Abstract
In Saccharomyces cerevisiae, the PUT1 and PUT2 genes are required for the conversion of proline to glutamate. The PUT1 gene encodes Put1p, a proline dehydrogenase (PRODH)1 enzyme localized in the mitochondrion. Put1p was expressed and purified from Escherichia coli and shown to have a UV-visible absorption spectrum that is typical of a bound flavin cofactor. A Km value of 36 mM proline and a kcat = 27 s−1 were determined for Put1p using an artificial electron acceptor. Put1p also exhibited high activity using ubiquinone-1 (CoQ1) as an electron acceptor with a kcat = 9.6 s−1 and a Km of 33 µM for CoQ1. In addition, knockout strains of the electron transfer flavoprotein (ETF) homolog in S. cerevisiae were able to grow on proline as the sole nitrogen source demonstrating that ETF is not required for proline utilization in yeast.
Keywords: proline metabolism, yeast, proline dehydrogenase, PUT1
Introduction
The oxidative conversion of proline to glutamate involves two enzymes, a flavin dependent proline dehydrogenase (PRODH) and a NAD+-dependent Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase (P5CDH). PRODH and P5CDH help organisms respond to changes in the nutritional environment by initiating the breakdown of proline as a source for nitrogen, carbon, and energy [1–6]. In Saccharomyces cerevisiae, Put1p and Put2p are the corresponding PRODH and P5CDH enzymes, respectively (Scheme 1) [7]. In yeast and other eukaryotes, PRODH and P5CDH are localized in the mitochondria [8, 9]. PRODH catalyzes the first step of proline oxidation by generating P5C, an intermediate in the proline catabolic pathway (see Scheme 1). The reaction involves two phases or two half-reactions. In the reductive half-reaction electrons are transferred from proline to the flavin adenine dinucleotide cofactor to form P5C. The second phase (i.e., oxidative half-reaction) of the catalytic cycle involves oxidation of reduced flavin (FADH2) to regenerate FAD. P5C is then hydrolyzed to γ-glutamic acid semialdehyde (GSA) which is subsequently oxidized by P5CDH to form glutamate and NADH.
Fig. 8.
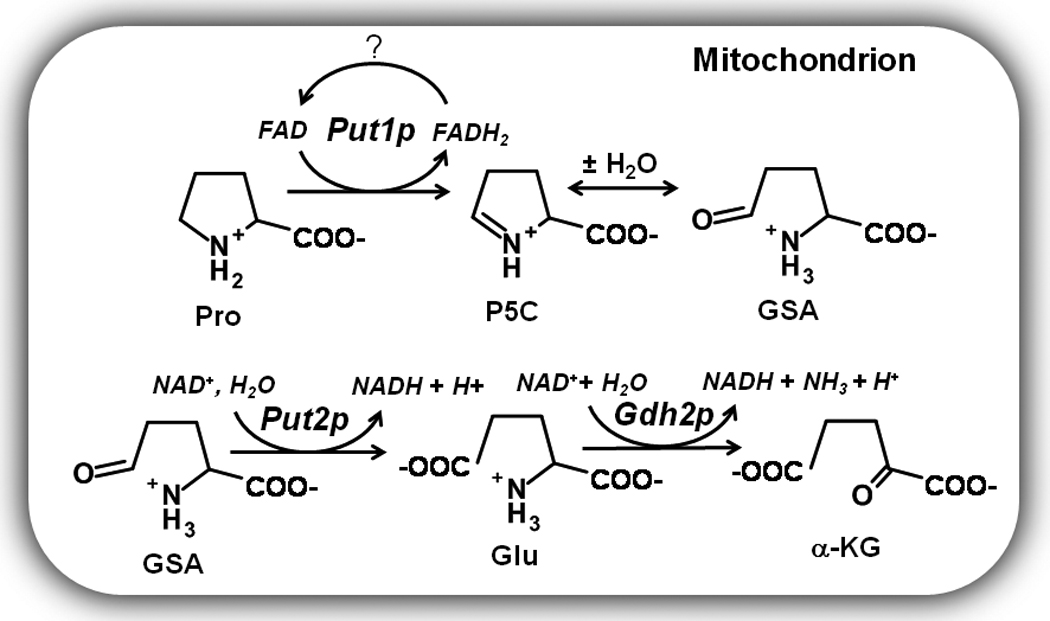
Expression of the PUT1 and PUT2 genes in yeast is sensitive to proline levels and nitrogen quality. In the presence of preferred nitrogen sources such as glutamine and ammonia, PUT gene expression is repressed [10]. In nitrogen limiting environments, PUT1 and PUT2 expression is upregulated coinciding with derepression of the PUT4 gene which encodes a proline specific transporter [10]. Activation of the PUT1 and PUT2 genes is regulated by Put3p, a transcriptional regulator which directly senses proline and belongs to the Zn(II)2Cys6 protein family [11–13]. Binding of proline to Put3p leads to a 20-fold activation of the PUT genes [12]. Put3p also induces small increases in PUT gene expression independently of proline in response to poor nitrogen sources [13]. The resulting Put3p activation of the PUT1 and PUT2 genes enables yeast to utilize proline as a nitrogen source in coordination with glutamate dehydrogenase (Gdh2p) which oxidatively deaminates glutamate to generate ammonia and α-ketoglutarate (Scheme 1) [10, 14].
Human PRODH (PRODH1), also known as proline oxidase (POX1), is of particular importance as it has been shown to have unique roles in regulating cell survival and apoptotic pathways [1, 15]. Expression of human PRODH is upregulated by the tumor suppressor p53 protein with increased PRODH activity leading to induction of cell death pathways and helping to prevent cancer progression [16, 17]. Using a mouse xenograft tumor model, Phang and colleagues recently showed that PRODH activity significantly reduces tumor formation [18]. The mechanism by which human PRODH contributes to apoptosis involves the generation of reactive oxygen species such as superoxide anions either directly at the enzyme active site or indirectly due to increased electron flux in the mitochondrial electron pathway.
Although PRODH has important roles in energy metabolism, nitrogen utilization, and in mammals, the programmed cell death pathway, detailed information about the biochemical and structural properties of PRODH from a eukaryotic organism are still lacking. In bacteria, the best characterized PRODH enzymes are the bifunctional PutA enzymes which encode PRODH and P5CDH on a single polypeptide and the monofunctional PRODH enzyme from Thermus thermophilus (TtPRODH) [19, 20]. X-ray crystal structures of the PRODH domain of PutA from Escherichia coli (EcPutA) and TtPRODH show a common (βα)8 barrel fold with a noncovalently bound FAD cofactor [19–21]. Human PRODH and Put1p most likely share the same (βα)8 barrel fold and sequence alignments with the bacterial enzymes show human PRODH and Put1p also share important residues for substrate and FAD binding [19].
To better understand PRODH enzymes from eukaryotic organisms, we sought to purify and characterize Put1p from S. cerevisiae. Particular focus was given to the oxidative half-reaction and Put1p reactivity with molecular oxygen as it may provide insight into the physiological role of human PRODH in superoxide production and apoptotic signaling pathways. The direct electron acceptor for Put1p in the oxidative half-reaction has not been demonstrated but a functional mitochondrial respiratory chain is necessary for S. cerevisiae to utilize proline as a nitrogen source [8]. Based on steady state kinetics and yeast genetic screens, we propose that Put1p directly couples proline oxidation to ubiquinone reduction in the electron transport chain.
