Abstract
The SWI/SNF complex is an ATP-dependent chromatin remodeling complex that plays pivotal roles in gene regulation and cell cycle control. In the present study, we explored the molecular functions of the BAF57 subunit of SWI/SNF in cell cycle control via transcription regulation of cell cycle-related genes. We affinity purified SWI/SNF from HeLa cells stably expressing FLAG-tagged BAF47/Ini1 with or without stable shRNA-mediated knockdown of BAF57. The subunit composition of the holo- and BAF57-depleted SWI/SNF complexes from these cells were determined using a quantitative SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture)-based proteomic approach. Depletion of BAF57 resulted in a significant co-depletion of BAF180 from the SWI/SNF complex without decreasing total cellular BAF180 levels. In biochemical assays of SWI/SNF activity, the holo- and BAF57/BAF180-depleted SWI/SNF complexes exhibited similar activities. However, in cell proliferation assays using HeLa cells, knockdown of BAF57 resulted in an accumulation of cells in the G2/M phase, inhibition of colony formation, and impaired growth in soft agar. Knockdown of BAF57 also caused transcriptional misregulation of various cell cycle-related genes, especially genes involved in late G2. Collectively, our results have a identified a new role for BAF57 within the SWI/SNF complex that is required for (1) maintaining the proper subunit composition of the complex and (2) cell cycle progression through the transcriptional regulation of a subset of cell cycle-related genes.
Keywords: SWI/SNF, BAF57, BAF180, Transcription, Cell Cycle, SILAC
Introduction
A variety of histone- and chromatin-modifying complexes have evolved to facilitate the access of nuclear proteins to genomic DNA in chromatin. These include ATPase-containing chromatin remodeling complexes, which mobilize or alter the structure of nucleosomes, the fundamental repeating units of chromatin (1). SWI/SNF is chromatin remodeling complex that contains either BRG1 or BRM as an ATPase catalytic core subunit, as well as a set of BRG1/BRM associated factors (BAFs) (2). SWI/SNF plays a key role in the regulation of gene expression and, as such, regulates diverse cellular processes, including the cell cycle (3-5). Loss of SWI/SNF function is associated with cancer development, and several subunits of the complex (e.g., BRG1, BRM, BAF47/Ini1/SMARCB1) have been shown to function as tumor suppressors (3, 6). Interestingly, many gene loci encoding SWI/SNF subunits are mutated or deleted in cancers (e.g., BAF47/SMARCB1/Ini1/SNF5, BAF57/SMARCE1, and BAF180/Polybromo/PB1) (3, 6, 7). Therefore, it is not surprising that SWI/SNF is a major contributor to tumorigenesis and malignancy. A greater understanding of how individual SWI/SNF subunits contribute to the cellular function of the complex, however, is needed.
Although biochemical assays have shown that BRG1 or BRM and three BAFs (i.e., BAF170, BAF155 and BAF47) are sufficient to remodel nucleosomes (8), the roles of the other BAFs are less clear. Recent studies have suggested roles for these remaining BAFs as modulators of SWI/SNF nucleosome remodeling activity or as a determinant of SWI/SNF specificity (2). Among the BAFs, BAF57 is only present in higher eukaryotes. It has been shown to cooperate with transcriptional coregulators, such as p160/steroid receptor coactivator (SRC1) (9, 10) and methyl-CpG binding protein (MeCP2) (11), as well as bind and regulate the activity of nuclear hormone receptors, such as estrogen receptor and androgen receptor (10, 12-15). BAF57 facilitates the recruitment of SWI/SNF to nuclear receptors bound at target promoters or enhancers in response to hormone to regulate transcription (16). In the studies described herein, we have addressed whether BAF57 functions simply an adapter molecule between SWI/SNF and transcriptional regulators, or if it might play a role in regulating the composition or activity of SWI/SNF at specific promoters.
Although BAF57 is unique to metazoan SWI/SNF (17), most other subunits are well conserved from yeast to mammals (2). The existence of two functionally distinct human SWI/SNF complexes with distinct subunit compositions is also well conserved from yeast to mammals: BAF, which contains BAF250, but not BAF180, and PBAF, which contains BAF180, but not BAF250 (2, 18-20). Interestingly, the BAF180-containing PBAF complex and its homologs have been shown to play a role in cell cycle regulation (4, 5, 21-26). Furthermore, recent studies have shown that the gene encoding BAF180 is often truncated in breast cancers, leading to disregulation of cell cycle-related genes (27, 28).
In the studies described herein, we have explored the physical and functional interactions between BAF57 and BAF180 in the control of cell proliferation using proteomic, biochemical, molecular, and cell-based assays. We have found that depletion of BAF57 results in a significant co-depletion of BAF180 from the SWI/SNF complex and the accumulation of cells in the G2/M phase of the cell cycle, as well as transcriptional misregulation of cell cycle-related genes involved in late G2 phase. Collectively, our studies have shed new light on the role of BAF57 in the transcriptional control of cell proliferation in cancer cells.
Materials and Methods
Additional details about the materials and methods for each of the sections below can be found in the Supplementary Data.
