Abstract
We demonstrate a fluorescence-based, label-free detection scheme that reports the presence of Hg(II) ion using photon upconverting nanoparticles. A single-stranded DNA containing a number of thymine bases to be used as the Hg2+-capturing element is covalently attached to the photon upconverting NaYF4:Yb3+,Tm3+ nanoparticles. Under the illumination of 980 nm laser, energy transfer takes place between the NaYF4:Yb3+,Tm3+ nanoparticles as the donor and SYBR green I, a DNA intercalating dye, as the acceptor. By monitoring the ratio of the acceptor emission to the donor emission, we can quantitatively detect the presence of the mercuric ions with a directly observed detection limit of 0.06 nM. The remarkably high signal-to-noise ratio of photon upconverting particles leads to very high sensitivity and specificity without the need of fluorophore labeling. The sensor also does not suffer from photobleaching.
Keywords: Photon upconversion, label-free detection, mercury ion, oligonucleotide, energy transfer
1. Introduction
Mercury is a pollutant with well-known toxicological effects (Hylander et al., 2006; Zahir et al., 2005). The inorganic mercuric ion is a carcinogenic material with high cellular toxicity (Zheng et al., 2007). In aquatic sediments, the mercuric ion can be converted into methyl mercury through microbial methylation. The subsequent accumulation of methyl mercury by humans through the food chain may lead to serious damage to the brain. Therefore, it is important for the routine detection of mercuric ion in the monitoring of larger bodies of water and the safety evaluation of aquatically derived food supplies.
Traditionally, the methods developed for the detection of mercuric ion include atomic absorption/emission spectroscopy, inductively coupled plasma atomic mass spectrometry and electrochemical sensor (Butler et al., 2006; Fong et al., 2007; Wang et al., 1998), which can achieve very low detection limit (<0.2 μg/L). Yet they tend to involve laborious procedures. In recent years, there have been increasing number of reports on alternative methods using colorimetric sensors and fluorescence sensors, with varying degree of success regarding the sensitivity, selectivity and stability (Ono et al., 2004; Zhu et al., 2006; Kim et al., 2006; Lee et al., 2007; Nolan et al., 2008; Che et al., 2008; Chiang et al., 2008; Wang et al., 2008). In light of the maximum allowable level for mercuric ion in drinkable water being 10 nM as set by the US Environmental Protection Agency (US EPA 2005), there is ongoing demand for development of detection methods that are sensitive, specific and ease-of-use. Here we report a luminescence-based method to detect mercuric ion using photon upconverting nanoparticles, which is highly sensitive and specific.
Photon upconversion is a phenomenon where higher energy photons are emitted after the material is excited by lower energy photons through multiphoton processes. NaYF4-based materials (doped with Yb3+ and Er3+ or Tm3+) have been recognized as one of the most efficient photon upconverting material. Because photon upconverting materials are much less common than their downconverting counterparts (Auzel 2004), they are ideal for sensing applications, as a significantly high signal-to-noise ratio can be achieved. There have been reports of using photon upconverting phosphor particles to detect single-stranded nucleic acids and proteins, either through direct labelling or through sandwich assay format (van de Rijke et al., 2001; Corstjens et al., 2003; Zhang et al., 2006; Kumar et al., 2009). We have previously demonstrated a design of label-free scheme to detect a DNA strand based on the energy transfer between the photon upconverting nanoparticles and an intercalating dye, with high sensitivity and specificity (Kumar and Zhang, 2009). In this report, we show that a variation of the method can detect mercuric ions in aqueous medium with very high sensitivity and specificity.
2. Materials and methods
2.1 Chemicals and materials
All chemicals were used as received without further purification. Sodium fluoride (NaF), Yttrium chloride (YCl3·6H2O), Ytterbium chloride (YbCl3·6H2O), Thulium nitrate (Tm(NO3)3·5H2O), diethylenetriaminepentaacetic acid (DTPA), and 2,6-pyridinedicarboxylic acid (PDCA) were purchased from Sigma-Aldrich. SYBR Green I was purchased from Invitrogen. 2-(4-Morpholino)-ethane sulfonic acid (MES), 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide HCl (EDC), barium chloride, cadmium nitrate, copper nitrate, lead nitrate, nickel nitrate, zinc nitrate were purchased from Fisher Scientific. DNA strands were purchased from Integrated DNA Technologies Inc (Coralville, IA). TEM grids were from Electron Microscopy Sciences, PA.