Materials and methods
Chemicals
Ubiquinone-1 (CoQ1), flavin adenine dinucleotide (FAD), L-tetrahydro-2-furoic acid (L-THFA), phenazine methosulfate, guanidinium hydrochloride, antibiotics and buffers were purchased from Sigma-Aldrich. 2,6-dichlorophenolindophenol (DCPIP), o-aminobenzaldehyde (o-AB), isopropyl β-D-1-thiogalactopyranoside (IPTG), sodium chloride and Tris-Cl were purchased from Fisher Scientific Inc. L-proline was purchased from Acros Organics Ltd. All other chemicals and buffers were purchased from Sigma-Aldrich, Inc. Restriction endonucleases and T4 DNA ligase were purchased from Fermentas and Promega, respectively. Molecular size standards used for calibrating size exclusion columns were purchased from Sigma. Bicinchoninic acid (BCA) reagents used for protein quantitation were obtained from Pierce. All experiments used Nanopure water.
Yeast strains and growth conditions
S. cerevisiae strains BY4741 parental wild-type (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), BY4741 PUT1 knockout (put1), BY4741 AIM45 knockout (aim45, EFT α homolog), and CIR1 knockout (cir1, ETF β homolog) were obtained from Open Biosystems Yeast Knockout Strains collection. Yeast strains were routinely cultured at 30 °C in YPD (1% yeast extract, 2% peptone, 2% dextrose) or synthetic dropout (SD) media with the appropriate supplements. Growth on different nitrogen sources was examined using either 0.5% ammonium sulfate or 0.5% proline.
Put1p construct
The PUT1 clone was a generous gift from Dr. Marjorie Brandriss at the University of Medicine and Dentistry of New Jersey-New Jersey Medical School [22]. The PUT1 gene was subcloned by PCR into a pET14b vector using NdeI and XhoI sites. Analysis (MitoProt II) of the primary structure of Put1p predicts the N-terminal 18 residues comprise a mitochondrial signaling peptide. The predicted mitochondrial signaling peptide of the N-terminal 18 amino acids was subsequently removed by site-directed mutagenesis by inserting an NdeI site immediately upstream of codon 19 using the oligonucleotide 5'-CTAAATCGCGCATACCCCATATGTGCTTTCCTTTGATAAAGAGG-3′ and its complement. Digestion with NdeI then resulted in the pET14b-PUT1Δ18 construct which codes for a N-terminal 6xHis tag fusion Put1p protein lacking the 18 residue mitochondrial signaling peptide (Put1pΔ18). PUT1Δ18 was also inserted into pET23b using NdeI and XhoI sites to make a pET23b-PUT1Δ18 construct with five additional amino acids and a 6xHis tag at the C-terminal end. Nucleic acid sequencing was performed to confirm the above constructs.
Purification and characterization of Put1p
Put1p lacking the predicted mitochondrial peptide (Put1pΔ18) was expressed and purified from E. coli strain BL21(DE3) pLysS with a N-terminal 6xHis tag using the pET14b-PUT1Δ18 construct or with a C-terminal 6xHis tag from a pET23b- PUT1Δ18construct as described here. The PUT1Δ18 constructs were transformed into E. coli BL21(DE3) pLysS. Transformed cells were plated onto Luria-Bertani (LB) agar containing chloramphenicol (34 µg/ml) and ampicillin (50 µg/ml). Resulting colonies were inoculated in 5 ml of LB broth containing the necessary antibiotics and grown to an optical density at 600 nm (OD600) of 1.0. 1 ml of the LB culture was then used to inoculate 1 L of Terrific Broth media containing chloramphenicol (34 µg/ml) and ampicillin (50 µg/ml). The 1 L cultures were incubated at 37°C with shaking (250 rpm) until OD600 of 0.8 at which point Put1p expression was induced with 0.5 mM IPTG overnight at 20°C.
The overnight cultures were centrifuged at 6000 rpm for 20 min at 4 °C. The resulting pellets were resuspended in a final 125 ml volume of binding buffer (20 mM Tris, 5 mM imidazole, 0.5 M NaCl, 10% glycerol, pH 7.9) supplemented with 1 mM FAD, protease inhibitors (3 mM ε-amino-N-caproic acid, 0.3 mM phenyl methyl sulfonyl chloride, 1.2 µM leupeptin, 48 µM N-p-tosyl-L-phenyl alanine chloromethyl ketone, 78 µM N-α-tosyl-L-lysine chloromethylketone) and 0.5% n-octyl-β-D-glucoside. The cell suspension was disrupted by sonication at 4 °C for a total of 5 min (5 sec pulse on, 15 sec pulse off, 40% power). The cell extract was centrifuged at 16000 rpm (4 °C) for 60 min. The supernatant (100 ml) was passed through a 0.8 µm filter (VWR) and applied to a Ni-NTA superflow (Qiagen) resin (25 ml bed volume in a 2.8 cm × 30 cm column) equilibrated with 1X binding buffer. Wash buffer (125 ml, 20 mM Tris, 60 mM imidazole, 0.5 M NaCl, 10% glycerol, pH 7.9) was then applied to the column followed by elution buffer (20 mM Tris, 500 mM imidazole, 0.5 M NaCl, 10% glycerol, pH 7.9) with a flow rate of 3 ml/min to elute Put1p. Fractions from the elution step were then analyzed by SDS-PAGE and pooled. Pooled Put1p was then dialyzed into 50 mM Tris buffer (pH 8.1) containing 50 mM NaCl, 10% glycerol and concentrated using an Amicon 30-kDa cutoff filter (Millipore). The 6xHis tag (N-terminal or C-terminal) was not removed after purification. No substantial differences in the spectroscopic and kinetic properties were observed between the N-terminal and C-terminal tagged Put1p proteins. The data reported here were generated with N-terminal 6xHis tagged Put1p unless stated otherwise.
The molar ratio of flavin to Put1p polypeptide was estimated by denaturing Put1p in 6 M guanidinium chloride. The spectrum was recorded from 600 to 250 nm. The total amount of polypeptide was determined using the molar extinction coefficient of denatured Put1p at 280 nm (å280= 45060 M−1 cm−1) estimated by the ProtParam tool (ExPASy Proteomics Server). The amount of flavin was determined using the molar extinction coefficient for free FAD at 450 nm (å450= 11700 M−1 cm−1) in guanidinium chloride [23]. The molar extinction coefficient of flavin bound to Put1p was estimated to be 10800 M−1 cm−1 at 451 nm. The concentration of total Put1p protein was determined using the BCA method while the concentration of flavin-bound Put1p was determined using the molar extinction coefficient for bound FAD (å451 = 10800 M−1 cm−1) [24].
Steady-state kinetic measurements
Put1p activity was measured at 25 °C using terminal electron acceptors DCPIP, oxygen and CoQ1. The concentrations of Put1p used for all of the kinetic assays are based on the amount of flavin-bound Put1p. Proline:DCPIP oxidoreductase assay was performed as described in a 1-ml reaction volume using Cary 100 and Cary 50 UV-visible spectrophotometers [25]. The DCPIP assay mixture contained 0.05 µM of Put1p enzyme, 0.27 mM phenazine methosulfate, 75 µM DCPIP and proline concentrations ranging from 0 to 300 mM in 20 mM Tris at pH 8.0. The rate of DCPIP reduction was measured at 595 nm (ε = 16100 cm−1 M−1) [25]. Proline:O2 oxidoreductase activity (1 ml assay volume) was measured in air saturated potassium phosphate buffer (50 mM, pH 8.0) using 1 µM Put1p, 4 mM o-AB and 0–300 mM proline at 443 nm by following the formation of the o-AB-P5C yellow complex (ε = 2590 cm−1 M−1) [26]. Oxygen utilization by Put1p was also measured using a Clark type Pt,Ag/AgCl-electrode. The reaction mixture contained 2.2 µM Put1p and 300 mM proline in 50 mM phosphate buffer at pH 8.0 (25 °C).
