Abstract
Aberrant glycosylation is a well-described hallmark of cancer. In a previous ovarian cancer case control study that examined polymorphisms in 26 glycosylation-associated genes, we found strong statistical evidence (P = 0.00017) that women who inherited two copies of a single-nucleotide polymorphism in the UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferase, GALNT1, had decreased ovarian cancer risk. The current study attempted to replicate this observation. The GALNT1 single-nucleotide polymorphism rs17647532 was genotyped in 6,965 cases and 8,377 controls from 14 studies forming the Ovarian Cancer Association Consortium. The fixed effects estimate per rs17647532 allele was null (odds ratio, 0.99; 95% confidence interval, 0.92–1.07). When a recessive model was fit, the results were unchanged. Test for hetero geneity of the odds ratios revealed consistency across the 14 replication sites but significant differences compared with the original study population (P = 0.03). This study underscores the need for replication of putative findings in genetic association studies.
Introduction
Glycosylation is a common posttranslational modification of proteins important for stability, solubility, secretion of signal, regulation of interactions, extracellular recognition, and folding (1). O-linked glycosylation involves the transfer of monosaccharide N-acetylgalactosamine (GalNAc) from UDP-GalNAc to the hydroxyl group of a serine or threonine residue on proteins and is catalyzed by GalNAc-transferases (ppGalNac-T or GALNT; ref. 2). In ovarian cancer cells, alterations in the O-glycosylation machinery result in aberrantly glycosylated proteins, which expose previously masked peptide motifs and new antigenic targets, thereby altering host immunogenic response (3).
We previously investigated associations between polymorphisms in 26 glycosylation-associated genes and epithelial ovarian cancer risk (4). Results based on 829 cases and 939 controls suggested that a polymorphism in the GALNT1 gene (rs17647532) was statistically significantly (P = 0.00017) inversely associated with epithelial ovarian cancer under a recessive model (4). To replicate this finding (5), we genotyped this GALNT1 variant in 14 independent study populations from the Ovarian Cancer Association Consortium (OCAC; ref. 6) and performed a pooled analysis.
Materials and Methods
Approval and Consent
All study participants provided written informed consent before the collection of biological samples or interview/clinical data. Each group involved in the OCAC has Institutional Review Board approval for this analysis and the Universities of Southern California and Duke have Institutional Review Board approval to serve as data coordinating centers for the OCAC.
Study Populations
The original study included the Mayo Clinic Ovarian Cancer Case Control Study (MAY) and the North Carolina Ovarian Cancer Study NCO-1 (Duke; ref. 4). The replication included non-Hispanic White subjects from 14 studies: the Australian cancer study and Australian ovarian cancer study (AUS); the Washington Ovarian Cancer case-control study (DOV); the German Ovarian Cancer case-control study (GER); the Hawaiian Ovarian Cancer study (HAW); the Hormones and Ovarian Cancer Prediction Study (HOP); the Danish Cancer Society MALOVA ovarian cancer case-control study (MAL); the North Carolina Ovarian Cancer Study (NCO-2); the New England-based Case-Control Study (NEC); the Polish Ovarian Cancer Study (POL); the SEARCH Ovarian Cancer Case-Control Study, Cambridge, United Kingdom (SEA); the Genetic Epidemiology of Ovarian Cancer Study, Stanford University (STA); the UC Irvine Ovarian Cancer Study (UCI); the UK Ovarian Cancer Population Study (UKO); and the USC/Los Angeles County Case-Control Studies of Ovarian Cancer (USC). Details of these studies are provided on the OCAC web portal26 and prior publications (7–16). Subjects (444 cases and 468 controls) from the NCO-1 that were included in the previous publication on GALNT1 were excluded from the replication analysis.
Genotyping and Quality Control
A single GALNT1 single-nucleotide polymorphism (SNP; rs17647532) was genotyped using either the iPlex Sequenom MassArray system (Sequenom, Inc.; Australian Cancer Study and Australian Ovarian Cancer Study) or 5′-nuclease TaqMan allelic discrimination assay (TaqMan, Applied Biosystems; all other sites). Laboratory procedures and quality control measures were described previously (4, 7–16). Call rates ranged from 96% to 99%, concordance across laboratories was 99%, and concordance between duplicate samples was 100%. No deviations from Hardy-Weinberg equilibrium (HWE) expectations were observed among the controls.
