Abstract
Through the shuffling of predefined modular zinc finger (ZF) domains with predictable target site recognition in vitro, we have generated a large repertoire of artificial transcription factors (ATFs) with five ZF domains (TFZFs). Here we report an effective strategy for the selection of ATF libraries through the coupling of the expression of transcriptional activators of the promoter of interest to the enhanced production of retroviral vector particles transferring the gene encoding the TFZF. Using this strategy, we successfully selected specific TFZFs that upregulate the expression of the γ-globin promoter. Selected transcription factors induced the expression of γ-globin when coupled to an activation domain and reduced expression when linked to a repression domain. This novel retroviral approach might be used to select other TFZFs but also might be generalized for the selection of other protein and small molecule interactions.
Keywords: polydactyl zinc finger, designer transcription factor, library selection, γ-globin reactivation
Introduction
Artificial transcription factors (ATFs) are proteins that are designed to specifically bind to DNA and modulate gene expression. Modular zinc finger (ZF) DNA-binding domains allow for the assembly of zinc finger transcription factors (TFZF) with predictable target site recognition in vitro. Typically a single zinc finger domain binds a 3-bp DNA sequence through the formation of specific contacts primarily within the major groove of the DNA. Our laboratory and others have successfully generated ATFs targeting unique sites within complex genomes 1-5. ATFs have been designed and constructed for the regulation of a variety of genes, such as ErbB2, ErbB3, VEGF, AP3 and EPO 2,4,6-8. While direct design and synthesis of TFZFs has been successful in many cases, this rational design strategy is often limited by a lack of information concerning both regulatory areas of the target gene and endogenous factors affecting transcription factor-DNA interactions; for example, endogenous TFs, chromatin structure and DNA accessibility which impact gene expression 4,8.
In order to overcome this limitation, we have generated large repertoires of zinc finger proteins through combination of the available ZF domains. When fused to a desired effector domain, these ZF libraries can be used as genome-wide screening tools for selection of novel functional TFZFs. Previously we reported construction and selection of TFZF libraries based on the modulation of cell surface markers 9,10. While effective, these screens were time-consuming and laborious, since after each of the several rounds of selection the selected library had to be recovered and re-cloned into an expression vector prior to the next screening cycle. Additionally, these selections were confined to the identification of activators of cell surface markers or a reporter system suitable for fluorescence activated cell sorting methods.
In this study, we describe a powerful strategy for the selection of potent activators and repressors of gene expression. The key feature of this strategy is the coupling of the expression of transcriptional activators of the promoter of interest to the enhanced production and release of retroviral vector particles that specifically package and transfer the gene encoding the TFZF that caused enhanced expression. We have applied this strategy to select for modulators of γ-globin expression.
The human β-globin locus, located on chromosome 11, contains five homologous globin genes. In the early stages of fetal development the embryonic (ε-) globin variant is predominantly expressed. During the later stages of fetal development, after the site of hematopoesis has switched to the fetal liver, fetal γ-globin is the predominant globin form produced. Shortly after birth, there is yet another switch in expression to the adult β-globin genes within the bone marrow. Transcriptional control of these genes is mediated by a complex interplay between cis and transacting regulatory elements 11-15. Upstream of these globin genes is a locus control region (LCR) that is necessary for the regulation of the entire locus 16,17.
The genetic diseases of β-thalassemias and sickle cell disease are associated with defective expression of β-globin or mutations within the β-globin product. These diseases can be mitigated by the expression of the fetal globin chain γ that forms tetrameric α2γ2 fetal hemoglobin (HbF). The hematologic benign condition known as “hereditary persistence of fetal haemoglobin” (HPFH) is characterized by an increase in the level of fetal haemoglobin. In adults, the fetal globin level is normally 1-2%; individuals with HPFH continue to express this gene in adult life wherein expression may be as high as 20 %. Patients with sickle cell disease and HPFH often do not have the serious life-threatening symptoms of those who have sickle cell anemia alone 18. Consequently, chemical inducers of fetal haemoglobin such as hydroxyurea, butyrate and others have been identified and used for the treatment of sickle cell disease and β-thalassemia 19. Treatment with these compounds, however, produces a range of serious side effects, preventing long-term treatment. A possible gene-based therapeutic intervention for these patients is the induction of fetal hemoglobin expression by an artificial TF 20-22. In our laboratory, we designed the artificial transcription factor gg1-VP64 and demonstrated that it increased the expression of endogenous γ-globin 21,23.
In this study, we selected for modulators of expression of the globin promoter from a library of TFZFs by coupling viral particle production with targeted promoter activation. We studied the coupling of a viral particle production phenotype with a defined phenotype of interest in order to explore the efficiency of this type of selection protocol. The selected transcription factors were then analyzed for their ability to modulate transcription of reporter constructs and cell lines that mimic endogenous human γ-globin gene expression profiles. Several of the selected transcription factors were able to bind to the γ-globin promoter directly and to upregulate human γ-globin gene expression in murine β-YAC cells.