The Km for proline was determined by measuring CoQ1 reduction using 0.1 µM Put1p in 50 mM phosphate buffer at pH 8.0 with 0–300 mM proline and 100 µM CoQ1. The Km for CoQ1 was determined by measuring CoQ1 reduction using 0.23 µM Put1p in 50 mM phosphate buffer at pH 8.0 with 300 mM proline and varying CoQ1 (0–200 µM). Reduction of CoQ1 was followed at 275 nm (ε=13700 cm−1 M−1) using a 0.15 cm path length on a Hi-Tech Scientific SF-61DX2 stopped-flow instrument at 25 °C [27]. The kinetic parameters Km and kcat were estimated by regression analysis of the initial reaction velocity versus proline concentration using the Michaelis-Menten equation and Lineweaver-Burk plot analysis [28]. The binding order of the Put1p proline:CoQ1 oxidoreductase reaction was evaluated using double reciprocal plots of reaction velocity versus variable substrate concentrations. Assays were performed in 50 mM potassium phosphate (pH 8.0) using 0.23 µM Put1p and with proline concentrations varied from 10–100 mM and CoQ1 concentrations varied 5–100 µM. Global curve fitting of the data to the equation for a classical ping-pong mechanism
| (eq 1) |
was performed using SigmaPlot 11 software. Inhibition studies with L-THFA were performed using the DCPIP assay as described previously [29]. For these assays, proline concentrations were varied from 0 – 200 mM and the final L-THFA concentrations were varied from 0 – 3 mM using 100X stock solutions of L-THFA in 50 mM Tris buffer (pH 8.0). The inhibition constant (Ki) for L-THFA was estimated by Lineweaver-Burk plot analysis. For all of the above experiments, the initial velocity values were the average from assays performed in triplicate.
The pH optimum for Put1p activity was determined using a mixed buffer system from pH 6.0–10.0 comprised of 20 mM each HEPES, MES, MOPS and TAB (80 mM total). Data were fit to the equation
| (eq 2) |
where vlim is the limiting velocity (µmol/min) and pKa1 and pKa2 represent the acidic and alkaline ionizations that contribute to the pH dependent curve of the reaction velocity. Assays were performed in duplicate.
Proline titrations of Put1p were performed at 20 °C in 50 mM Tris buffer (pH 8.0) containing 50 mM NaCl and 10% glycerol under anaerobic conditions. The Put1p-proline mixtures were equilibrated 5 min after each addition of proline prior to recording each spectrum on a Cary 100 spectrophotometer. A correction for turbidity that occurred during the experiment was applied by subtracting a light scattering spectrum. The amount of turbidity in each spectrum was determined by measuring the increase in absorbance at 700 nm due to scattering. The titration data were analyzed as previously described assuming the formation of a reduced Put1p-P5C complex [25, 30]. Although P5C is hydrolyzed to the open GSA form, the ring and open chain forms are rapidly reversible with P5C favored at pH > 7.0, indicating that P5C will be the predominant form at pH 8.0 [31].
Results
Properties of Put1p
Purified Put1p lacking the N-terminal residues 1–18 (Put1pΔ18) was shown by SDS-PAGE to have an apparent molecular weight of 50 kDa (Figure 1) which nearly matches the predicted molecular weight of 53665 Da for the product of the N-terminal 6xHis Put1pΔ18 construct. A smaller molecular weight species was also visible below the main Put1p band. Both bands were identified as Put1p polypeptides by mass spectral analysis. Presumably the lower molecular weight band is a degradation product of Put1p. The UV-visible spectrum of Put1p is shown in Figure 2. Absorption maxima are at 355 and 451 nm with a shoulder at 478 nm and are indicative of a bound flavin cofactor to the Put1p protein (Figure 2, Inset A). Quantitation of the polypeptide at 280 nm and the flavin at 451 nm indicates a flavin/polypeptide ratio of 0.43. The flavin/polypeptide ratios varied from 0.3–0.45 between different preparations. Thus, recombinant Put1p contains a significant amount of the apo-form. Addition of proline to Put1p resulted in reduction of the flavin cofactor in a concentration dependent manner (Figure 2, Inset B). No anionic semiquinone was observed indicating flavin semiquinone is not stabilized during proline reduction. 50% of the FAD in Put1p was reduced at 0.7 mM proline as estimated from the plot shown in Figure 2 (Inset C).
Fig. 1.
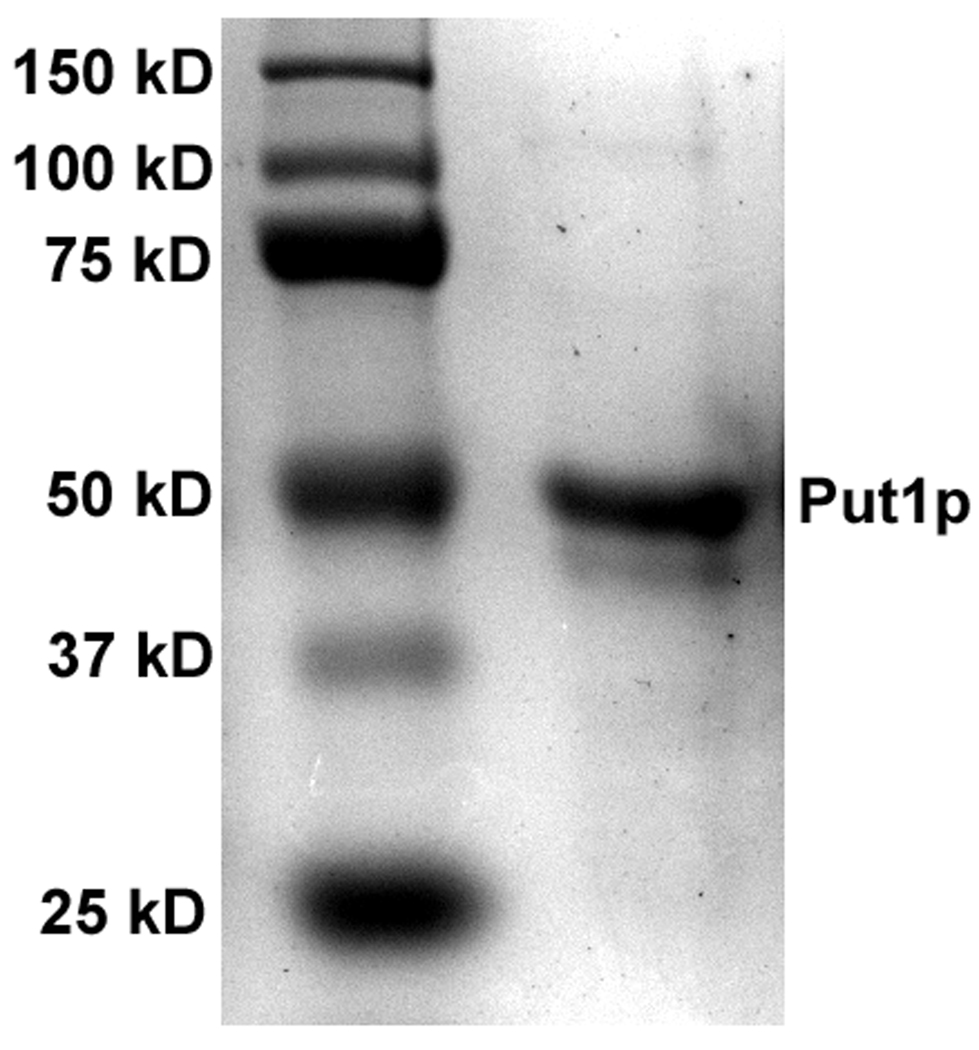
SDS-PAGE analysis of Put1p (3 µg) purified from E. coli. Predicted molecular weight of Put1p is 53665 Da.