Cell culture
HeLa-Ini1-11 cells (29) were obtained from the National Cell Culture Center in December 2006. The identity of the cells has regularly been re-confirmed throughout the course of these experiments by testing for the expression of FLAG-tagged BAF47/Ini1 by Western blotting (see for example Fig. 1A). The cells were maintained in Joklik-modified MEM medium (Sigma-Aldrich, Inc.) supplemented with 10% newborn calf serum. For the SILAC analyses, HeLa-Ini1-11 cells and derivatives were maintained in custom MEM medium supplemented with 10% dialyzed fetal bovine serum (Cambrex Corporation), non-essential amino acids, L-glutamine, L-leucine, penicillin/streptomycin, and either (1) “light” L-lysine and L-arginine or (2) heavy isotope-labeled amino acids, L-lysine-13C6, 15N2-HCl (Sigma-Aldrich, Inc./Isotec) and L-arginine-13C6, 15N4-HCl (Sigma-Aldrich, Inc./Isotec).
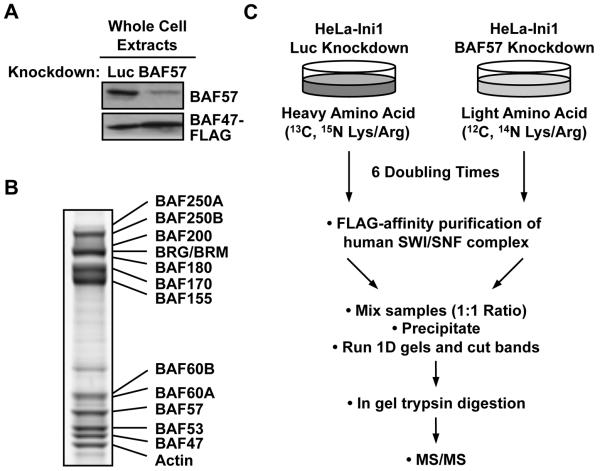
Figure. 1. Overview of SILAC and immunoaffinity purification of hSWI/SNF complex.
(A) Confirmation by Western blotting of BAF57 knockdown in whole cell extracts from HeLa-Ini1/LucKD cells and HeLa-Ini1/BAF57KD cells. (B) SDS-PAGE analysis of FLAG-affinity purified holo- and BAF57-depleted SWI/SNF complexes prepared under SILAC conditions, mixed in a 1:1 ratio, and visualized using Coomassie Blue G-250 staining. (C) Schematic diagram of the SILAC-based proteomic analysis scheme.
RNAi-mediated knockdown of BAF57 and BAF180
RNAi-mediated knockdown of BAF57 or BAF180 in HeLa-Ini1-11 cells was accomplished by retrovirus-mediated gene transfer of short hairpin RNAs (shRNAs), including control shRNAs targeting luciferase or GFP, using the pSuper.Retro system (Oligoengine, Inc.). Stable knockdown and outgrowth of cells under drug selection resulted in the HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cell lines. The target short hairpin RNA sequences used in this study are listed in the Supplementary Data.
Purification of FLAG-tagged hSWI/SNF complexes
Purification of human SWI/SNF complexes from HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells was performed by FLAG-M2 immunoaffinity purification as described previously (29).
Proteomic analysis of holo- and BAF57-depleted SWI/SNF complexes
Approximately 30 μg of purified holo- and BAF57-depleted SWI/SNF complexes mixed at 1:1 ratio were separated by SDS-PAGE and stained with Coomassie Blue G-250. In-gel trypsin digestion was performed as described previously (30). Peptides generated by in-gel digestion were analyzed by nanoflow liquid chromatography (31) using Aqua C18, 5 μm resin (Phenomenex, Inc.) for the trap column and ReproSil-Pur C18-AQ, 3 μm resin (Dr. Maisch GmbH, Ammerbuch, Germany) for the analytical column. The column eluent was analyzed by mass spectrometry, switching between MS and MS/MS acquisition. More details are provided in the Supplementary Data.
Protein identification and quantification
Raw data were converted to peak lists using Bioworks Browser 3.1 software (Thermo Finnigan, San Jose, CA). For protein identification, MS/MS data were submitted to the UniProtKB/Swiss-Prot 50.8 database using Mascot Version 2.1 (Matrix Science) using settings described in the Supplementary Data. Relative quantification ratios of identified proteins were derived by MSQuant (32). Conversion of 13C6-15N4-arginine to 13C5-15N1-proline, which was estimated as 22.5 percent, was corrected as described (33). The raw mass spectrometric data from these experiments can be downloaded from the ProteomeCommons.org Tranche Repository (34) using the following hash: A6l1v90bJmZ0FBJIikBwJD0bB0dUDniw0AfGWSoHpKZhNjyGbo2wR6DNCCB0d9+pV/khO SbbL67CmraUcSHCvWXodTwAAAAAAAAC2w==.
BAF57-BAF180 interaction assays
6xHis-BAF57 and untagged BAF180 were expressed either individually or together in Sf9 insect cells by using recombinant baculoviruses. Whole cell extracts were prepared by Dounce homogenization in 1% NP-40 lysis buffer [20 mM Tris-HCl (pH 7.9), 10% (v/v) glycerol, 500 mM NaCl, 1 mM EDTA, 1 mM DTT, plus protease inhibitors] and incubated with nickel-NTA resin (Qiagen, Inc.) at 4°C with mixing for 4 hours. The specifically bound proteins were eluted with elution buffer, subjected to immunoblotting with BAF57 and BAF180 antibodies.