Results
Coupling gene activation and retroviral particle production
We developed a library of zinc-finger transcription factors that each recognizes a 15-bp target site. The 1.7 × 107 member 5-ZF library provided a repertoire consisting of an equimolar mixture of a subset of defined zinc-finger DNA sequences specific for binding to GNH (H = A, C, or T) and ANN subsites at each finger position. The defined pool excludes GNG domains that in some cases have been implicated in target site overlap interactions that decrease zinc finger domain modularity. The resulting 5-ZF DNA binding protein coding sequence was fused to the VP16-derived transactivation domain VP64 and cloned into the retroviral transfer vector pMX-5ZF-Lib-VP64, which expressed TFZF via the viral long terminal repeat (LTR) and green fluorescent protein (GFP) from an internal CMV promoter.
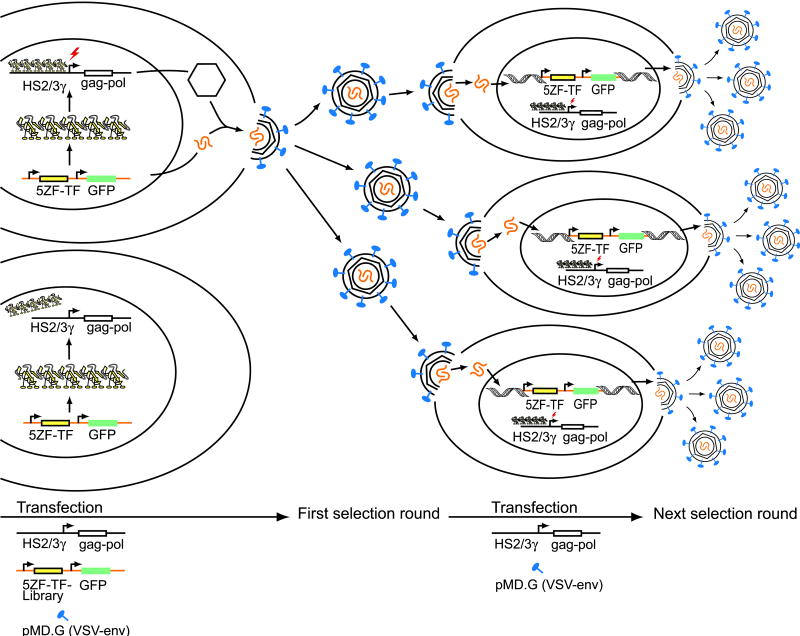
To select for activators of the γ-globin gene, we constructed a plasmid (pPur-γ-gp) in which murine leukaemia virus (MLV) gag-pol expression is driven by the γ-globin promoter. This plasmid encodes the γ-globin promoter fragment sequence from -206 to +47 relative to the start of transcription. Also included in pPur-γ-gp were the DNAse I-hypersensitive sites 2 and 3 from the β-globin LCR. This portion of the LCR contains enhancer elements important for high-level activation of the entire β-globin locus 24. MLV gag-pol genes encode the major structural and enzymatic proteins necessary for formation of virions when expressed with an envelope gene. In this case VSV-envelope was used to pseudotype viral particles, as it enables high infectivity. The virions package a viral transfer vector, in this case pMX-5ZF-Lib-VP64, thus introducing the TFZF gene into the genome of target cells (Figure 1).
Figure 1.
Schematic illustration of selection strategy. For the selection of γ-globin promoter activators, cells were transfected with VSV-G envelope and a 5-ZF-VP64-library expressed by a retroviral transfer vector. Cells were cotransfected with the pPur-γ-gp plasmid encoding the MLV structural genes gag and pol under the control of γ-globin promoter and enhancer elements. Cells that expressed an activator of the γ-globin promoter showed increased expression of MLV gag/pol genes which lead to increased transduction efficiency of target cells. To start the next selection round, target cells were transfected directly with pPur-γ-gp and VSV-G envelope for the production of new viral particles. Five rounds of selection were carried out.
The key feature for this selection is the coupling of the expression of transcriptional activators of the γ-globin promoter to the enhanced production and release of retroviral vector particles that specifically package and transfer the gene encoding the TFZF responsible for activation. Cells that express an activator of the γ-globin promoter show increased expression of MLV gag/pol genes leading to increased viral particle production and quantitatively greater transduction of target cells.
Selection of transactivators of the γ-globin promoter
To begin the selection, pPur-γ-gp, pMX-5ZF-Lib-VP64 and a VSV-envelope (pMD.G) plasmid were transfected into 293T cells. Two days later, the supernatant was used for transduction of fresh 293T cells. After expansion, these cells were transfected again with pPur-γ-gp and VSV-envelope (see Materials and Methods for details). For the next selection round, supernatants were used for transduction of fresh 293T cells. After harvesting of the supernatant, the producer cells were collected and genomic DNA analyzed (Figure 1). The retrovirally integrated ZF pools were PCR amplified and recloned into pMX-VP64 vector. Five rounds of selection were carried out. The resulting pMX-5ZF-VP64 library constructs were transfected into HEK-293T cells for characterization of the selected zinc-finger variants using a transduction efficiency test. The number of GFP expressing cells was analyzed by flow cytometry. Included in each experiment was a positive control pMX-gg1-VP64-CMV-GFP, a designed zinc-finger protein previously shown to strongly activate γ-globin expression 21.