Fig. 2.
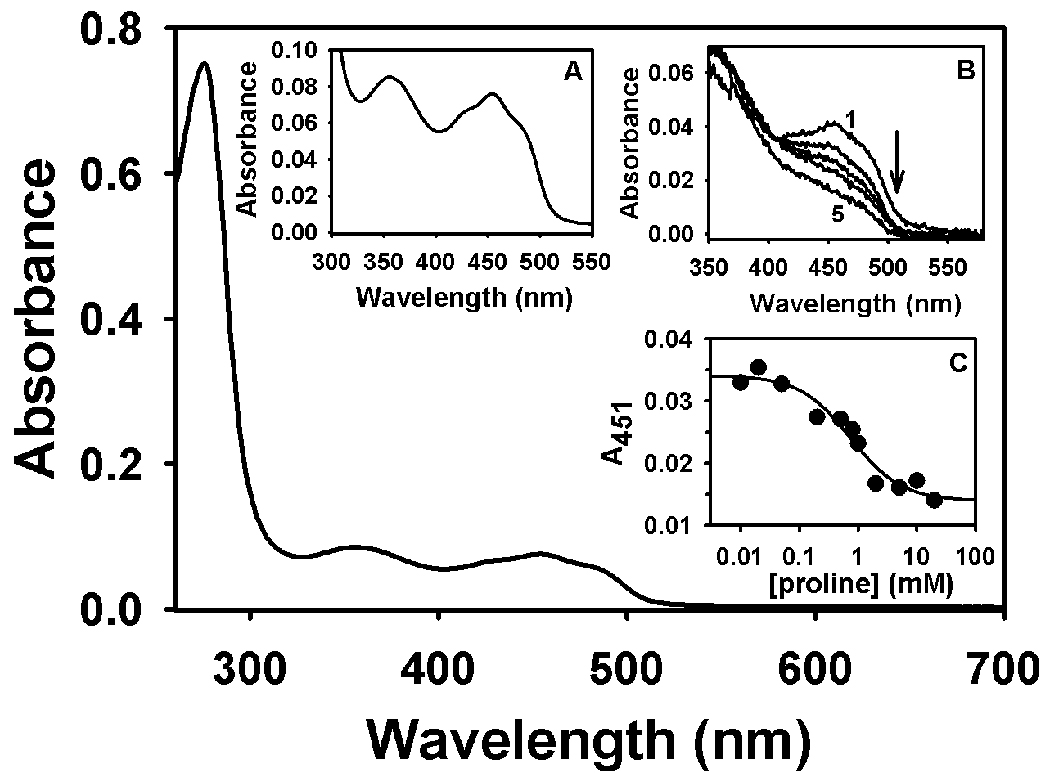
UV-visible spectrum of purified Put1p (C-terminal 6xHis tagged form) in 20 mM Tris-HCl buffer (pH 8.0) containing 100 mM NaCl and 0.25% octyl-glucoside. Concentrations of total and flavin-bound Put1p protein are 16.1 µM and 7 µM, respectively. Inset A, Flavin spectrum from 550-300 nm of the same Put1p protein shown below. Inset B, Titration of Put1p (3.8 µM flavin-bound form) with proline in 50 mM Tris-NaCl buffer (pH 8.0). Curves 1–5 are selected spectra from the titration at 0, 0.01, 0.200, 1.0, and 5 mM proline. Inset C, Best fit analysis of the absorbance decrease at 451 nm during the titration with proline assuming the formation of a Put1p-P5C complex from which a Keq of 1.4 mM−1 proline was estimated.
Kinetic parameters for Put1p using the proline:DCPIP oxidoreductase assay were Km = 36 ± 5 mM proline and kcat = 27 s−1. The Km value is similar to that reported for the monofunctional TtPRODH enzyme (Km = 27 mM proline, kcat = 13 s−1) but the turnover number is about 2-fold higher [20]. For PutA enzymes, Km values range from 100–150 mM proline with turnover numbers around 5–12 s−1 [32]. The kinetic parameters for Put1p along with TtPRODH may indicate that monofunctional enzymes generally exhibit higher turnover numbers and a lower Km values for proline than bifunctional PutA enzymes. The pH optimum for the Put1p reaction was determined by following proline:DCPIP oxidoreductase activity over the pH range 6–10. Figure 3 shows a symmetrical and bell-shaped curve for Put1p activity with a pH optimum of 8.6. By fitting the data to equation 2, pKa values of 7.2 ± 0.1 (pKa1) and 10.3 ± 0.15 (pKa2) were estimated for the two ionization events observed in the pH dependence of Put1p activity.
Fig. 3.
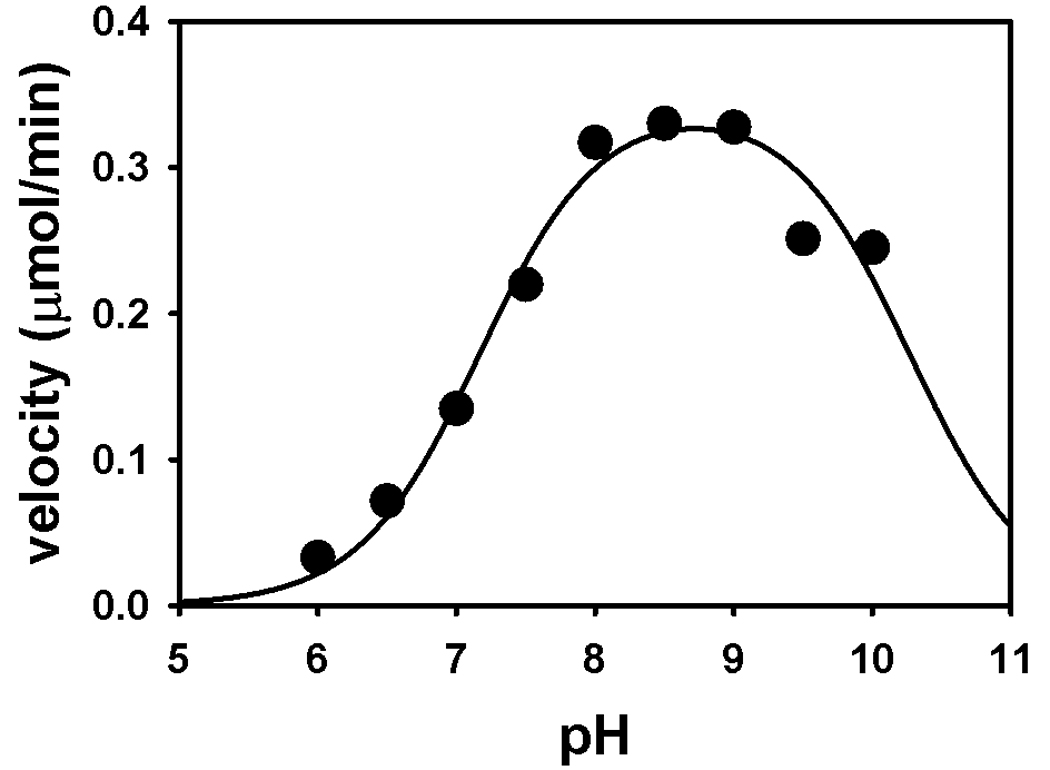
pH dependence of Put1p proline:oxidoreductase activity. Shown is the average activity from measurements performed in duplicate with 10 µg of Put1p in a 80 mM mixed buffer system from pH 6.0 – 10.0 at 25 °C. The pKa values estimated by fitting the data to Eq. 1 are 7.2 ± 0.1 (pKa1) and 10.3 ± 0.15 (pKa2) for two ionizable groups that influence the observed pH-dependence of Put1p activity.