Mononucleosome remodeling assays
Monucleosomes were assembled on a PCR-generated 571 bp double-stranded DNA fragment containing the 601 nucleosome positioning element as previously described (35). Mononucleosome remodeling reactions contained 32P-labeled mononucleosomes, 3 mM ATP, 1 U HhaI, and purified holo- or BAF57-depleted hSWI/SNF as indicated (0.6 to 2.4 nM). The DNA was (1) recovered by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation and (2) analyzed on a native 4% polyacrylamide gel run followed by autoradiographic detection.
Cell proliferation and soft agar growth assays
For the proliferation assays, HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells were plated at a density of 1 × 105 cells per 6 cm plate and counted at the indicated two-day intervals. For the soft agar growth assays, HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD were resuspended in a 0.3% soft agar matrix (Sigma Type VII) containing MEM medium with 5% calf serum. They were plated at a density of 4 × 103 cells per well in a 6-well dish (9.6 cm2 per well) on a pre-solidified 0.7% soft agar matrix containing basal layer. Colony formation was observed under a microscope after 14 days.
Colony formation assays
HeLa-Ini1-11 cells were plated at a density of 1 × 105 cells per well in a 6-well plate. At day 0, the cells were infected with recombinant retroviruses expressing shRNAs against GFP (control), BAF57, or BAF180 and selected with 0.5 μg/ml puromycin for 7 days. Colonies were fixed, stained with Giemsa, and subjected to automated counting using Quantity One Software (BioRad, Inc.).
Cell synchronization and cell cycle analyses
HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells were plated at a density of 1 × 106 cells per 10 cm plate and treated with either 100 nM of nocodazole or 5 μg/ml of aphidicolin for 16 hours. The cells were released from arrest, harvested by trypsinization at various time points, fixed with 70% ethanol, stained with propidium iodide, and analyzed by flow cytometry using BD FACSAria™ (BD-Biosciences, Inc.).
Gene expression analyses by reverse transcription-quantitative PCR (RT-qPCR)
HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells were arrested with aphidicolin and released from arrest as described above, then collected for RNA expression analyses. Changes in the expression of cell cycle-related genes were analyzed as described previously (36). The qPCR primer sets are listed in Supplementary Table S1.
Chromatin immunoprecipitation (ChIP) assays
ChIP-qPCR assays in HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells were performed as described previously (36). The qPCR primer sets are listed in Supplementary Table S2.
Results
BAF57 promotes the stable association of BAF180 with the hSWI/SNF complex
In this study, we used a proteomic approach in conjunction with a variety of cell-based assays to systematically address the functions of BAF57 in more detail. Specifically, we explored the role of BAF57 in determining the subunit composition of the hSWI/SNF complex. To this end, we used short hairpin RNA (shRNA) technology to stably knockdown BAF57 in HeLa-Ini1-11 cells (Fig. 1A), a HeLa-derived cell line stably expressing a FLAG epitope-tagged version of BAF47/Ini1 (29). The FLAG-tagged BAF47/Ini1 subunit allows rapid, single-step immunoaffinity purification of essentially native hSWI/SNF complex from the cells (Fig. 1B). We then used SILAC coupled with mass spectrometry to quantitatively determine the subunit composition of the complex without or with BAF57 knockdown (holo- and BAF57-depleted SWI/SNF, respectively). As illustrated in Fig. 1C, control cells (i.e., HeLa-Ini1-11 cells expressing an shRNA against luciferase; HeLa-Ini1/LucKD cells) were grown in medium containing heavy isotope-labeled L-arginine and L-lysine, whereas BAF57 knockdown cells (HeLa-Ini1/BAF57KD) were grown in the same medium containing light L-arginine and L-lysine. After six doubling times, the SWI/SNF complexes purified from the two cell lines were combined in a 1:1 ratio based on the amount of the FLAG-tagged BAF47/Ini1 subunits and then subjected to mass spectrometry analysis.
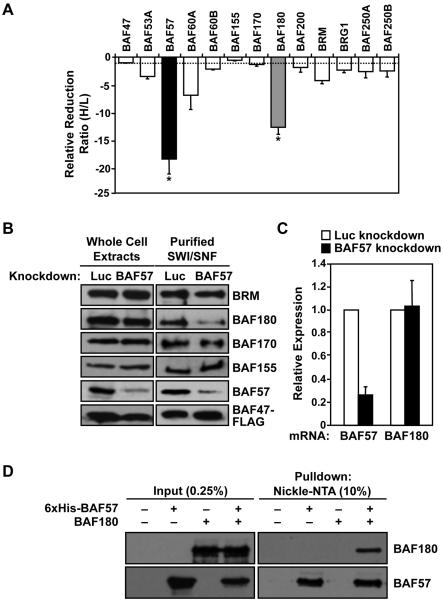
All SWI/SNF subunits were detected with high mass coverage and a large number of unique peptides from both complexes (Supplementary Table S3). Using this information, the “heavy:light” ratio of each peak pair was determined and an overall enrichment ratio for each subunit was calculated. In this study, a high ratio of heavy isotope to light isotope (>1) for a given polypeptide indicates a reduced level of the subunit in the BAF57-depleted complex, whereas a low ratio (<1) indicates an elevated level of the subunit in the BAF57-depleted complex. For example, BAF57 had a heavy:light ratio of 18.4, indicating considerably more BAF57 polypeptide in the holo-SWI/SNF complex (heavy) compared to the BAF57-depleted complex (light). Thus, as expected, BAF57 levels were reduced considerably upon knockdown (Fig. 2A). BAF57 knockdown also had a striking and statistically significant effect on the levels of BAF180, which had a heavy:light ratio of 12.7 (i.e., it was reduced by 12.7-fold in the complex upon BAF57 knockdown) (Fig. 2A). Although a number of other subunits also showed a modest reduction in levels upon BAF57 knockdown, these were not statistically significant (Fig. 2A).