Statistical Analysis
The variables included study site, age at diagnosis for cases or interview for controls, tumor behavior, and histology (serous, mucinous, clear cell, and endometrioid). Unconditional logistic regression was used to model the association between the SNP and risk of ovarian cancer adjusted for age group, fitting both log-additive and recessive models. Goodness-of-fit P values were calculated to evaluate heterogeneity across the study populations. Statistical analyses were carried out using PLINK (17) and SAS version 9.1 (SAS, Inc.). All statistical significance levels (P values) presented are two-sided.
Results
A total of 6,965 non-Hispanic White invasive epithelial ovarian cancer cases and 8,377 non-Hispanic White controls were included in the replication analysis (Table 1). The mean ages were 55.6 and 55.9 years, respectively. More than 79% of the cases had an invasive tumor behavior and 53.5% had a serous histology.
Table 1.
Distribution of demographic and clinicopathologic characteristics for 15,342 OCAC non-Hispanic Caucasian subjects
| Variable | Cases (n = 6,965) | Controls (n = 8,377) |
|---|---|---|
| Age (y), mean (SD) | 55.6 (11.8) | 55.9 (11.1) |
| Age group (y), n (%) | ||
| <40 | 642 (9.2) | 586 (7.0) |
| 40–49 | 1,422 (20.4) | 1,969 (23.5) |
| 50–59 | 2,182 (31.3) | 2,459 (29.4) |
| 60–69 | 1,872 (26.9) | 2,284 (27.3) |
| >70 | 847 (12.2) | 1,079 (12.8) |
| Site, n (%) | ||
| AUS | 930 (13.4) | 1,064 (12.7) |
| DOV | 620 (8.9) | 617 (7.4) |
| GER | 251 (3.6) | 428 (5.1) |
| HAW | 90 (1.3) | 158 (1.9) |
| HOP | 307 (4.4) | 594 (7.1) |
| MAL | 440 (6.3) | 794 (9.5) |
| NCO | 250 (3.6) | 202 (2.4) |
| NEC | 982 (14.1) | 1,050 (12.5) |
| POL | 275 (3.9) | 597 (7.1) |
| SEA | 1,092 (15.7) | 1,213 (14.5) |
| STA | 369 (5.3) | 181 (2.2) |
| UCI | 404 (5.8) | 418 (5.0) |
| UKO | 467 (6.7) | 564 (6.7) |
| USC | 488 (7.0) | 497 (5.9) |
| Histology | ||
| Serous | 3,718 (53.5) | |
| Mucinous | 919 (13.2) | |
| Endometroid | 906 (13.1) | |
| Clear cell | 489 (7.0) | |
| Mixed cell | 158 (2.3) | |
| Other | 755 (10.9) | |
| Behavior | ||
| Borderline/LMP | 1,237 (17.8) | |
| Invasive | 5,520 (79.2) | |
| Unknown | 208 (3.0) | |
Abbreviation: LMP, low malignant potential.
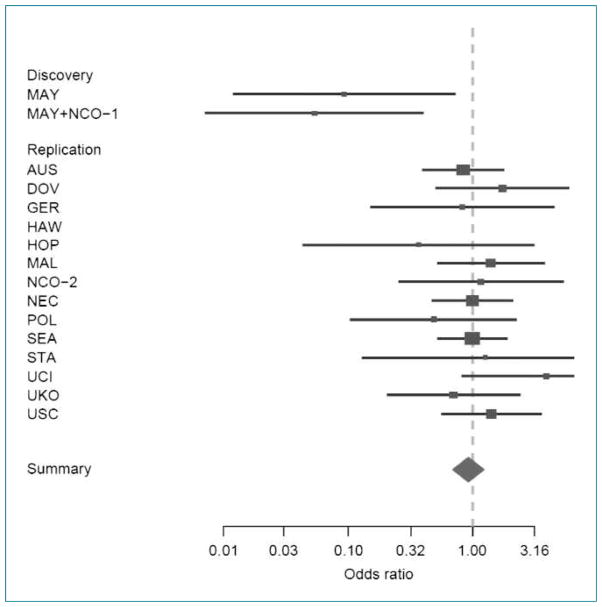
Across the studies, the minor allele frequencies varied from 9% to 12% among controls (Table 2). There was no association of the variant with cancer risk on a log-additive scale for any of the individual studies or across all OCAC studies combined. When the previously published data were included, the association remained null [odds ratio (OR), 0.98; 95% confidence interval (95% CI), 0.91–1.05] in the additive model. Because the earlier report (4) found the strongest results under a recessive model of transmission, additional models were fit to the data. Although the estimated ORs at five of the 14 replication sites were in the same inverse direction as the earlier report, none of these were statistically significant and the combined results across sites suggest no association with risk (Fig. 1). A test for heterogeneity of the ORs by site suggests no significant differences (P = 0.65). However, when we tested for heterogeneity of the ORs again, with the earlier results from Mayo and Duke included, the results were statistically significant (P = 0.03).