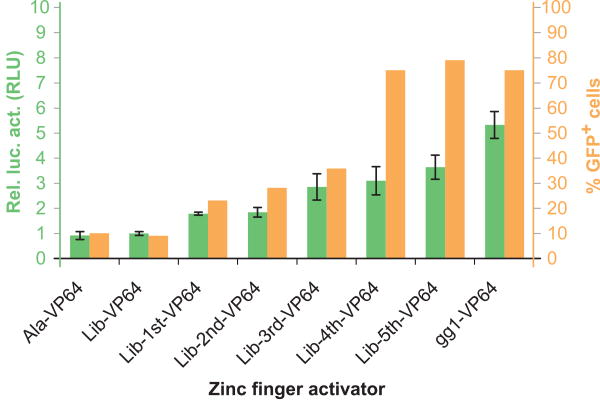
With each selection round, the number of GFP-expressing cells increased. After the fifth selection round, 79% of cells expressed GFP as shown in Figure 2. This was a nearly 9-fold increase compared to cells transfected with the starting library (9% GFP-expressing cells) or the non-DNA-binding control-ZF (Ala-VP64, 10% GFP-expressing cells). When transfected with positive control, pMX-gg1-VP64, 75% of cells expressed GFP (Figure 2).
Figure 2.
Selected TFZF libraries show increased GFP+ transduced cells after each round of TFZF selection. Retroviral particles were produced in 293T cells co-transfected with pPur-γ-gp and selected TFZF-libraries (Lib 1st – Lib 5th), used for transduction of fresh 293T target cells and compared to cells transduced with the non-DNA-binding control-ZF (Ala-VP64), cells transduced with unselected pMX-5ZF-library (Lib) and the positive control gg1-VP64. Three days after transduction, cells were analyzed by FACS to assess GFP expression. Percentage of GFP+ cells is depicted in yellow (right scale). Selected TFZF libraries show increased transcriptional activation on γ-globin promoter after each selection round. HeLa cells were transfected with control-ZFs (Ala-VP64 and gg1-VP64) and TFZF libraries (Lib 1st – Lib 5th) together with a luciferase reporter plasmid expressing firefly luciferase under the control of γ-globin and Renilla luciferase under the control of the β-globin promoter. Specific activation of γ-globin promoter was calculated by normalizing to Renilla luciferase values. Normalized luciferase activity of the selected TFZF libraries was compared to that of unselected pMX-5ZF-library (Lib = 1) and the positive control gg1-VP64. The normalized luciferase activities are shown in green (± s.d.) (right scale). For Lib-VP64 vs Lib-1st-VP64, p= 0.00015; for Lib-VP64 vs Lib-5th-VP64, p=0.00073.
To further analyze the ability of the selected zinc-finger variants to activate expression of the γ-globin promoter, we analyzed their ability to upregulate luciferase expression in the luciferase reporter construct γβ-luc. Like the pPur-γ-gp vector, γβ-luc encodes the γ-globin promoter fragment from -206 to +47 relative to the start of transcription and also includes the DNAse I-hypersensitive sites 2 and 3 from the β-globin LCR 21. This plasmid also encodes a β-globin promoter fragment consisting of the sequence from -206 to +47 relative to start of transcription, which drives expression of Renilla luciferase. Specific γ-globin promoter activation is determined by dividing firefly luciferase values with Renilla luciferase values. The pMX-VP64 plasmids, expressing the selected transcriptional activators, were co-transfected with γβ-luc into HeLa cells. With each selection round, luciferase activity increased. As shown in Figure 2, the pool of the selected variants after the fifth selection round showed a 3.6-fold increase in luciferase activity compared to cells transfected with starting library or the non-DNA binding control Ala-VP64. This was slightly less expression than was observed in cells transfected with the positive control pMX-gg1-VP64, which produced a 5.3-fold increase relative to untransfected cells. Thus, we were able to select for zinc fingers that strongly upregulated expression from the γ-globin promoter and enhancer region.
Characterization of single clones of selected zinc-finger transactivators
A total of 15 individual clones from the fifth round of selection were further analyzed. Transfection with seven of the individual clones resulted in more than double the number of GFP-positive 293T cells when compared to the non-DNA binding control Ala-ZF. Clones #γA-VP64 and #γL-VP64 increased the number of GFP-positive cells 7.3- and 7.8-fold, respectively, a level comparable to that of the positive control pMX-gg1-VP64 (Supplementary Table 1).
Next, we investigated whether or not the selected TFZFs were able to modulate expression from the γ-globin promoter in the luciferase reporter system. The strongest activators in this assay, #γA-VP64, #γK-VP64, #γL-VP64, and #γM-VP64, showed between 2.9- and 3.9-fold increase in luciferase activity when compared to the non-DNA binding control Ala-ZF-VP64; the positive control pMX-gg1-VP64, increased expression 5.8 fold (Supplementary Table 1). This data demonstrates that our selection system can be used to select clones that strongly upregulate the γ-globin promoter.
Binding of selected ZF to γ-globin promoter target site
The γ-globin promoter and the DNAse I-hypersensitive sites 2 and 3 from the β-globin LCR as well as the plasmid backbone of the pPur-γ-gp vector were analyzed using the Zinc Finger Tools software (available at http://www.zincfingertools.org) to identify putative DNA binding sites. For six of the zinc finger proteins that strongly activated expression from the γ-globin promoter, no binding site was identified with less than four mismatches in the γ-globin promoter and the DNAse I-hypersensitive sites 2 and 3 or the pPur-γ-gp vector backbone.