Figure. 2. Knockdown of BAF57 alters the subunit composition of the hSWI/SNF complex by promoting co-depletion of the BAF180 subunit.
(A) SILAC-based proteomic analysis of the composition of FLAG-affinity purified holo- and BAF57-depleted SWI/SNF complexes. The ratios of heavy to light isotope for each peptide peak pair for each SWI/SNF subunits were determined by mass spectrometry and are expressed as the relative reduction, which is assigned a negative value to emphasize the reduction. Each bar = mean ± the range from two biological replicates. * = significant at p < 0.05, Student’s t-test. (B) Confirmation by Western blotting of changes in the subunit composition of SWI/SNF upon BAF57 knockdown. (C) Analysis of BAF57 and BAF180 mRNA levels in HeLa-Ini1/Luc cells and HeLa-Ini1/BAF57KD cells by qRT-PCR. Each bar = mean + SEM, n ≥ 5. (D) Interaction between recombinant BAF57 and BAF180 in cells in the absence of other hSWI/SNF subunits. 6xHis-BAF57 and untagged BAF180 were expressed either individually or together in Sf9 cells using recombinant baculoviruses. Interactions were assessed by nickel-NTA affinity chromatography and Western blotting with BAF57 and BAF180 antibodies.
To confirm the results of the proteomics analysis, we performed Western blot analyses with whole cell extracts and purified SWI/SNF from the HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cell lines (Fig. 2B). These results also showed a reduction in BAF180 levels within the complex upon BAF57 knockdown. In addition, BAF57 depletion did not change the total cellular levels of BAF180 (Fig. 2B; see whole cell extract) or affect BAF180 mRNA levels (Fig. 2C), indicating a direct effect of BAF57 on the association BAF180 with the SWI/SNF complex. We also found BAF180 to be depleted from SWI/SNF immunoprecipitated from BT549 human breast cancer cells, which do not express a functional BAF57 protein (9) (Supplementary Fig. 1). Furthermore, recombinant human BAF57 and human BAF180 co-expressed in insect cells interacted specifically and robustly in the absence of other human SWI/SNF subunits (Fig. 2D and Supplementary Fig. S2), suggesting a direct interaction between the two proteins. Together, our quantitative proteomic analysis and follow-up experiments indicate that BAF57 is required for the stable association of BAF180 with the hSWI/SNF complex. Given the interesting biology of BAF180, as well as its dramatic response to BAF57 knockdown, we decided to focus on the functional interplay between BAF57 and BAF180 in the remaining studies.
Knockdown of BAF57 and co-depletion of BAF180 from the SWI/SNF does not affect the nucleosome remodeling activity of SWI/SNF
To determine if knockdown of BAF57 and the co-depletion of BAF180 affects the nucleosome remodeling activity of SWI/SNF, we performed in vitro nucleosome remodeling assays using SWI/SNF complex purified from the HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cell lines. In this assay, mononucleosomes were assembled using a 571 bp 32P-end-labeled linear DNA template containing the 601 nucleosome positioning element (Fig. 3A). Upon addition of purified active hSWI/SNF and ATP, nucleosome remodeling results in the exposure of an HhaI restriction enzyme site. The extent of cleavage by HhaI, which releases a 242 bp 32P-labeled DNA fragment, is indicative of the extent of nucleosome remodeling. As shown in Figs. 3B and 3C, both the holo- and BAF57-depleted SWI/SNF complexes had similar nucleosome remodeling activities, suggesting that depletion of BAF57 and co-depletion of BAF180 does not affect the remodeling activity of the complex. This result fits well with the observation that a core SWI/SNF sub-complex comprising BRG1/BRM, BAF170, BAF155, and BAF47 retains nearly full remodeling activity (8).
Figure. 3. BAF57 depletion does not affect hSWI/SNF nucleosome remodeling activity.
(A) Schematic of the 571 bp mononucleosome containing a 601-nucleosome positioning sequence. (B) Mononucleosome remodeling assays. A two-fold titration series of intact (i.e., from HeLa-Ini1/LucKD cells) and BAF57-depleted (i.e., from HeLa-Ini1/BAF57KD cells) hSWI/SNF complexes was assessed for remodeling activity. The extent of HhaI cutting was assessed and quantified using native PAGE followed by autoradiographic imaging and phosphoimaging analysis. (C) Quantification of the mononucleosome remodeling assays like those shown in panel B. Each bar = mean + SEM, n = 3.