Table 2.
Summary OR and 95% CI for the GALNT1 SNP rs17647532 and risk of invasive epithelial ovarian cancer among non-Hispanic Caucasians
| Site | N | Case/control |
MAF* | PHWE† | Additive model,‡ OR (95% CI) | Recessive model,‡ OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | ||||||
| Discovery | ||||||||
| MAY | 846 | 1/12 | 70/71 | 314/378 | 0.10 | 0.0003 | 0.88 (0.64–1.21) | 0.14 (0.02–1.14) |
| NCO-1 (Duke) | 922 | 0/8 | 80/91 | 364/379 | 0.10 | 0.35 | 0.78 (0.57–1.06) | NA |
| Replication | ||||||||
| AUS | 1,994 | 12/16 | 185/207 | 733/841 | 0.11 | 0.44 | 1.00 (0.82–1.21) | 0.84 (0.4–1.8) |
| DOV | 1,237 | 7/4 | 109/133 | 504/480 | 0.11 | 0.16 | 0.85 (0.66–1.11) | 1.75 (0.51–6.03) |
| GER | 679 | 2/4 | 43/75 | 206/349 | 0.10 | 1.00 | 0.96 (0.66–1.39) | 0.83 (0.15–4.58) |
| HAW | 248 | 2/0 | 18/39 | 70/119 | 0.12 | 0.13 | 0.97 (0.54–1.74) | NA |
| HOP | 901 | 1/5 | 69/124 | 237/465 | 0.11 | 0.41 | 1.05 (0.76–1.46) | 0.37 (0.04–3.19) |
| MAL | 1,234 | 7/9 | 77/152 | 356/633 | 0.11 | 1.00 | 0.96 (0.73–1.25) | 1.4 (0.51–3.83) |
| NCO-2 | 452 | 4/3 | 46/41 | 200/158 | 0.12 | 0.74 | 0.92 (0.61–1.38) | 1.17 (0.26–5.36) |
| NEC | 2,032 | 11/8 | 97/92 | 380/397 | 0.10 | 0.09 | 1.11 (0.91–1.35) | 1.00 (0.48–2.08) |
| POL | 872 | 2/9 | 47/95 | 226/493 | 0.09 | 0.35 | 0.98 (0.70–1.38) | 0.49 (0.1–2.27) |
| SEA | 2,305 | 19/20 | 196/245 | 877/948 | 0.12 | 0.33 | 0.93 (0.77–1.12) | 1.00 (0.52–1.92) |
| STA | 550 | 3/1 | 76/37 | 290/143 | 0.11 | 0.70 | 1.01 (0.66–1.53) | 1.27 (0.13–12.43) |
| UCI | 822 | 8/2 | 79/83 | 317/333 | 0.10 | 0.29 | 1.13 (0.83–1.54) | 3.92 (0.82–18.65) |
| UKO | 1,031 | 6/8 | 81/100 | 380/456 | 0.10 | 0.36 | 0.96 (0.71–1.31) | 0.71 (0.21–2.43) |
| USC | 985 | 14/15 | 192/184 | 776/851 | 0.11 | 0.18 | 1.12 (0.86–1.47) | 1.41 (0.56–3.55) |
| Overall§ | 15,342 | 98/104 | 1,315/1,607 | 5,552/6,666 | 0.11 | 0.76 | 0.99 (0.92–1.07) | 1.12 (0.85–1.49) |
| Cumulative summary|| | 17,108 | 99/124 | 1,465/1,768 | 6,230/7,422 | 0.11 | 0.11 | 0.98 (0.91–1.05) | 0.94 (0.72–1.23) |
NOTE: The discovery set includes two sites (MAY and NCO-1) and the replication set includes 14 OCAC sites (AUS, DOV, GER, HAW, HOP, MAL, NCO-2, NEC, POL, SEA, STA, UCI, UKO, and USC). The study site nomenclature is described in Materials and Methods.
Abbreviation: MAF, minor allele frequency.
MAF estimated among controls.
P value for testing departure from HWE among controls.
ORs and 95% CIs in models adjusted for age groups.
Summary results across all 14 OCAC replication sites.
Cumulative summary including previously published data from Mayo and Duke (NCO-1).
