining strains of Tomato yellow leaf curl virus, which result from mutation or recombination phenomena followed by host-specific adaptation.
Figure 1.
Occurrence over the last 20 years of scientific articles using the terms ‘emerging infectious diseases’ for all (solid line) or viral (dashed line) diseases. Data are from ISI Web of Science covering 1980–2009. Solid line: Topic = (Emerg* infect* diseas*) AND Year Published = (year). Dashed line: Topic = (Emerg* infect* diseas*) AND Topic = (virus) AND Year Published = (year).
Even for cases of well-characterized viral diseases such as mumps, foot-and-mouth disease, rabies, AIDS or hepatitis C, efforts to control and eradicate them have been of quite limited success. This lack of success is a consequence of the great evolvability of viral populations, owing to their very large population size, short generation times and compacted genomes, all associated with error-prone replication mechanisms. Immune escape strains and strains resistant to antivirals may arise soon after challenged by the immune system or drugs, and viruses may jump the species barrier from their natural reservoir host to a naive one. Altogether, the picture is quite pessimistic, since the perspectives for future eradications would be overbalanced by the emergence and re-emergence of new viruses. Classical pharmacological approaches have largely ignored the dynamic evolution of viruses. New approaches that take into consideration the evolution of viral populations at both the within- and between-host levels are thus very much needed.
The study on emerging viral infectious diseases must begin with evaluating the relative importance of the several factors influencing viral evolution. Understanding the ecological and genetic mechanisms behind the genesis, maintenance and fate of viral diversity, and the interaction of viral populations with their standard and putative hosts has become pivotal for the development of new antiviral strategies or epidemiological control methods. Note that such approaches can be developed in parallel for the control of animal and plant viruses, owing to the fact that their evolution mostly results from similar evolutionary pressures on the different hosts (i.e. mutation, selection, drift and migration).
Today, comprehensive research programmes are conducted to gain insights into the mechanisms of viral emergence. Theoreticians have been developing increasingly complex models to account for the peculiarities of viral populations that aim to predict virus behaviour and, consequently, to propose control strategies that might be robust to viral evolution. Unfortunately, a gap exists between these models and observations which is a consequence of lack of communication between empiricists and theoreticians. Therefore, it is thus of great interest to bring theoreticians and experimentalists together in the same room so that cross-talk will enhance interactions and result, on the one side, in a stronger biological component in future mathematical models and, on the other side, in allowing the design of new experiments that will test specific predictions.
This Theme Issue results from a Jacques Monod Conference (CNRS) organized by us entitled ‘Understanding emergence of infectious diseases: new experimental and theoretical approaches to virus evolution’ that took place in Roscoff (France), 26–30 September 2009. The Jacques Monod Conference gathered 112 scientists from 11 different countries (Brazil, France, Germany, Ireland, Italy, South Africa, Spain, Switzerland, The Netherlands, UK and USA). There are 13 articles in this Theme Issue, covering a few of the many topics treated during the conference. Experts in areas such as virology, pathology, population genetics, physics, ecology and epidemiology have written these articles.
Amore et al. (2010) describe the ecological and epidemiological dynamics of West Nile virus in an area of recent introduction. Related to this topic, Frost & Volz (2010) present a unifying framework for epidemiological and phylogenetic methods to infer the number of infected individuals at the beginning of an epidemic. Duffy & Seah (2010) question the definition of ‘new’ and ‘emergent’ plant viruses by distinguishing single and multiple introductions of the same pathogen into a country based on looking at the percentages of sequence identity. In her article, Roossinck (2010) calls our attention to the existence of persistent plant viruses (i.e. viruses that are asymptomatic, transmitted vertically and not moving between cells) that may serve as reservoirs for future emerging ones. Froissart et al. (2010) review empirical investigations reporting to what extent within-host viral accumulation determines the transmission rate and the virulence of vector-borne plant viruses. Ogbunugafor et al. (2010) used an epidemiological model containing observations of pathogen productivity in reservoirs, as a means to predict which pathogens should be most prone to emerge in a primary host such as humans. Related to these two articles, Steinmeyer et al. (2010) describe new nested mathematical models studying the epidemiology of a viral disease with dose-dependent replication and transmission by nesting a differential-equation model of the within-host viral dynamics inside a between-host epidemiological model.
Lethal mutagenesis has been proposed as a potential way of curing viral infections (Anderson et al. 2004a). Two articles, Manrubia et al. (2010) and Martin & Gandon (2010), provide two different theoretical frameworks to explain the phenomenon of lethal mutagenesis. The first one emphasizes the therapeutic applications of the concept but highlighting their weaknesses. The second one describes the epidemiological implications of lethal mutagenesis. Using HIV-1 as an example, Das & Berkhout (2010) review causes for the observed high genetic variation of RNA viruses and how these variations jeopardize antiviral treatments. The fate of mutations in populations depend on their effect on fitness, Sanjuán (2010) compares the effect that mutations have on the fitness of five RNA or ssDNA viruses infecting bacteria, plants or animals with widely different genetic organizations and draws some general conclusions on virus evolution.