Knockdown of BAF57 reduces cell proliferation and growth in soft agar
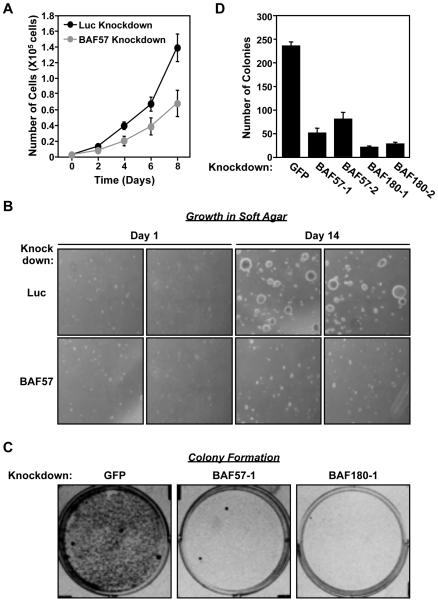
As a follow-up to the biochemical assays, we assessed the role of BAF57 in the control of cell growth and proliferation. First, we compared the rate of cell proliferation of HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells over an 8 day time course. The proliferation rate of the BAF57 knockdown cell line was reduced by approximately two-fold compared to the Luc knockdown control cell line (Fig. 4A). Next, we compared the growth of HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells in soft agar over a 14 day time course. This is a common method to monitor anchorage- independent growth, which is one of the hallmarks of cellular transformation. The size, but not the number, of foci was considerably smaller in the BAF57 knockdown cell line compared to the Luc knockdown control cell line (Fig. 4B).
Figure. 4. BAF57 knockdown decreases cell proliferation, growth in soft agar, and colony formation.
(A) Effect of stable BAF57 knockdown on the proliferation of HeLa-Ini1 cells. The data shown are for HeLa-Ini1/LucKD cells versus HeLa-Ini1/BAF57KD cells. Each point = mean ± SEM, n = 6. (B) Effect of BAF57 knockdown on the growth of HeLa-Ini1-11 cells in soft agar. The data shown are for HeLa-Ini1/LucKD cells versus HeLa-Ini1/BAF57KD cells. Two panels for each condition are shown. n = 2. (C) Effect of BAF57 or BAF180 knockdown on colony formation in HeLa-Ini1-11 cells. The cells were plated and then infected with recombinant retroviruses expressing shRNAs directed against GFP (as a control), BAF57, or BAF180, followed by selection with puromycin for 7 days. (D) Quantification of the colony formation assays like those shown in panel C. Each bar = mean + SEM, n = 3.
BAF180 knockdown is phenotypically similar to BAF57 knockdown in colony formation assays
We were unable to determine the effect of BAF180 knockdown in the proliferation and soft agar growth assays described above since HeLa-Ini1 cells and other cell lines are not viable when subjected to stable knockdown of BAF180 (data not shown). Thus, to compare the effects of BAF57 or BAF180 knockdown on cell growth in side-by-side experiments, we performed colony formation assays in HeLa-Ini1-11 cells with shRNA-mediated knockdown of BAF57 or BAF180 under appropriate drug selection, but without stable propagation of the knockdown cells. We tested two different targeting sequences for each factor and both significantly reduced colony formation when compared to control cells expressing an shRNA targeting GFP (Fig. 4C and 4D). Similar results were observed in MCF-7 human breast cancer cells (Supplemental Fig. S3). Together, the results of these assays indicate an important role for both BAF57 and BAF180 in supporting cell proliferation.
Knockdown of BAF57 and co-deletion of BAF180 from SWI/SNF promotes the accumulation of cells in G2/M phase
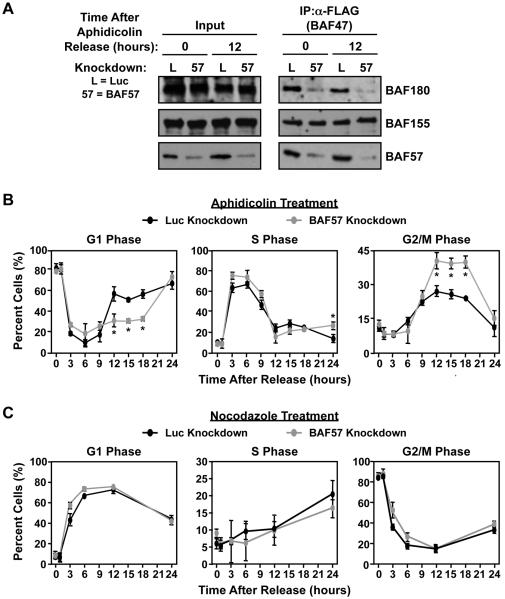
The cell growth and proliferation assays shown above (Fig. 4) suggest that depletion of BAF57 or BAF180 slows or inhibits progression through the cell cycle. To explore this possibility in more detail, we performed cell cycle analyses of HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells synchronized using aphidicholin or nocodazole, which arrest cells at the G1/S transition or G2/M transition (or early M phase), respectively (37, 38). Immunoprecipitation experiments from cells arrested with aphidicholin and released confirmed the depletion of BAF57 and BAF180 at 0 and 12 hours post-release (Fig. 5A).
Figure. 5. BAF57 knockdown increases the accumulation of cells in G2/M.
HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells were arrested either at G1/S phase using aphidicolin (5 μg/ml) (panels A and B) or at G2/M phase using nocodazole (100 nM) (panel C)(A) Knockdown of BAF57 causes the depletion of BAF180 from SWI/SNF at multiple time points after release from arrest. At the time points indicated, SWI/SNF was immunoprecipitated from nuclear extracts prepared from the cells and the levels of different subunits were analyzed by Western blotting. (B and C) After release from arrest, the cells were fixed, stained with propidium iodide followed, and analyzed by flow cytometry at the time points indicated. Each point = mean ± SEM, n = 3. * = significant at p < 0.01, Student’s t-test.