A target site search using the ZF domains of #γA identified the site 5′-GAT GCC GTT TGA GGT-3′ on the reverse strand 301 nucleotides upstream of the site of transcription initiation; this site contains a 3-base mismatch. To determine the binding affinity of #γA to this target site, we performed an electrophoretic mobility shift assay (EMSA) with a purified #γA-maltose-binding fusion protein (MBP). The calculated KD value of #γA with this DNA target was 3.2 nM (data not shown).
Because the predicted 15-bp target site of the #γA-protein was not the optimal site for this ZF, the 5-ZF protein was optimized for specific binding to this particular site using rational design. Three out of the five ZF domains were replaced with domains determined to have better specificity for this sequence. The modified protein, #γA-opt, was composed of the following α-helical ZF domains: F1-GGT (TSGHLVR), F2-TGA (QAGHLAS), F3-GTT (TSGSLVR), F4-GCC (DCRDLAR) and F5-GAT (TSGNLVR). This represents a ZF-clone that had not been present in the starting library which did not contain zinc-finger DNA sequences specific for binding TNN subsites. The KD of purified MBP-#γA-opt for the target site DNA was determined to be 7.1 nM (data not shown). A slight reduction in binding affinity was expected since the incorporated domains were chosen based on specificity rather than affinity. In transduction efficiency and luciferase activity assays, #γA-opt-VP64 induced an increase in activity similar to the parental #γA-VP64 construct (Supplementary Table 1).
Downregulation of γ-globin promoter
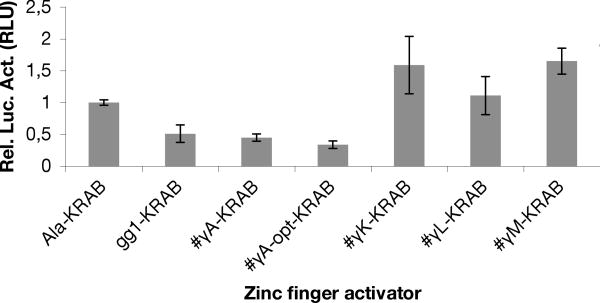
To analyze the ability to also repress expression of the γ-globin promoter, the four selected ZF-clones showing strongest activation (#γA, #γL, #γK #γM) in the reporter assays, gg1, Ala and #γA-opt were subcloned into the pMX retroviral vector as fusions to the transcriptional repressor Krüppel-associated box (KRAB). The resulting constructs (#γA-KRAB, #γL-KRAB, #γA-KRAB, #γA-opt-KRAB, #γL-KRAB, #γK-KRAB and #γM-KRAB gg1-KRAB) were tested for repression in luciferase assays. #γL-KRAB, #γK-KRAB and #γM-KRAB did not repress expression from the γ-globin promoter when compared to the non-DNA-binding Ala-ZF-KRAB. In contrast, #γA, which may bind directly to a sequence in the γ-globin promoter, showed a 55% reduction in luciferase activity, similar to that of the positive control pMX-gg1-KRAB (Figure 3). Interestingly, the optimized TFZF #γA-opt-KRAB showed an even stronger reduction with 64% decreased luciferase activity when compared to Ala-ZF-KRAB (Figure 3). Thus, this data shows that exchange of the transactivator domain VP64 with the transcriptional repressor domain KRAB produced repressors of the γ-globin promoter indicating direct regulation through binding of the target sequence.
Figure 3.
Selected TFZFs shows repression of γ-globin promoter expression with when fused to the repressor domain KRAB. HeLa cells were transfected with TFZFs fused to KRAB (#γA-KRAB, #γA-opt-KRAB, #γL-KRAB, #γK-KRAB and #γM-KRAB gg1-KRAB) and a luciferase reporter plasmid expressing firefly luciferase under the control of γ-globin and Renilla luciferase under the control of the β-globin promoter. Specific activation of the γ-globin promoter was calculated by normalizing to Renilla luciferase values. Error bars represent standard deviation from the mean. Normalized luciferase activity of the selected TFZFs was compared to that of cells transfected with the non-DNA binding negative control Ala-KRAB and the positive control gg1-KRAB. For Ala-KRAB vs #γA-KRAB, p= 0.00018; For Ala-KRAB vs #γA-opt-KRAB, p= 9.6E-05.