2.2 Synthesis of photon upconverting nanoparticles
The NaYF4:Yb3+,Tm3+ nanoparticles were synthesized using a hydrothermal method as describe previously (Kumar and Zhang, 2009). In brief, 815 μl of YCl3, 170 μl of YbCl3, and 15 μl of TmCl3 (all of 0.2 M) were mixed together in a Teflon container along with 1 ml of 0.2 M DTPA and 10 ml of DI water. After 25 minutes 3 ml of 0.83 M NaF was added under continuous stirring. The Teflon container was then placed inside an autoclave, and heated at around 160 °C for 20 hours. The resulting nanoparticles were collected by adding large volume of acetone and then centrifuging at 14000 rpm for 20 minutes. Collected nanoparticles were washed several times with acetone and dried before further use.
2.3 Characterizations of the photon upconverting nanoparticles
The NaYF4:Yb3+,Tm3+ nanoparticles were first suspended in methanol through sonication to ensure the extraction of representative dispersion volumes. Then a drop of suspension was deposited on a Formvar-covered carbon-coated copper grid (Electron Microscopy Sciences, PA) and let dried at room temperature. The nanoparticles were analyzed using a JEOL TEM at an accelerating voltage of 80 kV.
2.4 FTIR measurement
A drop of nanoparticle suspension in ethanol was deposited on a plate of sodium chloride crystal, and allowed to dry at room temperature. Infrared absorption spectra were taken on a Nicolet Nexus 8700 FTIR spectrometer.
2.5 Attachment of DNA to the photon upconverting nanoparticles
All DNA were synthesized by IDT Technologies, Inc. (Coralville, IA). An amine-modified single-stranded DNA (5′-AmineC6-TTTGGTTTGTTCCCTTCTTTCCTTT-3′) is bound covalently to the NaYF4:Yb3+,Tm3+ nanoparticle through their –COO− groups following the widely used EDC method (Walsh et al., 2001). Procedures to attach the amine-modified DNA to the photon upconverting nanoparticles with -COOH groups were as follows: 55 mg of nanoparticles were mixed with 54 mg of EDC in 550 μL of MES buffer (100 mM, pH 4.5) under stir. 1080 μL 50 μM of theee amine-modified DNA was then added to the mixture. The mixture was continuously stirred for 2 hours before 500 μL of L of 100 mM phosphate buffer (pH 8.0) was added. Large amount of acetone was next added to the mixture to precipitate the nanoparticles out of the solution and collected via centrifuge. The conjugated nanoparticles were subsequently washed with 100 mM phosphate buffer (pH 8.0) for three to four times, before left dried in the oven overnight and weighed. The weighed conjugated particles were then suspended in 600 μL of 100 mM phosphate buffer solution (pH 7.0, 1 mM EDTA) and stored at 4 °C for later use.
2.6 Photoluminescence Measurements
The excitation/emission wavelengths for SYBR Green I are 490/515 nm, respectively, as measured on our spectrofluorometer. The photoluminescence measurements using DNA/nanoparticle complex were carried out at 50 °C using a spectrofluorometer (Photon Technologies International) equipped with a Hamamatsu R928 photomultiplier tube (PMT) operating at 1kV. Instead of using the built-in Xenon lamp as the illumination source, a 980 nm diode laser (CrystaLaser, NV) was attached to the side port of the spectrofluorometer, and used to excite the sample. The output of the laser was set at 400 mW for all measurements. The emission slit was set at 4 nm for all measurements.
For each run, 100 μL of DNA/nanoparticle complex solution was taken into a cuvet after sonicating the stock solution to ensure homogenous suspension of nanoparticles in phosphate buffer. Then 250 μL of phosphate buffer (10 mM, pH 8.0) solution and 10 μL of 75 μM SYBR Green I solution were added to the cuvet. The mixture was treated as blank, and photoluminescence spectrum was taken. An increment of the targets (50 nM of mercuric ion or other metal ions, mixed with 6.8 mM of PDCA) was then added to the blank, and spectra were taken. There was a 10-minute waiting period between every addition of the targets and the measurement, to allow for complete hybridization.
3. Results and discussion
A TEM image of photon upconverting nanoparticles is shown in Figure 1. A photoluminescence spectrum of the photon upconverting nanoparticles is shown in Figure 2. A time course measurement of nanoparticle emission excited by the intense 980 nm laser is shown in Figure S1, which demonstrates that such photon upconverting nanoparticles are not susceptible to photobleaching.