Figure 1.
Forest plot for study-specific risk and 95% CIs for the association between ovarian cancer risk and the GALNT1 SNP (rs17647532) in the discovery set and the 14 studies in the replication set using a recessive model. The study site nomenclature is described in Materials and Methods.
The GALNT1 SNP associations were not observed in histologic cell type–specific subgroup analyses or in borderline compared with invasive ovarian cancer cases (data not shown).
Discussion
We previously reported an association between a GALNT1 SNP and ovarian cancer: Homozygous carriers of the variants were observed to have >90% lower risk than noncarriers (4), but the current study does not replicate this finding. Because the variant is rare, only 1.2% of the population would be expected to be homozygous carriers. Thus, the initial report was based on few cases and controls. In addition, there was deviation from HWE among controls in the original finding (P = 0.002 in combined data). In the current analysis with nearly 7,000 cases and more than 8,400 controls, there were only 98 cases and 104 controls homozygous for the variant. A logical conclusion is that the original observation represents a false-positive finding (18). This conclusion is underscored by the test for heterogeneity of the ORs across all study sites: No heterogeneity was observed among the 14 OCAC replication sites, but inclusion of the original findings yielded statistical evidence for heterogeneity. However, we cannot rule out the possibility that other as yet unidentified variants at the locus influence ovarian cancer risk.
In summary, the present analysis fails to replicate an earlier reported association of a GALNT1 variant with risk of ovarian cancer. This study highlights the need to replicate putative findings in genetic association studies.
Acknowledgments
Grant Support
Genotyping was supported by a grant from the Ovarian Cancer Research Fund provided by the family and friends of Kathryn Sladek Smith (Principal Investigator: A. Berchuck), NIH grant R01 CA 86888 (Principal Investigator: T. Sellers), and NIH grant R01 CA114343 (Principal Investigator: T. Sellers).
Support was provided by:
AOCS: U.S. Army Medical Research and Material Command under grant DAMD 17-01-1-0729, the Cancer Council Tasmania, Cancer Foundation of Western Australia (Australian Ovarian Cancer Study), and The National Health and Medical Research Council of Australia (grant 199600; Australian Cancer Study). The full AOCS Study Group is listed on http://www.AOCStudy.org/. G. Chene-vix-Trench, P.M. Webb, and D.C. Whiteman are supported by fellowships from the National Health and Medical Research Council.
DOV: NIH grants R01CA112523 and R01 CA87538.
GER: The German Ovarian Cancer Study was supported by the German Federal Ministry of Education and Research of Germany, Programme of Clinical Biomedical Research grant 01 GB 9401, the genotyping in part by the state of Baden-Württemberg through Medical Faculty of the University of Ulm (P.685), and data management by the German Cancer Research Center. We thank Ursula Eilber and Tanja Koehler for competent technical assistance.
HAW: The Hawaiian Ovarian Cancer Study is supported by NIH grants R01-CA58598 and N01-PC35137.
MAL: MALOVA was supported by grants from Mermaid 1, The Danish Cancer Society, and National Cancer Institute, Bethesda, MD (grant R01 CA61107).
MAY: The Fraternal Order of Eagles Cancer Research Fund and the Minnesota Ovarian Cancer Alliance (Mayo Clinic Ovarian Cancer Case-Control Study).
NCO: NIH grant R01-CA-76016.
NEC: NIH grants R01-CA54419 and P50-CA105009.
POL: Intramural Funds from the National Cancer Institute, NIH, Bethesda, MD. We thank Drs. Louise Brinton and Mark Sherman of the National Cancer Institute, Prof. Neolina Szeszenia-Dabrowska and Dr. Beata Peplonska of the Nofer Institute of Occupational Medicine (Lodz, Poland), Prof. Witold Zatonski of the M. Sklodowska-Curie Cancer Center and Institute of Oncology (Warsaw, Poland), and Pei Chao and Jane Wang (IMS, Silver Spring, MD) for their contribution to the Polish Ovarian Cancer Study. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Division of Cancer Epidemiology and Genetics.
SEA: SEARCH is funded by a program grant from Cancer Research UK. We thank the SEARCH team and the Eastern Cancer Registration and Information Centre for patient recruitment.
STA: The Roswell Park Alliance and the National Cancer Institute (grant CA71766 and Core Grant CA16056).
UCI: NIH, National Cancer Institute grants CA-58860 and CA-92044 and Lon V Smith Foundation grant LVS-39420.