Substrate specificity and inhibition
The ability of Put1p to use alternative substrates such as trans-4-hydroxy-L-proline and L-pipecolic acid was tested. Only very low activity (0.008 U/mg) was observed with 4-hydroxy-L-proline which is more than 2000-fold lower than the specific activity of Put1p with proline (18 U/mg). Put1p also exhibited very low activity (0.004 U/mg) with L-pipecolic acid and no activity with D-proline. Thus Put1p has a strong substrate specificity for L-proline.
A potent inhibitor of PRODH activity is L-THFA, an isostructural analog of proline. Previously, TtPRODH was shown to be inhibited by L-THFA with a Ki of 1 mM [20]. Figure 4 shows a Line-weaver Burk plot of the inhibition of Put1p activity by L-THFA. The plot is consistent with L-THFA acting as a competitive inhibitor of Put1p with a Ki of 5.3 mM. Most likely L-THFA binds to the active site of Put1p similarly to that observed in the X-ray crystal structures of the E. coli PRODH domain complexed with L-THFA [33].
Fig. 4.
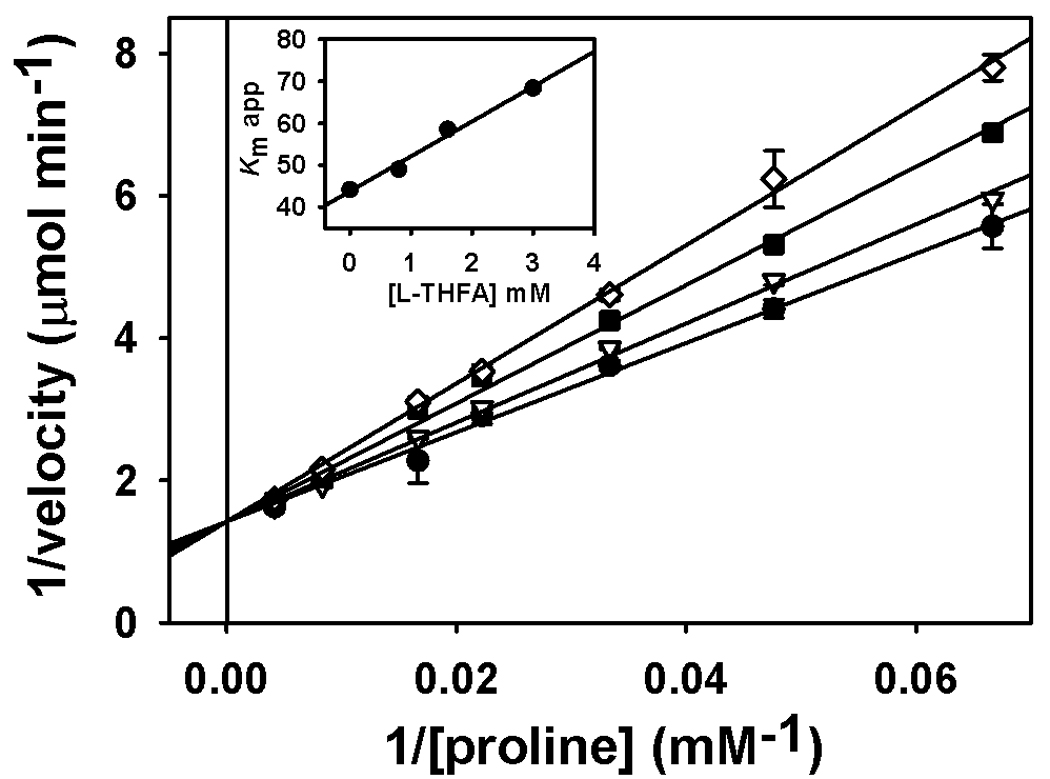
Inhibition of Put1p (C-terminal 6xHis tagged form) activity by L-THFA. A Ki value of 5.3 mM THFA was estimated by Lineweaver-Burk plot analysis of the reaction velocity versus L-THFA (0, 0.8, 1.6, and 3.0 mM) at various proline concentrations (0–200 mM). Data are shown as mean values ± standard errors of the mean from triplicate measurements. Inset is the plot of the apparent Km vs inhibitor concentration (R2 = 0.987).
Oxidative half-reaction
Because upregulation of human PRODH1 (or POX) activity leads to the formation of reactive oxygen species and has important roles in apoptosis, we sought to characterize the reactivity of Put1p with oxygen. The reactivity of Put1p with molecular oxygen or oxidase activity was evaluated at different proline concentrations in air-saturated buffer by monitoring P5C production with o-AB. Kinetic parameters for Put1p oxidase activity were kcat = 0.8 min−1 and Km = 50 mM proline. Put1p displayed similar activity in assays using an O2 electrode in which Put1p depleted oxygen at a rate of 0.54 min−1. The turnover number with molecular oxygen is 15-fold lower for Put1p than TtPRODH (12 min−1) [20]. Put1p was also tested for reactivity with sulfite. Flavoenzymes that react with molecular oxygen during catalytic turnover typically form reversible sulfite-FAD adducts [34]. Upon incubation of Put1p with sulfite, no bleaching of the absorbance at 450 nm was observed indicating no reactivity with sulfite which is consistent with Put1p displaying minimal turnover with proline and molecular xygen (data not shown).
The Put1p oxidative half-reaction was then characterized using CoQ1 as the electron acceptor. The kcat and Km values determined with CoQ1 were 9.6 ± 0.5 s−1 and 33 ± 5 µM, respectively. The kinetic parameters for proline with CoQ1 concentration held constant (100 µM) were estimated to be kcat = 13.4 ± 0.2 s−1 and Km = 59 ± 3 mM proline. The high turnover rate of Put1p with CoQ1 suggests ubiquinone is the most likely physiological electron acceptor for Put1p. The order of the reaction was then analyzed by varying proline concentration at different CoQ1 concentrations. Figure 5 shows double reciprocal plots of the reaction velocity versus proline (Figure 5A) and CoQ1 (Figure 5B). The data in both plots exhibited parallel trends that could be fit to equation 1 for a ping-pong mechanism indicating that a ternary complex is not an obligate step during catalytic turnover.
Fig. 5.