Figure 1.
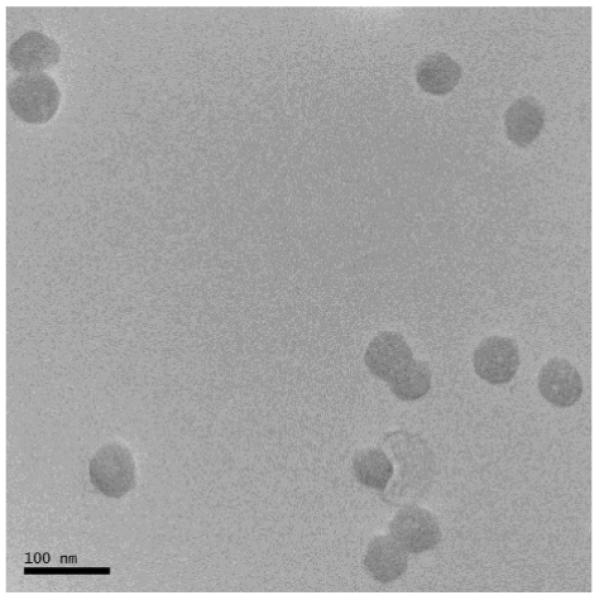
TEM image of the NaYF4:Yb3+,Tm3+ nanoparticles.
Figure 2.
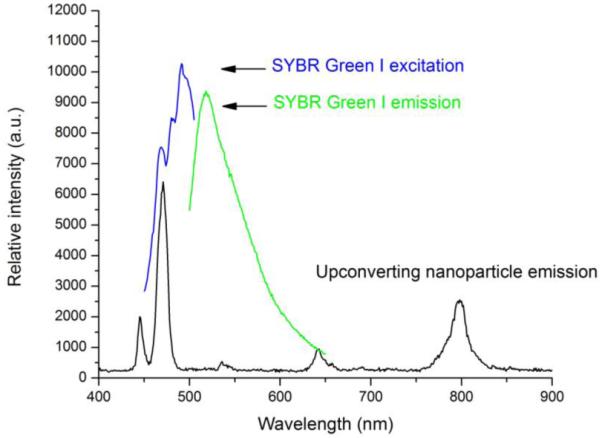
Photoluminescence spectrum of the NaYF4:Yb3+,Tm3+ nanoparticles, excited by a 980 nm diode laser. The excitation and emission spectra of SYBR Green I are included for comparison.
Most of the photon upconverting nanoparticles reported in the literature are hydrophobic. There have been a lot of interests and efforts in converting them to hydrophilic for biologically related applications. Typically, SiO2 (Hilderbrand et al., 2009; Zhang et al., 2007) or polymer coating (Wang et al., 2005; Naccache et al., 2009) and ligand exchange (Yi et al., 2006; Boyer et al., 2010) are adopted for this purpose. In this regard, the synthetic method used in this work demonstrates the advantage in that the resulting photon upconverting nanoparticles are highly water dispersible after the synthesis without the need of surface modification, probably due to the presence of the surface carboxylate groups. The FTIR spectrum of the photon upconverting nanoparticles is shown in Figure S2. The 1589 and 1400 cm−1 bands are most likely associated with the asymmetric and symmetric stretching vibrations of the surface carboxylate group. The surface carboxylate groups not only render the nanoparticles hydrophilic but also facilitate further functionalization readily if other agents are to be attached to the nanoparticle surface, as is the case in this study.
Thymine-Hg2+-Thymine (T-Hg2+-T) chemistry has been actively explored in the development of Hg2+ sensors, as T-T mismatch displays high selectivity toward Hg2+ against many other metal ions (Miyake et al., 2006). Our detection scheme is also based on this chemistry, as shown in Figure 3. The underlying principle for the detection is luminescence resonance energy transfer. NaYF4:Yb3+,Tm3+ nanoparticles are used as the energy donor. Carboxyl groups are crafted onto the surface of these NaYF4:Yb3+,Tm3+ nanoparticles during the synthesis. Upon excitation by an infrared (~980 nm) laser, strong visible bands appear around 477, 650 and 800 nm. These bands correspond to the transitions from 1G4, 3F2 and 3F4 to 3H6 of Tm3+, respectively. The 477-nm emission matches well with the absorption of an intercalating dye, SYBR Green I, used as the energy acceptor.