The distribution of cells at different points in the cell cycle was analyzed by flow cytometry at 0, 1, 3, 6, 9, 12, 15, 18, and 24 hours after release from arrest. Cells synchronized with aphidicholin and released from G1 arrest showed significant differences in the cell cycle progression profile between the Luc and BAF57 knockdown cells at the 12, 15, and 18 hour time points (Fig. 5B). Specifically, the BAF57 knockdown cells showed a significant reduction in G1 phase (~50% of control cells) and a significant accumulation in G2/M phase (~1.7-fold compared to the control cells). In contrast, cells synchronized using nocodazole and released from G2/M arrest did not show significant differences between the Luc and BAF57 knockdown cells (Fig. 5C). This may be due to the fact that nocodazole blocks polymerization of microtubules in early M phase (38). If BAF57 knockdown promotes accumulation in G2 phase, as suggested by Fig. 5B, then nocodazole acts after BAF57, which would allow the BAF57 cells an opportunity to “catch up” to the control cells upon arrest and abrogate any obvious effect. Together, the results of these assays show that BAF57 has a specific effect on cell cycle progression at late G2 phase.
Collectively, the results from our proteomic analyses, cell proliferation assays, and colony formation assays suggest that BAF57 and BAF180 function together within the SWI/SNF complex to support cell growth, proliferation, and progression through the cell cycle.
Knockdown of BAF57 and co-depletion of BAF180 from SWI/SNF alters the expression cell cycle-related genes
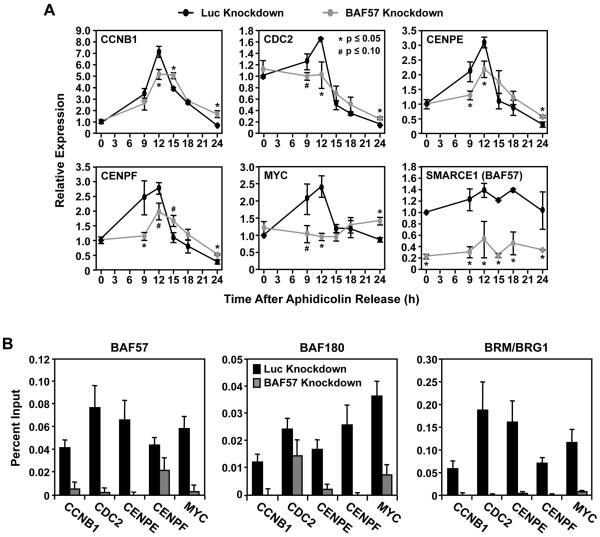
A major function of SWI/SNF is to regulate the transcriptional activation or repression of a subset of target genes (1, 2, 4). Based on the results of our cell cycle analyses shown in Fig. 5, we hypothesized that BAF57 might be required for the expression of genes involved in cell cycle progression. We compared the expression of a panel of more than 30 cell cycle-related genes between the HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells at various time points after release from aphidicholin arrest (Fig. 6A and data not shown). Interestingly, we found that BAF57 knockdown delays or inhibits the expression of a subset of genes required for progression through G2/M phase, including CCNB1, CDC2, CENPE, CENPF, CCNF, CCNG1, and CENPA (Fig. 6A and data not shown). These genes encode factors required at late G2 phase for cell cycle progression by regulating spindle checkpoint activation (see Discussion in Supplementary Data). MYC, a gene that encodes the oncogenic transcription factor c-Myc, is also down-regulated in BAF57 knockdown cells. c-Myc has positive effect on cell growth and tumorigenic transformation (39). The levels of the cognate protein products encoded by these genes are also altered during the cell cycle upon BAF57 knockdown in a manner that, for the most part, reflects the expression of the genes (Supplementary Fig. S4), although such an analysis does not account for alternate regulatory mechanims (e.g., microRNAs, intrinsic RNA stability). In contrast, many of the G1/S phase-regulating genes that we screened (e.g., CCND1, CCNE1) were slightly up-regulated or unchanged in the BAF57 knockdown cells compared to the control cells (data not shown), indicating that cell cycle progression is not compromised at G1/S phase, as expected from the cell cycle analyses.
Figure. 6. BAF57 knockdown alters the expression of genes required for G2/M progression, as well as the recruitment of SWI/SNF to target gene promoters.
(A) The expression of cell cycle-related genes in HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells was analyzed using qRT-PCR at the time points indicated post-release from aphidicolin arrest. The results are expressed relative to the 0 hour time point in the HeLa-Ini1/LucKD cells. Each bar = mean + SEM, n ≥ 5. * and # = significant at p ≤ 0.05 and p ≤ 0.10, respectively, by Student’s t-test. (B) ChIP-qPCR analysis of the recruitment of SWI/SNF subunits at the promoters of cell cycle-regulated genes. Each bar = mean + SEM, n ≥ 3.