Analysis of human γ-globin gene expression in murine β-YAC cells
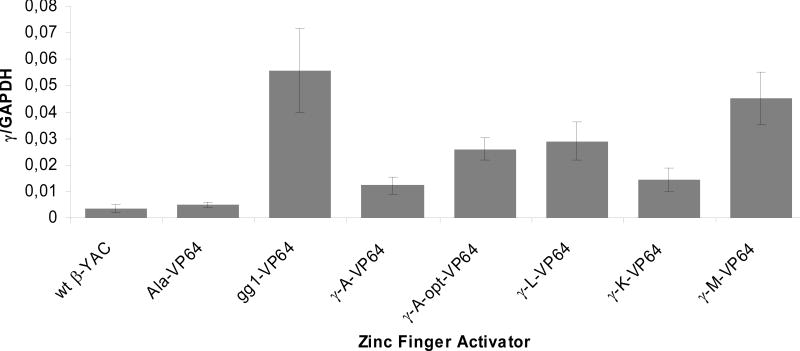
Encouraged by the high level of promoter activation in the reporter assays, we investigated the regulation of the endogenous γ-globin gene by the selected ATFs. Full-length cDNAs of #γA, #γL, #γK #γM and control ZFs were fused to the activation domain VP64 under the control of a CMV promoter (in a pcDNA3.1-hygro-context). These constructs were transfected into established immortalized cells derived from the bone marrow of β-YAC transgenic mice expressing exclusively β-globin from the YAC; YAC derived γ-globin is not detectable either in these mice or the derivative cells. In these cells, human γ-globin expression can be reactivated by various treatments and the endogenous murine globin genes maintain their native expression profiles. Pools of β-YAC cells stably expressing the TFZFs were generated and assessed for γ-globin expression by qRT-PCR. Levels of TFZF transcription ranged from approximately 0.4 to 1.2 of GAPDH gene expression as measured by RT-PCR (data not shown). In each of these cell-lines, γ-globin expression was observed. Human β-globin was not increased in cells expressing any of the TFZFs (data not shown); in fact in some instances it decreased reciprocally with the increase in γ-globin, as expected when γ- and β-globin genes compete for interaction with the LCR. Total β-like globin chain balance was maintained. Mouse α-globin levels also were unchanged (data not shown). In cells stably expressing #γM-VP64, γ-globin was expressed at a level similar to that in cells expressing the positive control gg1-VP64 (Figure 4). These experiments indicate that all 4 of the 15 TFZFs analyzed reactivated endogenous γ-globin gene transcription.
Figure 4.
Activation of human γ-globin in β-YAC BMCs containing selected zinc-fingers measured by RT-PCR. β-YAC BMCs were transfected with TFZFs (Ala-VP64, #γA-VP64, #γA-opt-VP64, #γL-VP64, #γK-VP64 and #γM-VP64 gg1-VP64) expressed in pcDNA-hygro. TFZF-expressing cells were selected for hygromycin resistance. RNA was isolated and γ-globin expression was measured by RT-PCR normalized to GAPDH expression. The data shown are the averages of duplicates or triplicates from at least two independent experiments for each sample. Error bars represent standard deviation from the mean. For Ala-VP64 vs γA-opt-VP64, p= 0.0012.
Discussion
We report here a novel strategy for the selection of zinc finger transcription factor regulators of the promoter and enhancer region of γ-globin. This system is based on coupling of the activation of a promoter to the increased production of retroviral particles that encode the regulatory zinc finger protein. After 5 rounds of selection, we successfully selected specific modulators of γ-globin expression from a library of 1.7 × 107 TFZFs. The TFZFs that were selected after the fifth round of selection showed in the transduction efficiency test activation of the γ-globin promoter comparable to that of the rationally designed activator of the γ-globin promoter gg1-VP64. Thus, the number of selection rounds performed seemed sufficient for the selection carried out in this study. However, more rounds of selection might lead to an increase in selection efficiency. Many of the selected clones from the 5-finger ZF library induced significant levels of luciferase as compared to controls. Some of the selected clones, when fused to the activation domain VP64 showed levels of activation similar to that of gg1-VP64. Additionally, the selected TFZFs were able to significantly induce human γ-globin expression in adult bone marrow cells derived from β-YAC transgenic mice. These experiments suggest that selected TFs, should they ultimately prove specific, could be used to treat patients suffering from sickle cell or thalassemic diseases. The efficacy of zinc finger-based therapeutics agents has been demonstrated in murine models and human studies are ongoing 25,26.
Previously, we selected TFZF libraries for transcriptional activators of different cell surface markers 9,10. The selection method we used was time-consuming and laborious, as after each round of selection the selected library had to be recovered from genomic DNA and then re-cloned into an expression vector; each selection round took two to three weeks. The selection system presented here eliminates the need for a laborious cloning process after each selection round, reducing the time needed for each round to less than one week. As transduced cells are used directly as producer cells for the next selection round, cloning and PCR steps that introduce mutations are avoided. Moreover this method is not limited to selection of activators of cell surface markers, and thus should be generally useful. The promoter region of any transcript of interest, including promoters of non-coding RNAs, such as microRNAs, might, in principle, be used in the selection. This method differs significantly from the OPEN bacterial selection approach 27 that has been shown to work for the selection of 3-finger zinc finger proteins that bind defined 9-bp target sites and our phage based approach 28 that can be used to select proteins against larger DNA fragments. In this new approach selections are performed in a mammalian cell background taking into account chromatin structure and the potential to select for indirect activators that act elsewhere in the mammalian genome.
One of the strongest activators of the γ-globin promoter, the 5-finger ZF #γA, interacted directly with a fragment of the γ-globin promoter encompassing positions 301–315 relative to transcriptional start site. Despite a predicted 3-bp mismatch, the selected ZF bound to the promoter derived oligonucleotide with an affinity of 3.1 nM. This affinity is slightly weaker than the affinity of the rationally designed 6-finger TFZF gg1 that was shown to bind with a Kd of 0.7 nM to its target site in the γ-globin promoter.21 The 5-finger library used here excluded CNN domains thereby preventing reselection of gg1 target site-binding TFZFs. When the #yA ZF was linked to the repressor domain KRAB, luciferase expression from the γ-globin was repressed. Thus, the method described here can also be used for the selection of activators or repressors of a promoter of interest. In addition to using this approach to identify TFZFs that modulate gene expression, this approach might be used to identify zinc finger proteins that target the activities of zinc finger enzymes such as methylases, nucleases and recombinases 23,29-34.