Concerning host–virus interactions, Pagán et al. (2010) tested the hypothesis of plant–virus coevolution in the wild. Finally, Lalić et al. (2010) show that virus adaptation to a partially susceptible host plant genotype allowed the virus to successfully infect, replicate and induce symptoms in other host genotypes that were fully resistant to the ancestral virus.
Acknowledgements
We want to express our thanks to all the authors for their willingness to contribute to this Theme Issue and to the reviewers for devoting their time in the difficult task of providing useful suggestions to improve the already fantastic manuscripts.
Footnotes
One contribution of 14 to a Theme Issue ‘New experimental and theoretical approaches towards the understanding of the emergence of viral infections’.
References
- Amore G., Bertolotti L., Hamer G. L., Kitron U. D., Walker E. D., Ruiz M. O., Brawn J. D., Goldberg T. L.2010Multi-year evolutionary dynamics of West Nile virus in suburban Chicago, USA, 2005–2007. Phil. Trans. R. Soc. B 365, 1871–1878 (doi:10.1098/rstb.2010.0054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. P., Daifuku R., Loeb L. A.2004aViral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol 5, 183–205 (doi:10.1146/annurev.micro.58.030603.123649) [DOI] [PubMed] [Google Scholar]
- Anderson P. K., Cunningham A. A., Patel N. G., Morales F. J., Epstein P. R., Daszak P.2004bEmerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends. Ecol. Evol. 19, 535–544 (doi:10.1016/j.tree.2004.07.021) [DOI] [PubMed] [Google Scholar]
- Das A. T., Berkhout B.2010HIV-1 evolution: frustrating therapies, but disclosing molecular mechanisms. Phil. Trans. R. Soc. B 365, 1965–1973 (doi:10.1098/rstb.2010.0072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Seah Y. M.201098% identical, 100% wrong: per cent nucleotide identity can lead plant virus epidemiology astray. Phil. Trans. R. Soc. B 365, 1891–1897 (doi:10.1098/rstb.2010.0056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froissart R., Doumayrou J., Vuillaume F., Alizon S., Michalakis Y.2010The virulence–transmission trade-off in vector-borne plant viruses: a review of (non-)existing studies. Phil. Trans. R. Soc. B 365, 1907–1918 (doi:10.1098/rstb.2010.0068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost S. D. W., Volz E. M.2010Viral phylodynamics and the search for an ‘effective number of infections’. Phil. Trans. R. Soc. B 365, 1879–1890 (doi:10.1098/rstb.2010.0060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. E., Patel N. G., Levy M. A., Storeygard A., Balk D., Gittelman J. L., Daszak P.2008Global trends in emerging infectious diseases. Nature 451, 990–993 (doi:10.1038/nature06536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalić J., Agudelo-Romero P., Carrasco P., Elena S. F.2010Adaptation of tobacco etch potyvirus to a susceptible ecotype of Arabidopsis thaliana capacitates it for systemic infection of resistant ecotypes. Phil. Trans. R. Soc. B 365, 1997–2007 (doi:10.1098/rstb.2010.0044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrubia S. C., Domingo E., Lázaro E.Pathways to extinction: beyond the error threshold. Phil. Trans. R. Soc. B 365, 1943–1952 (doi:10.1098/rstb.2010.0076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Gandon S.2010Lethal mutagenesis and evolutionary epidemiology. Phil. Trans. R. Soc. B 365, 1953–1963 (doi:10.1098/rstb.2010.0058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbunugafor C. B., Basu S., Morales N. M., Turner P. E.2010Combining mathematics and empirical data to predict emergence of RNA viruses that differ in reservoir use. Phil. Trans. R. Soc. B 365, 1919–1930 (doi:10.1098/rstb.2010.0075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán I., Fraile A., Fernandez-Fueyo E., Montes N., Alonso-Blanco C., García-Arenal F.2010Arabidopsis thaliana as a model for the study of plant–virus co-evolution. Phil. Trans. R. Soc. B 365, 1983–1995 (doi:10.1098/rstb.2010.0062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck M. J.2010Lifestyles of plant viruses. Phil. Trans. R. Soc. B 365, 1899–1905 (doi:10.1098/rstb.2010.0057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R.2010Mutational fitness effects in RNA and single-stranded DNA viruses: common patterns revealed by site-directed mutagenesis studies. Phil. Trans. R. Soc. B 365, 1975–1982 (doi:10.1098/rstb.2010.0063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmeyer S. H., Wilke C. O., Pepin K. M.2010Methods of modelling viral disease dynamics across the within- and between-host scales: the impact of virus dose on host population immunity. Phil. Trans. R. Soc. B 365, 1931–1941 (doi:10.1098/rstb.2010.0065) [DOI] [PMC free article] [PubMed] [Google Scholar]