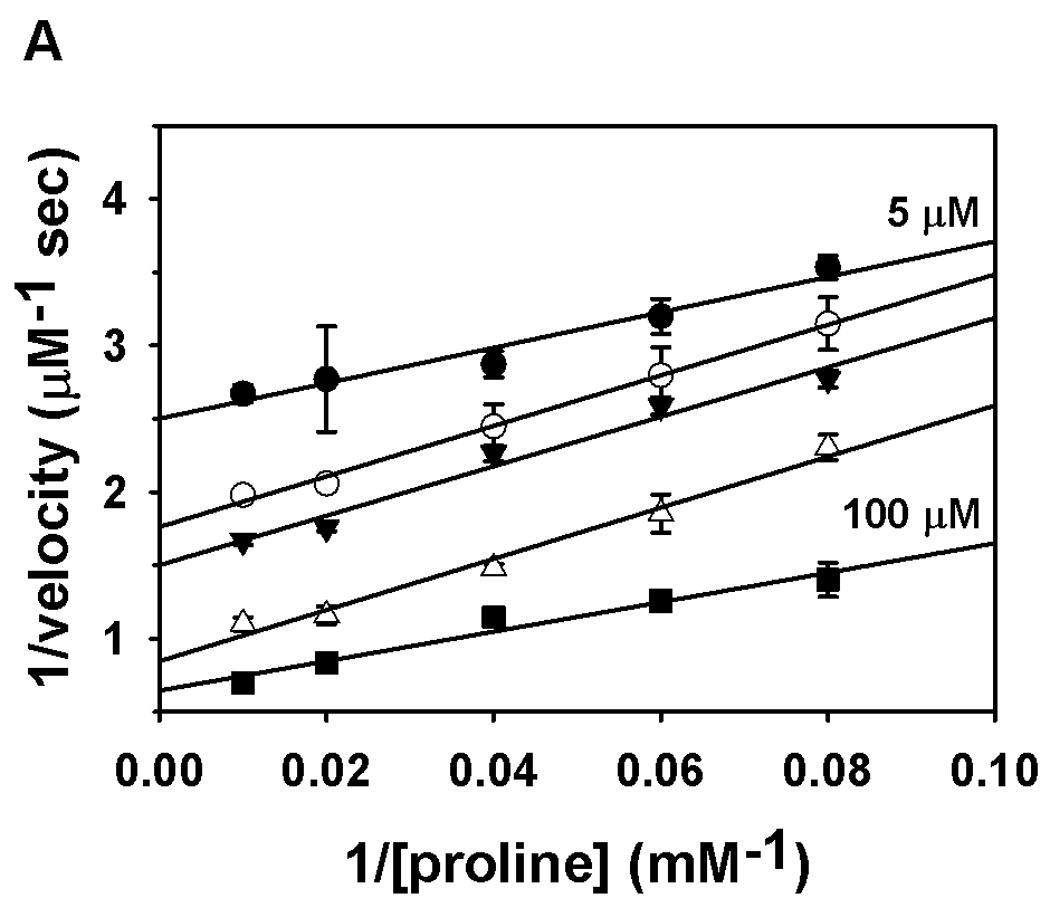
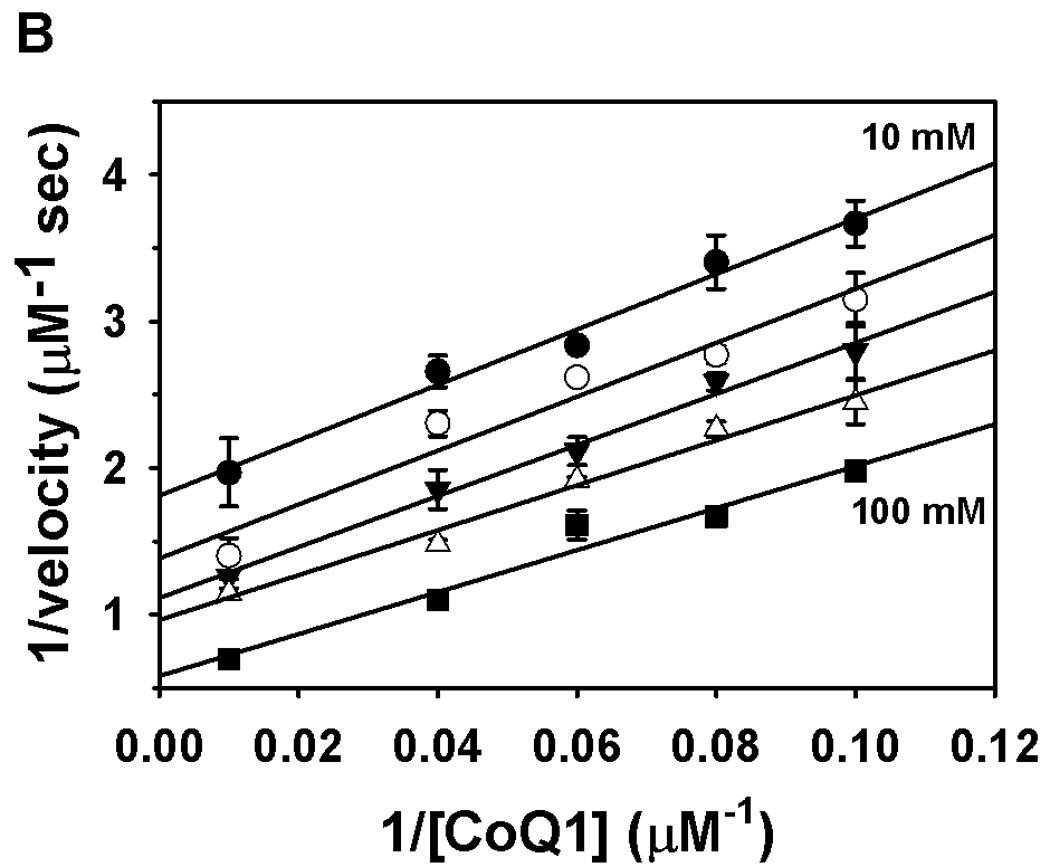
Binding order analysis of Put1p with proline and CoQ1. Panel A shows a double reciprocal plot of velocity versus proline at five different CoQ1 concentrations (5, 10, 12.5, 25, and 100 µM). Panel B shows a double reciprocal plot of velocity versus CoQ1 at five different proline concentrations (10, 12.5, 16.7, 25 and 100 mM). Assays were performed in 50 mM potassium phosphate (pH 8.0) at 25 °C. Data are shown as mean values ± standard errors of the mean from triplicate measurements. Data were fit globally to the equation for a ping-pong mechanism (R2 values of 0.97 and 0.98).
We also considered the possibility that the electron transfer flavoprotein (ETF) may act as an electron acceptor for Put1p and mediate electron transfer to the mitochondrial respiratory chain. ETF along with ETF ubiquinone oxidoreductase couples electron transfer from several enzymes involved in the oxidation of fatty acids and amino acids to the mitochondrial respiratory chain [35, 36]. Previously, high proline levels were found in the plasma of glutaric acid type II patients resulting in a condition called hyperprolinemia [37, 38]. The glutaric acid type II patients had a defect in the either ETF or ETF ubiquinone oxidoreductase. These findings suggest that proline oxidation may be coupled with the mitochondrial respiratory chain via ETF and ETF ubiquinone oxidoreductase. To test this possibility in yeast, we screened ETFα and ETFβ homolog knockout strains of S. cerevisiae for the ability to use proline as a nitrogen source. As mentioned previously, Put1p activity and the mitochondrial respiratory chain are required for S. cerevisiae to utilize proline as a source of nitrogen in ammonium deplete medium. Thus, disrupting the coupling between proline oxidation and the electron transport chain should interfere with the ability of S. cerevisiae to utilize proline. Figure 6 shows the growth phenotype of a PUT1 knockout strain versus the parental wild-type strain. Clearly the put1 strain is deficient for growth on proline as a nitrogen source relative to the wild-type strain. ETF knockout strains were also tested for growth on proline as a nitrogen source. Figure 6 shows that the ETF knockout strains (aim45 and cir1) grow similarly to the wild-type strain and have no apparent phenotype with proline as the primary nitrogen source. These results indicate that ETF is not required for proline metabolism in yeast and confirms that Put1p reduces ubiquinone directly in the mitochondrial membrane.
Fig. 6.