For many of the selected clones we were unable to define a definitive binding site on the γ -globin promoter fragment used in the selection. It is possible that these clones recognize degenerate sequences in the promoter region or regulate a gene elsewhere in the genome of the cell that ultimately impacts γ-globin expression. Using reliable parameters for prediction of the ZF target sites, the strongest activator in luciferase reporter assays, #γL, was not predicted to bind to the γ-globin promoter. This TFZF might regulate a gene involved in γ-globin expression or regulate a gene involved in post-transcriptional modification and therefore this approach might be applicable to the identification of regulatory genes of a defined promoter. TFZFs that were direct regulators of the γ-globin promoter were identified and shown to induce expression of the endogenous γ-globin gene in immortalized cells derived from the bone marrow of β-YAC transgenic mice that otherwise exclusively express β-globin. Thus, these TFZFs are potential candidates for the development of TFZF therapy of sickle cell patients.
In conclusion, we have developed a novel and efficient method to identify functional zinc finger proteins that regulate a targeted promoter by screening large combinatorial libraries of TFZFs. The method couples the increased expression from a promoter of interest to the enhanced production and release of retroviral vector particles that specifically package and transfer the gene responsible for upregulation. This method should be amenable to adaption to other types of selections. For example, selection of receptor activators that lead to induction of a specific promoter could be coupled to virus production and thus could be used for the identification of proteins other than transcription factors that function within the context of particular mammalian cell type.
Methods
Plasmids
The γβ-luc plasmid used for luciferase reporter assays was previously described21. Each member of the 5-ZF library of artificial transcription factors recognizes a 15-bp DNA site. We constructed the 5-ZF library by combinatorial assembly of five different zinc finger repertoires, connected by the canonical TGEKP linker. Each repertoire consisted of an equimolar mixture of a subset of defined zinc-finger DNA sequences previously selected in vitro by phage display for specific binding to each of the possible GNH/ANN subsites 2,5. Construction of the library is described in detail in Gonzalez et al. 35 The resulting 5-ZF DNA binding protein coding sequences were fused to the VP16-derived transactivation domain VP64 and the 5-ZF-VP64 library was cloned into the retroviral vector pMX-5ZF-Lib-VP64 by SfiI digest as previously described 9,10. The pMX-5ZF-Lib-VP64 plasmid expressed TFZF via the viral long terminal repeat (LTR) and green fluorescent protein (GFP) from an internal CMV promoter. To fuse the 5-ZF DNA binding domains to the Krüppel-associated box (KRAB) repressor domain, the ZFs were excised from pMX-5ZF-VP64 by SfiI digest and inserted into a corresponding pMX-KRAB9,10.
The construction of pPur-γ-gp is described in detail in supplementary methods. Briefly, this plasmid expresses murine leukemia virus (MLV) gag and pol genes under the control of γ-globin promoter and enhancer region.
The construction of the modified protein, #γA-opt is described in detail in supplementary methods. The #γA-opt zinc finger was composed of the following α-helical ZF domains: F1-GGT (TSGHLVR), F2-TGA (QAGHLAS), F3-GTT (TSGSLVR), F4-GCC (DCRDLAR) and F5-GAT (TSGNLVR).
Cell lines
The human embryonic kidney (HEK) 293T cells (CRL-11268), HeLa cells (ATCC: CCL-2) were obtained from ATCC. CID-dependent β-YAC bone marrow cells (BMCs) were maintained and transfected with various plasmid DNAs as described 23.
Selection of activators of γ-globin regulatory elements
HEK-293T cells (3.5 × 106) were cotransfected with pMD.G plasmid encoding the vesicular stomatitis virus-G envelope protein, pMX-5ZF-VP64 library DNA and pPur-γ-gp plasmid. Cell-free and concentrated Virus was then incubated with HEK-293T target cells in presence of 8 μg mL-1 polybrene. Cells were expanded and the next selection cycle was initiated by cotransfecting with pMD.G and pPur-γ-gp plasmid as described above. After transduction of the target cells, the DNA encoding the pool of TFZFs was recovered by PCR from the producer cells using the primers pMXf2 (forward) 5′-TCAAAGTAGACGGCATCG-3′ and VP64AscB (reverse) 5′-TCGTCCAGCGCGCGTCGGCGCG-3′ and cloned into the pMX vector. Selections were repeated for five rounds.
Luciferase assays
For each sample pMX-5ZF-VP64 or pMX-5ZF-KRAB was cotransfected with reporter plasmid γβ-luc. Cell lysates were analyzed for firefly and Renilla luciferase expression using the dual luciferase reporter assay system (Promega). Firefly luciferase values were normalized to the Renilla luciferase activity to determine specific activation of the γ-globin promoter. Data shown represents the average of three experiments. Ala-ZF, (which does not bind to DNA) was used as a negative control and gg1-ZF (which has a target site in the γ-globin promoter, GTC AAG GCA AGG CTG GCC) was used as a positive control.