A potential role of the BAF subunits of SWI/SNF is to promote gene-specific recruitment of the complex to a subset of genes. To determine how BAF57 knockdown and concomitant loss of BAF180 from the SWI/SNF complex can affect SWI/SNF recruitment to promoters to alter gene expression, we performed chromatin immunoprecipitation (ChIP) assays with antibodies against BRM/BRG1, BAF57, and BAF180 in HeLa-Ini1/LucKD and HeLa-Ini1/BAF57KD cells, focusing on the promoters of the G2/M-related genes noted above. As expected, knockdown of BAF57 abolished the BAF57 signal at the promoters of these genes (Fig. 6B). Also as expected, knockdown of BAF57 inhibited the recruitment of BAF180 to the same promoters (Fig. 6B). Moreover, knockdown of BAF57 blocked the recruitment of BRM/BRG1 (Fig. 6B). These results indicate that although a BRM/BRG1-containing complex remains intact in the cells upon BAF57 knockdown and co-depletion of BAF180 (Fig. 2B), it loses its ability to be properly recruited to the promoters of key SWI/SNF target genes (Fig. 6B). Together, the results of our gene regulation assays suggest that BAF57 knockdown and co-depletion of BAF180 from the SWI/SNF complex affects the expression of a subset of genes that are required for progression through G2 phase.
Discussion
In this study, we have examined the role of the BAF57 subunit in transcriptional regulation and cell proliferation using a variety of biochemical, proteomic, molecular, and cell-based assays. We have found that RNAi-mediated depletion of BAF57 from cells: (i) alters the composition of the SWI/SNF complex by promoting the dissociation (or preventing the association) of BAF180, (ii) decreases the rate of cell proliferation by promoting the accumulation of cells in the late G2 phase, (iii) alters the composition or prevents the association of SWI/SNF at target gene promoters, and (iv) promotes the down-regulation of a subset of gene that are required for cell cycle progression from G2 to M phase. These findings highlight the functional interplay between SWI/SNF subunits within the complex that regulate biological outcomes, such as cell proliferation.
A role for BAF57 in the regulation of gene expression and cell proliferation by SWI/SNF
SWI/SNF regulates the transcription of subsets of genes controlling key biological processes, including the cell cycle and tumorigenesis (4-6). The ability of SWI/SNF to be (i) targeted to specific promoters by sequence-specific DNA binding transcriptional regulators and (ii) mobilize or structurally alter promoter nucleosomes to make it ideally suited for its role in transcriptional regulation (1, 2). Although a core SWI/SNF complex containing BRG1 or BRM and three BAFs (i.e., BAF170, BAF155 and BAF47) retains its ability to remodel nucleosomes (8), the remaining BAFs likely function as modulators of SWI/SNF nucleosome remodeling activity or as a determinant of SWI/SNF specificity (2).
BAF57, a SWI/SNF subunit unique to higher eukaryotes (17), plays a role in transcriptional regulation by binding to (i) DNA through its HMG-like domain and (ii) transcriptional regulators through protein-protein interactions (10, 17, 40). Our results from HeLa cells indicate that BAF57-dependent gene expression is required for normal progression through the cell cycle. Likewise, previous studies have shown that inhibition of BAF57 with an amino-terminally-deleted BAF57 dominant negative mutant or an inhibitory polypeptide reduces androgen receptor-dependent cell proliferation (14, 15). In contrast to these studies showing a positive role for BAF57 in cell cycle progression, other studies have suggested a negative role. For example, re-expression of BAF57 in a BAF57-deficient cell line induces cell cycle arrest and restores contact inhibition, which is accompanied by the induction of a wide variety of genes (41). These contrasting roles of BAF57 - both as a promoter and inhibitor of cell proliferation - suggest that BAF57 function may be context-specific or may require careful regulation of BAF57 levels within a narrowly defined range.
A role for BAF57 in determining the composition of SWI/SNF: Effects on BAF180
Our results showing that BAF57 (i) interacts with BAF180 and (ii) is required for the stable association of BAF180 with SWI/SNF in multiple cell types provides new insights about the assembly and function of the complex. The loss of BAF57 results in the loss of BAF180 from the complex and a loss of SWI/SNF recruitment to target gene promoters. The link that we have uncovered between BAF57 and BAF180 is particularly interesting because the latter is the signature subunit of the PBAF complex, one of two functionally distinct mammalian SWI/SNF complexes that have distinct subunit compositions (2). Previous studies of BAF180 within PBAF have demonstrated a role in cell cycle regulation at G2/M phase, possibly through a mechanism involving the localization of PBAF to the kinetochores of mitotic chromosomes (21). BAF180 has also been implicated in the transcriptional regulation of cell cycle-related genes, such as p21 (27). These results fit well with our observations that knockdown of BAF57 and co-depletion of BAF180 from the complex leads to the misregulation of a subset of genes that are required for cell cycle progression from G2 to M phase.
Collectively, our results have a identified a new role for BAF57 within the SWI/SNF complex that is required for the proper regulation of the cell cycle through the transcriptional regulation of a subset of cell cycle-related genes. In addition, our results suggest that the effects of BAF57 are mediated, at least in part, through the association of BAF180 with the complex, which might play a role in directing the assembly of the PBAF versus BAF complexes. Interestingly, the cellular levels of BAF57 protein are precisely regulated by other subunits in the complex (42), which suggests a complex regulatory network controlling the composition and activity of SWI/SNF.
Supplementary Material
Acknowledgements
The authors would like to thank Marcel G.T. Winter for technical support, Weidong Wang for the BAF57 cDNA, Ramon Parsons for the BAF180 cDNA, Robert Kingston and James Kadonaga for recombinant baculoviruses, Hans Salamanca for technical assistance with the soft agar growth assays, and Raga Krishnakumar for critical reading of this manuscript.