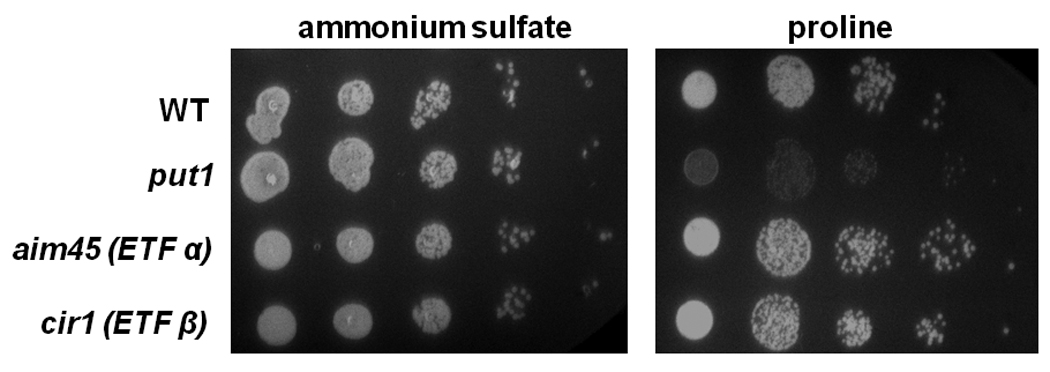
Growth of yeast strains on proline as a nitrogen source. Yeast strains BY4741 (wild-type, WT), BY4741 PUT1 knockout (put1), BY4741 AIM45 knockout (aim45, ETF α homolog), and BY4741 CIR1 knockout (cir1, ETF β homolog) were grown overnight in YC medium containing ammonium sulfate. Cells were diluted to an OD600 of 0.1 and were then serially diluted in 5-fold dilution and plated on YC plates supplemented with either 0.5% ammonium sulfate (left panel) or 0.5% proline (right panel) as a sole nitrogen source. Growth was monitored after 2–3 days incubation at 30 °C.
Discussion
Sequence comparisons among bacterial PutA/PRODHs, Put1p, and human PRODH predict that a number of residues important for substrate and FAD binding in the PRODH catalytic core are conserved [20]. The sequence similarity between Put1p (476 amino acids) and the PRODH domain of EcPutA (1–669 residues) is 39%. Put1p and human PRODH1 have 25% sequence identity and 41% similarity consistent with a shared PRODH domain and a catalytic core structure. Nine conserved motifs important for substrate (1–3, 7–9) and FAD binding (4–6) have been identified in the PRODH domain of bacterial PutA and monofunctional PRODH enzymes [20]. Of these, motifs 3, 8, and 9 are strongly conserved in Put1p and human PRODH1. Figure 7 shows a sequence alignment of motifs 3, 8 and 9 in Put1p, human PRODH1 and EcPutA. Conserved residues in these motifs which are critical for proline binding in EcPutA include Asp370, Tyr540, Arg555, and Arg556 (Asp257, Tyr430, Arg445, and Arg446 in Put1p). Important residues for FAD binding are also conserved in Put1p and human PRODH1. These residues are found in motifs 4–6 and in EcPutA include Gln404 (Gln 294, Put1p) that forms a hydrogen bond with the FAD O(2), Arg431 which hydrogen bonds to the N(5) of FAD (K321, Put1p), and His487 (His377, Put1p) that interacts with the FAD pyrophosphate [20]. Another important residue for FAD binding in EcPutA is Glu559 (motif 9) which hydrogen bonds to the ribityl 3′-OH and is conserved in Put1p (Glu449). Last, Put1p is predicted to have the α5a helix that is a unique feature of the (βα)8 barrel in EcPutA. In EcPutA, Trp438 (motif 5) from the α5a helix stacks against the adenine ring of the FAD. The corresponding residues in Put1p and human PRODH1 are Ile328 and Leu363, respectively, which would serve a similar purpose of providing non-polar interactions with the adenine ring.
Fig. 7.
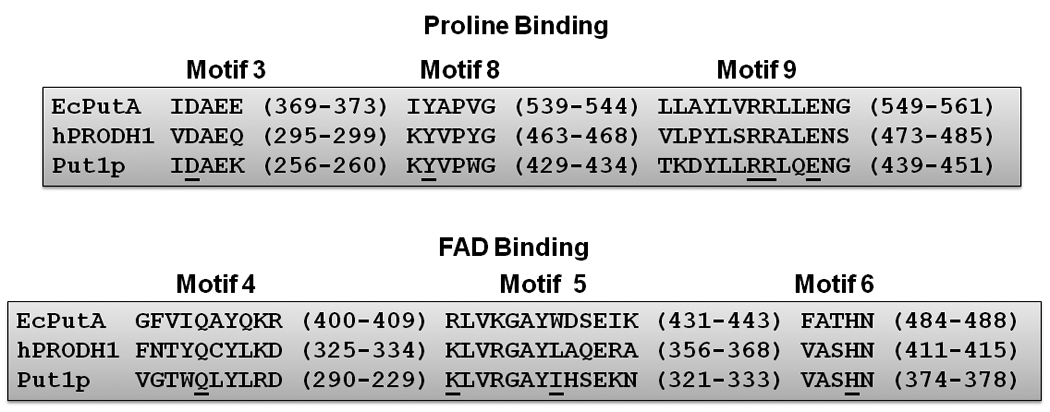
Alignment of PRODH domain sequence motifs between E. coli PutA (EcPutA), human PRODH1 (hPRODH1) and Put1p. Conserved residues with critical roles in proline and FAD interactions as mentioned in the text are underlined. Alignment was performed using GeneDoc software and the Put1p (NP_013243), hPRODH1 (O43272) and EcPutA (AAB59985) sequences.
Put1p activity with other secondary amine compounds was tested to explore how strongly L-proline is the preferred substrate. Humans have two PRODH enzyme forms that share 45% sequence identity, PRODH1 (NM_016335) which is specific for L-proline, and PRODH2, (NM_021232) which is specific for trans-4-hydroxy-L-proline. PRODH2 (OH-PRODH) catalyzes the oxidation of trans-4-hydroxy-L-proline to Δ1-pyrroline-3-hydroxy-5-carboxylate [39, 40]. Humans also have the enzyme, L-pipecolic acid oxidase, which converts L-pipecolic acid to Δ1-piperideine-6-carboxylic acid. Because S. cerevisiae encodes only one PRODH gene and does not have a gene for L-pipecolic acid oxidase, we were interested to see whether Put1p exhibited a broad substrate specificity that could accommodate variations in the five-membered pyrrolidine ring. Conserved among all bacterial PutA/PRODH enzymes is an active site Tyr residue (Tyr540 in EcPutA) that helps shape the proline binding site [39]. Interestingly, human PRODH1 and HO-PRODH differ at this corresponding residue with the Tyr replaced by a smaller Ser residue in HO-PRODH [39]. Recently, Ostrander et al. provided evidence that Tyr540 in EcPutA imposes spatial constraints in the active site that determines the preference for proline over hydroxyproline [39]. Put1p is predicted to share the corresponding Tyr residue (Tyr430, motif 8) and accordingly exhibits a strict preference for proline over hydroxyproline (2000-fold) consistent with the active site Tyr residue helping to define substrate specificity. We also did not observe significant activity with L-pipecolic acid, a result that is similar to previous studies on bacterial PRODH enzymes [29]. The strict preference for proline by Put1p further confirms the inability of S. cerevisiae to metabolize pipecolic acid and hydroxyproline.