Transduction efficiency test
The transduction efficiency test was similar to the first round of selection. HEK-293T cells were cotransfected with pMD.G plasmid, pMX-5ZF-VP64 DNA and pPur-γ-gp plasmid. The product retroviral particles were collected and then incubated with HEK-293T target cells in presence of 8 μg mL-1 Polybrene (Sigma). Three days post transduction, cells were analyzed by flow cytometry for GFP expression.
Cloning, expression, purification and characterization of DNA binding of the selected ZFs
All of the assembled five-finger coding regions were digested with the restriction endonuclease SfiI and cloned into pMal-CSS, a derivative of the bacterial expression vector pMal-C2 (New England Biolabs). Fusion proteins were purified to >90% homogeneity by using the Protein Fusion and Purification System (New England Biolabs) as per the manufacturer's instructions except that ZBA with 5 mM DTT was used as the column buffer. Target oligonucleotides were labeled at their 5′ or 3′ ends with [32P]ATP and gel purified. Eleven 3-fold serial dilutions of protein (starting with 1.6 μM) were incubated in 20 μL of binding reaction solution (1× binding buffer,10% glycerol (v/v),1 pM target oligonucleotide) for 3 hr at room temperature, then resolved on a 5% polyacrlyamide gel in 0.5× TBE buffer (90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA, pH 8.3). Quantitation of dried gels was performed by using a Phosphoimager and IMAGEQUANT software (Molecular Dynamics) and the KDs were determined by Scatchard analysis.
Theoretical analysis of selected ZFs
Theoretical binding analysis of the selected zinc fingers to the γ-globin locus was performed using the Zinc Finger Tools software (http://www.zincfingertools.org) 36.
Analysis of endogenous γ-globin gene expression
Total RNA was isolated from CID-dependent β-YAC bone marrow cells (BMCs), either stably transfected with linear fragments purified from plasmid constructs or not, using the Promega RNAgents Kit according to the manufacturer's instructions. For reverse transcription, 1 μg total RNA was used to synthesize cDNA with SuperScript II Reverse Transcriptase (Invitrogen). Samples were analyzed by quantitative real-time PCR with iQ SYBR Green Supermix (BioRad). PCR conditions for both γ-globin and GAPDH primers were: 95°C for 3 min, followed by 40 cycles of 95°C, 10 s and 60°C, 25 s. The melting curve was obtained from measurements taken every 0.5 s from 60-90°C. A series of dilutions of samples from -117 Greek HPFH β-YAC BMCs was utilized to generate a standard curve. Non-transfected wild-type β-YAC BMC samples were included as controls in all runs. Primer sequences were Mouse GAPDH forward, AGGTTGTCTCCTGCGACTTCA; GAPDH reverse, CCAGGAAATGAGCTTGACAAAG; human γ-globin forward, GTATTGCTTGCAGAATAAAGCC; and γ-globin reverse, ACCGTTTTGGCAATCCATTTC. The data shown are the averages of duplicates or triplicates from at least two independent experiments for each sample. Error bars represent standard deviation from the mean. TFZF transcription was measured by RT-PCR and compared to GAPDH transcription within the same cell pools using a pair of primers that universally amplified TFZF transcripts, regardless of the zinc finger sequence present. Primer sequences were TFZF forward, ATGGCCCAGGCGGCCCTCG; and TFZF reverse, AAAGTCATCGAGGGCATCAG.
Supplementary Material
Acknowledgments
Thanks to Flavia C. Costa and Renee Neades for technical assistance with the BMC transfections and expression assays.
Funding: This study was supported by the Skaggs Institute for Chemical Biology and in part by National Institutes of Health Grants RO1 DK61803 and R01GM065059 (CFB) and R01 DK061804 (KRP).
Abbreviations
- ZF
zinc finger
- TF
transcription factor
- TFZFs
ZF transcription factor
- ATF
artificial transcription factor
- GFP
green fluorescent protein
- KRAB
Krüppel-associated box
- EMSA
electrophoretic mobility shift assay
- LCR
locus control region
- HbF
fetal hemoglobin
- HPFH
hereditary persistence of fetal haemoglobin
- β-YAC
β-globin locus yeast artificial chromosome
- LTR
long terminal repeat
- MLV
murine leukaemia virus
- MBP
maltose-binding fusion protein
Footnotes
Author Contributions: U.T, K.P. and C.B. designed the research; U.T., K.P., B.G., and H.F. performed the experiments; U.T, K.P. and C.B wrote the manuscript, which all authors commented on.