References
- 1.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 2.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–42. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 4.Moshkin YM, Mohrmann L, van Ijcken WF, Verrijzer CP. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol Cell Biol. 2007;27:651–61. doi: 10.1128/MCB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muchardt C, Yaniv M. When the SWI/SNF complex remodels…the cell cycle. Oncogene. 2001;20:3067–75. doi: 10.1038/sj.onc.1204331. [DOI] [PubMed] [Google Scholar]
- 6.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 7.Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang W, Weissman BE. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol. 2001;186:136–45. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–53. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 9.Kiskinis E, Garcia-Pedrero JM, Villaronga MA, Parker MG, Belandia B. Identification of BAF57 mutations in human breast cancer cell lines. Breast Cancer Res Treat. 2006;98:191–8. doi: 10.1007/s10549-005-9149-9. [DOI] [PubMed] [Google Scholar]
- 10.Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. Embo J. 2002;21:4094–103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harikrishnan KN, Chow MZ, Baker EK, et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet. 2005;37:254–64. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Pedrero JM, Kiskinis E, Parker MG, Belandia B. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J Biol Chem. 2006;281:22656–64. doi: 10.1074/jbc.M602561200. [DOI] [PubMed] [Google Scholar]
- 13.Huang CY, Beliakoff J, Li X, et al. hZimp7, a novel PIAS-like protein, enhances androgen receptor-mediated transcription and interacts with SWI/SNF-like BAF complexes. Mol Endocrinol. 2005;19:2915–29. doi: 10.1210/me.2005-0097. [DOI] [PubMed] [Google Scholar]
- 14.Link KA, Burd CJ, Williams E, et al. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25:2200–15. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Link KA, Balasubramaniam S, Sharma A, et al. Targeting the BAF57 SWI/SNF subunit in prostate cancer: a novel platform to control androgen receptor activity. Cancer Res. 2008;68:4551–8. doi: 10.1158/0008-5472.CAN-07-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Kinyamu HK, Archer TK. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol. 2006;20:1–13. doi: 10.1210/me.2005-0192. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree GR. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci U S A. 1998;95:492–8. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohrmann L, Langenberg K, Krijgsveld J, Kal AJ, Heck AJ, Verrijzer CP. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol Cell Biol. 2004;24:3077–88. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie Z, Xue Y, Yang D, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–88. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr Top Microbiol Immunol. 2003;274:143–69. doi: 10.1007/978-3-642-55747-7_6. [DOI] [PubMed] [Google Scholar]
- 21.Xue Y, Canman JC, Lee CS, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A. 2000;97:13015–20. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu JM, Huang J, Meluh PB, Laurent BC. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol. 2003;23:3202–15. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Hsu JM, Laurent BC. The RSC nucleosome-remodeling complex is required for Cohesin’s association with chromosome arms. Mol Cell. 2004;13:739–50. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 24.Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Y, Cairns BR, Kornberg RD, Laurent BC. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–34. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns BR, Lorch Y, Li Y, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–60. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 27.Xia W, Nagase S, Montia AG, et al. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667–74. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekine I, Sato M, Sunaga N, et al. The 3p21 candidate tumor suppressor gene BAF180 is normally expressed in human lung cancer. Oncogene. 2005;24:2735–8. doi: 10.1038/sj.onc.1207694. [DOI] [PubMed] [Google Scholar]
- 29.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–51. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 31.Meiring H, Van der Heeft GJtH E., Jong APJMd. Nanoscale LC–MS(n): Technical design and applications to peptide and protein analysis. Journal of Separation Science. 2002;25:557–68. [Google Scholar]
- 32.MSQuant [Internet] Center for Experimental Bioinformatics (CEBI) at the University of Southern Denmark; Denmark: [cited 2010 March 27]. Available from: http://msquant.alwaysdata.net/ [Google Scholar]
- 33.Mousson F, Kolkman A, Pijnappel WW, Timmers HT, Heck AJ. Quantitative proteomics reveals regulation of dynamic components within TATA-binding protein (TBP) transcription complexes. Mol Cell Proteomics. 2008;7:845–52. doi: 10.1074/mcp.M700306-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Proteome Commons Tranche Repository [Internet] ProteomeCommons.org; Ann Arbor (MI): [cited 2010 March 27]. Available from: https://proteomecommons.org/tranche/ [Google Scholar]
- 35.Lee KM, Narlikar G. Assembly of nucleosomal templates by salt dialysis. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb2106s54. Chapter 21: Unit 21 6. [DOI] [PubMed] [Google Scholar]
- 36.Kininis M, Chen BS, Diehl AG, et al. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27:5090–104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lalande M. A reversible arrest point in the late G1 phase of the mammalian cell cycle. Exp Cell Res. 1990;186:332–9. doi: 10.1016/0014-4827(90)90313-y. [DOI] [PubMed] [Google Scholar]
- 38.Burke DJ. Complexity in the spindle checkpoint. Curr Opin Genet Dev. 2000;10:26–31. doi: 10.1016/s0959-437x(99)00040-4. [DOI] [PubMed] [Google Scholar]
- 39.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–48. [PubMed] [Google Scholar]
- 40.Pal S, Yun R, Datta A, et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–87. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Baiocchi RA, Pal S, Mosialos G, Caligiuri M, Sif S. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol Cell Biol. 2005;25:7953–65. doi: 10.1128/MCB.25.18.7953-7965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Archer TK. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol Cell Biol. 2005;25:9016–27. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.