The oxidative half-reaction of Put1p was investigated to provide molecular insights into the mitochondrial functions of human PRODH. Put1p exhibited low reactivity with molecular oxygen and high activity with ubiquinone during catalytic turnover with proline. In addition, ETF knockout strains of S. cerevisiae did not have a growth phenotype with proline as a nitrogen source demonstrating that S. cerevisiae does not require ETF for proline oxidation. Thus, in S. cerevisiae, Put1p directly couples proline oxidation to reduction of ubiquinone. How adequately these results address the properties of human PRODH are not yet clear as Put1p and human PRODH may have fundamental differences in the oxidative half-reaction step.
Acknowledgements
This research was supported by National Institutes of Health Grant GM061068 and the National Center for Research Resources Grant P20 RR-017675. This research is a contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. We thank Dr. Nandakumar Madayiputhiya in the Proteomics/Metabolomics Core Facility for performing mass spectral analysis of Put1p.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: FAD, flavin adenine dinucleotide; put, proline utilization; NAD+, nicotinamide adenine dinucleotide; PRODH, proline dehydrogenase; POX, proline oxidase; P5C, Δ1-pyrroline-5-carboxylate; P5CDH, P5C dehydrogenase; THFA, tetrahydro-2-furoic acid; DCPIP, dichlorophenolindophenol; EDTA, ethylenediaminetetraacetic acid; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; GSA, γ-glutamic acid semialdehyde; MES, 2-(N-morpholino)ethanesulfonic acid; TAPS, N-[Tris(hydroxymethyl)methyl]-3-aminopropanesulfonic acid; PIPES, piperzaine-N,N’-bis(2-ethanesulfonic acid); HEPES, 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid; ubiquinone-1, CoQ1; ETF, electron transfer flavoprotein; o-AB, o-aminobenzaldehyde; IPTG, isopropyl β-D-1-thiogalactopyranoside.
References
- 1.Phang JM, Pandhare J, Liu Y. J. Nutr. 2008;138:2008S–2015S. doi: 10.1093/jn/138.10.2008S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takagi H. Appl. Microbiol. Biotechnol. 2008;81:211–223. doi: 10.1007/s00253-008-1698-5. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan N, Doster AR, Duhamel GE, Becker DF. Infect. Immun. 2008;76:3037–3044. doi: 10.1128/IAI.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis J, Shearer G, Kohl DH. Plant Physiol. 2004;136:3313–3318. doi: 10.1104/pp.104.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohl DH, Schubert KR, Carter MB, Hagedorn CH, Shearer G. Proc. Natl. Acad. Sci. USA. 1988;85:2036–2040. doi: 10.1073/pnas.85.7.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamour N, Riviere L, Coustou V, Coombs GH, Barrett MP, Bringaud F. J. Biol. Chem. 2005;280:11902–11910. doi: 10.1074/jbc.M414274200. [DOI] [PubMed] [Google Scholar]
- 7.Brandriss MC, Magasanik B. J. Bacteriol. 1979;140:498–503. doi: 10.1128/jb.140.2.498-503.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S-S, Brandriss MC. Mol. Cell. Biol. 1987;7:4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. J. Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 10.Magasanik B, Kaiser CA. Gene. 2002;290:1–18. doi: 10.1016/s0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- 11.Brandriss MC, Magasanik B. J. Bacteriol. 1979;140:504–507. doi: 10.1128/jb.140.2.504-507.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sellick CA, Reece RJ. Embo J. 2003;22:5147–5153. doi: 10.1093/emboj/cdg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leverentz MK, Campbell RN, Connolly Y, Whetton AD, Reece RJ. J. Biol. Chem. 2009;284:24115–24122. doi: 10.1074/jbc.M109.030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller SM, Magasanik B. J. Bacteriol. 1990;172:4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Borchert GL, Surazynski A, Hu CA, Phang JM. Oncogene. 2006;25:5640–5647. doi: 10.1038/sj.onc.1209564. [DOI] [PubMed] [Google Scholar]
- 16.Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, Phang JM. Cancer Res. 2001;61:1810–1815. [PubMed] [Google Scholar]
- 17.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Borchert GL, Donald SP, Diwan BA, Anver M, Phang JM. Cancer Res. 2009;69:6414–6422. doi: 10.1158/0008-5472.CAN-09-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanner JJ. Amino Acids. 2008;35:719–730. doi: 10.1007/s00726-008-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White TA, Krishnan N, Becker DF, Tanner JJ. J. Biol. Chem. 2007;282:14316–14327. doi: 10.1074/jbc.M700912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ. Nat. Struct. Biol. 2003;10:109–114. doi: 10.1038/nsb885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S-S, Brandriss MC. Mol. Cell. Biol. 1986;6:2638–2645. doi: 10.1128/mcb.6.7.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorpe C, Matthews RG, Williams CHJ. Biochemistry. 1979;18:331–337. doi: 10.1021/bi00569a016. [DOI] [PubMed] [Google Scholar]
- 24.Brown RE, Jarvis KL, Hyland KJ. Anal. Biochem. 1989;180:136–139. doi: 10.1016/0003-2697(89)90101-2. [DOI] [PubMed] [Google Scholar]
- 25.Becker DF, Thomas EA. Biochemistry. 2001;40:4714–4722. doi: 10.1021/bi0019491. [DOI] [PubMed] [Google Scholar]
- 26.Mezel VA, Knox WE. Anal. Biochem. 1976;74:430–440. doi: 10.1016/0003-2697(76)90223-2. [DOI] [PubMed] [Google Scholar]
- 27.Grauschopf U, Fritz A, Glockshuber R. Embo J. 2003;22:3503–3513. doi: 10.1093/emboj/cdg356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lineweaver H, Burk D. J. Am. Chem. Soc. 1934;56:658–666. [Google Scholar]
- 29.Zhu W, Gincherman Y, Docherty P, Spilling CD, Becker DF. Arch. Biochem. Biophys. 2002;408:131–136. doi: 10.1016/s0003-9861(02)00535-0. [DOI] [PubMed] [Google Scholar]
- 30.Brown ED, Wood JM. J. Biol. Chem. 1993;268:8972–8979. [PubMed] [Google Scholar]
- 31.Bearne SL, Wolfenden R. Biochemistry. 1995;34:11515–11520. doi: 10.1021/bi00036a026. [DOI] [PubMed] [Google Scholar]
- 32.Krishnan N, Becker DF. Biochemistry. 2005;44:9130–1939. doi: 10.1021/bi050629k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, White TA, Schuermann JP, Baban BA, Becker DF, Tanner JJ. Biochemistry. 2004;43:12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey V, Muller F, Feldberg R, Schuman M, Sullivan PA, Howell LG, Mayhew SG, Matthews RG, Foust GP. J. Biol. Chem. 1969;244:3999–4006. [PubMed] [Google Scholar]
- 35.Toogood HS, Leys D, Scrutton NS. FEBS J. 2007;274:5481–5504. doi: 10.1111/j.1742-4658.2007.06107.x. [DOI] [PubMed] [Google Scholar]
- 36.Swanson MA, Usselman RJ, Frerman FE, Eaton GR, Eaton SS. Biochemistry. 2008;47:8894–8901. doi: 10.1021/bi800507p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Przyrembel H, Wendel U, Becker K, Bremer HJ, Bruinvis L, Ketting D, Wadman SK. Clin. Chim. Acta. 1976;66:227–239. doi: 10.1016/0009-8981(76)90060-7. [DOI] [PubMed] [Google Scholar]
- 38.Freneaux E, Sheffield VC, Molin L, Shires A, Rhead WJ. J. Clin. Invest. 1992;90:1679–1686. doi: 10.1172/JCI116040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostrander EL, Larson JD, Schuermann JP, Tanner JJ. Biochemistry. 2009;48:951–959. doi: 10.1021/bi802094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams E, Frank L. Annu. Rev. Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]