References
- 1.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–41. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 2.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–78. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 3.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–61. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 4.Liu PQ, et al. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J Biol Chem. 2001;276:11323–34. doi: 10.1074/jbc.M011172200. [DOI] [PubMed] [Google Scholar]
- 5.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci U S A. 1999;96:2758–63. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerli RR, Dreier B, Barbas CF., 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97:1495–500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stege JT, Guan X, Ho T, Beachy RN, Barbas CF., 3rd Controlling gene expression in plants using synthetic zinc finger transcription factors. Plant J. 2002;32:1077–86. doi: 10.1046/j.1365-313x.2002.01492.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, et al. Synthetic zinc finger transcription factor action at an endogenous chromosomal site. Activation of the human erythropoietin gene. J Biol Chem. 2000;275:33850–60. doi: 10.1074/jbc.M005341200. [DOI] [PubMed] [Google Scholar]
- 9.Blancafort P, Magnenat L, Barbas CF., 3rd Scanning the human genome with combinatorial transcription factor libraries. Nat Biotechnol. 2003;21:269–74. doi: 10.1038/nbt794. [DOI] [PubMed] [Google Scholar]
- 10.Magnenat L, Blancafort P, Barbas CF., 3rd In vivo selection of combinatorial libraries and designed affinity maturation of polydactyl zinc finger transcription factors for ICAM-1 provides new insights into gene regulation. J Mol Biol. 2004;341:635–49. doi: 10.1016/j.jmb.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 11.Chada K, Magram J, Costantini F. An embryonic pattern of expression of a human fetal globin gene in transgenic mice. Nature. 1986;319:685–9. doi: 10.1038/319685a0. [DOI] [PubMed] [Google Scholar]
- 12.Chada K, et al. Specific expression of a foreign beta-globin gene in erythroid cells of transgenic mice. Nature. 1985;314:377–80. doi: 10.1038/314377a0. [DOI] [PubMed] [Google Scholar]
- 13.Kollias G, Wrighton N, Hurst J, Grosveld F. Regulated expression of human A gamma-, beta-, and hybrid gamma beta-globin genes in transgenic mice: manipulation of the developmental expression patterns. Cell. 1986;46:89–94. doi: 10.1016/0092-8674(86)90862-7. [DOI] [PubMed] [Google Scholar]
- 14.Peterson KR. Hemoglobin switching: new insights. Curr Opin Hematol. 2003;10:123–9. doi: 10.1097/00062752-200303000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Rutherford T, Nienhuis AW. Human globin gene promoter sequences are sufficient for specific expression of a hybrid gene transfected into tissue culture cells. Mol Cell Biol. 1987;7:398–402. doi: 10.1128/mcb.7.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–86. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–13. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 18.Murray N, Serjeant BE, Serjeant GR. Sickle cell-hereditary persistence of fetal haemoglobin and its differentiation from other sickle cell syndromes. Br J Haematol. 1988;69:89–92. doi: 10.1111/j.1365-2141.1988.tb07607.x. [DOI] [PubMed] [Google Scholar]
- 19.Swank RA, Stamatoyannopoulos G. Fetal gene reactivation. Curr Opin Genet Dev. 1998;8:366–70. doi: 10.1016/s0959-437x(98)80095-6. [DOI] [PubMed] [Google Scholar]
- 20.Blouin MJ, et al. Genetic correction of sickle cell disease: insights using transgenic mouse models. Nat Med. 2000;6:177–82. doi: 10.1038/72279. [DOI] [PubMed] [Google Scholar]
- 21.Graslund T, Li X, Magnenat L, Popkov M, Barbas CF., 3rd Exploring strategies for the design of artificial transcription factors: targeting sites proximal to known regulatory regions for the induction of gamma-globin expression and the treatment of sickle cell disease. J Biol Chem. 2005;280:3707–14. doi: 10.1074/jbc.M406809200. [DOI] [PubMed] [Google Scholar]
- 22.May C, et al. Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature. 2000;406:82–6. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 23.Blau CA, et al. {gamma}-Globin gene expression in chemical inducer of dimerization (CID)-dependent multipotential cells established from human {beta}-globin locus yeast artificial chromosome ({beta}-YAC) transgenic mice. J Biol Chem. 2005;280:36642–7. doi: 10.1074/jbc.M504402200. [DOI] [PubMed] [Google Scholar]
- 24.Collis P, Antoniou M, Grosveld F. Definition of the minimal requirements within the human beta-globin gene and the dominant control region for high level expression. EMBO Journal. 1990;9:233–40. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powars DR, Chan L, Schroeder WA. The influence of fetal hemoglobin on the clinical expression of sickle cell anemia. Ann N Y Acad Sci. 1989;565:262–78. doi: 10.1111/j.1749-6632.1989.tb24174.x. [DOI] [PubMed] [Google Scholar]
- 26.Rebar EJ, et al. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat Med. 2002;8:1427–32. doi: 10.1038/nm1202-795. [DOI] [PubMed] [Google Scholar]
- 27.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lund CV, Blancafort P, Popkov M, Barbas CF., 3rd Promoter-targeted phage display selections with preassembled synthetic zinc finger libraries for endogenous gene regulation. J Mol Biol. 2004;340:599–613. doi: 10.1016/j.jmb.2004.04.057. [DOI] [PubMed] [Google Scholar]
- 29.Gordley RM, Smith JD, Graslund T, Barbas CF., 3rd Evolution of programmable zinc finger-recombinases with activity in human cells. J Mol Biol. 2007;367:802–13. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–85. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 31.Nomura W, Barbas CF., 3rd In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J Am Chem Soc. 2007;129:8676–7. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 32.Szczepek M, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–93. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 33.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 34.Xu GL, Bestor TH. Cytosine methylation targetted to pre-determined sequences. Nat Genet. 1997;17:376–8. doi: 10.1038/ng1297-376. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez B, et al. Modular System for Construction of Zinc-Finger Libraries and Proteins (in preparation) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandell JG, Barbas CF., 3rd Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Research. 2006;34:W516–23. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.