UKO: The work of S.A. Gayther and S.J. Ramus, was undertaken at UCLH/UCL, which received a proportion of funding from the Department of Health NIHR Biomedical Research Center funding scheme.
USC: California Cancer Research Program grants 00-01389V-20170 and 2110200; U.S. Public Health Service grants CA14089, CA17054, CA61132, CA63464, N01-PC-67010, and R03-CA113148; and California Department of Health Services subcontract 050-E8709.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Bennett EP, Weghuis DO, Merkx G, van Kessel AG, Eiberg H, Clausen H. Genomic organization and chromosomal localization of three members of the UDP-N-acetylgalactosamine: polypeptide N-acetylgalactosaminyltransferase family. Glycobiology. 1998;8:547–55. doi: 10.1093/glycob/8.6.547. [DOI] [PubMed] [Google Scholar]
- 2.Hanisch FG. O-glycosylation of the mucin type. Biol Chem. 2001;382:143–9. doi: 10.1515/BC.2001.022. [DOI] [PubMed] [Google Scholar]
- 3.Li B, An HJ, Kirmiz C, Lebrilla CB, Lam KS, Miyamoto S. Glycoproteomic analyses of ovarian cancer cell lines and sera from ovarian cancer patients show distinct glycosylation changes in individual proteins. J Proteome Res. 2008;7:3776–88. doi: 10.1021/pr800297u. Epub 2008 Jul 22. [DOI] [PubMed] [Google Scholar]
- 4.Sellers TA, Huang Y, Cunningham J, et al. Association of SNPs in glycosylation genes with risk of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:397–404. doi: 10.1158/1055-9965.EPI-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–82. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 6.Berchuck A, Schildkraut JM, Pearce CL, Chenevix-Trench G, Pharoah PD. Role of genetic polymorphisms in ovarian cancer susceptibility: development of an international ovarian cancer association consortium. Adv Exp Med Biol. 2008;622:53–67. doi: 10.1007/978-0-387-68969-2_5. [DOI] [PubMed] [Google Scholar]
- 7.Pearce CL, Near AM, Van Den Berg DJ, et al. Validating genetic risk associations for ovarian cancer through the international Ovarian Cancer Association Consortium. Br J Cancer. 2009;100:412–20. doi: 10.1038/sj.bjc.6604820. Epub 2009 Jan 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmieri RT, Wilson MA, Iversen ES, et al. Polymorphism in the IL18 gene and epithelial ovarian cancer in non-Hispanic white women. Cancer Epidemiol Biomarkers Prev. 2008;17:3567–72. doi: 10.1158/1055-9965.EPI-08-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramus SJ, Vierkant RA, Johnatty SE, et al. Consortium analysis of 7 candidate SNPs for ovarian cancer. Int J Cancer. 2008;123:380–8. doi: 10.1002/ijc.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce CL, Wu AH, Gayther SA, et al. Progesterone receptor variation and risk of ovarian cancer is limited to the invasive endometrioid subtype: results from the Ovarian Cancer Association Consortium pooled analysis. Br J Cancer. 2008;98:282–8. doi: 10.1038/sj.bjc.6604170. Epub 2008 Jan 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gayther SA, Song H, Ramus SJ, et al. Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67:3027–35. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 12.Garcýa-Closas M, Brinton LA, Lissowska J, et al. Ovarian cancer risk and common variation in the sex hormone-binding globulin gene: a population-based case-control study. BMC Cancer. 2007;7:60. doi: 10.1186/1471-2407-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearce CL, Hirschhorn JN, Wu AH, et al. Clarifying the PROGINS allele association in ovarian and breast cancer risk: a haplotype-based analysis. J Natl Cancer Inst. 2005;97:51–9. doi: 10.1093/jnci/dji007. [DOI] [PubMed] [Google Scholar]
- 14.Quaye L, Dafou D, Ramus SJ, et al. Functional complementation studies identify candidate genes and common genetic variants associated with ovarian cancer survival. Hum Mol Genet. 2009;18:1869–78. doi: 10.1093/hmg/ddp107. [DOI] [PubMed] [Google Scholar]
- 15.Schildkraut JM, Goode EL, Clyde MA, et al. Single nucleotide polymorphisms in the TP53 region and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2009;69:2349–57. doi: 10.1158/0008-5472.CAN-08-2902. Epub 2009 Mar 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song H, Ramus SJ, Krüger Kjaer S, et al. Association between invasive ovarian cancer susceptibility and 11 best candidate SNPs from breast cancer genome-wide association study. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. Epub 2007 Jul 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]