Figure 3.
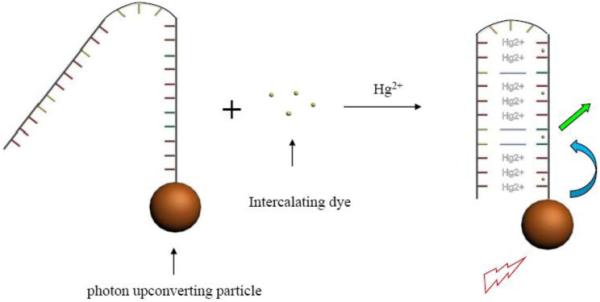
Schematic of mercuric ion detection.
SYBR Green I is mixed in the solution containing the DNA-nanoparticle complex. If there is no mercuric ion in the solution, the quantum yield of SYBR Green I, either free in solution or bound to the single-stranded DNA, is rather low. Although SYBR Green I may be close to the NaYF4:Yb3+,Tm3+ nanoparticles through binding to the single-stranded DNA, the emission of SYBR Green I would be insignificant under infrared illumination. In the presence of mercuric ion, the single-stranded DNA forms a hairpin structure due to the T-Hg2+-T binding. The long stem of the hairpin, being double-stranded DNA, can intercalate SYBR Green I and significantly increase its quantum yield. Under infrared illumination, the emission of the NaYF4:Yb3+,Tm3+ nanoparticles will be transferred to the nearby SYBR Green I, leading to the emission of SYBR Green I. By monitoring the emission of SYBR Green I, one can detect the presence of mercuric ions. Note that SYBR Green I do not bind covalently to neither DNA strands nor nanoparticles in this scheme. “Label-free” detections do not exclude the use of fluorophores, nor do they exclude the fact that target-capturing elements are attached to some kinds of surface (e.g. DNA probes immobilized on surface in SPR technique). In this case, the Hg2+-capturing DNA strands are covalently attached to the surface of the photon upconverting nanoparticles.
All assays were carried out on a spectrofluorometer at 50 °C. Briefly, a certain amount of DNA-nanoparticle complex was first mixed with SYBR Green I in phosphate buffer (final concentration of SYBR Green I being ~2 μM) containing an excess amount (~6.8 mM) of 2,6-pyridinedicarboxylic acid (PDCA). The mixture was illuminated by an IR laser of 980 nm, and photoluminescence spectrum was taken. The ratio of SYBR Green I emission to the NaYF4:Yb3+,Tm3+ nanoparticles emission, denoted as Iblank, was measured. Stock solution of mercuric ion (50 nM) in small increments was then added to the mixture. After 10 minutes of hybridization, the same ratio of emissions, denoted as Isignal, was again measured while the sample was being illuminated by the IR laser. The photoluminescence spectra of the mixture before and after the addition of mercuric ion are shown in Figure 4. The energy transfer between the NaYF4:Yb3+,Tm3+ nanoparticles and SYBR Green I is indicated by the simultaneous decrease of the 477-nm emission of the nanoparticles and the increase of signals corresponding to SYBR Green I emission (490nm-520nm region).
Figure 4.
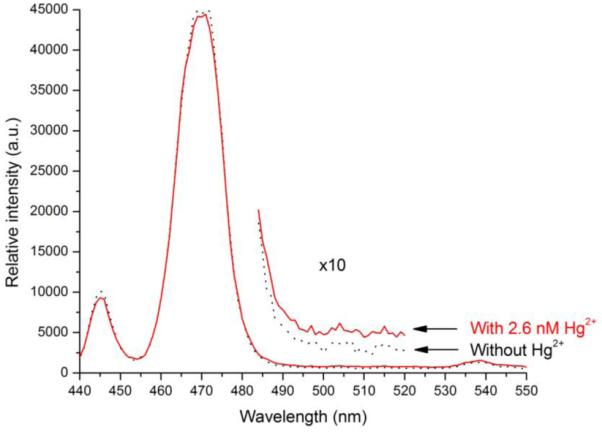
Photoluminescence spectra of the photon upconverting nanoparticles before (dark) and after (red) the addition of 2.6 nM of Hg ions, in the presence of SYBR Green I (~2 μM) and PDCA (~6.8 mM).
The titration curve is shown in Figure 5, where Isignal/Iblank is plotted against the mercuric ion concentration. We have determined the sensitivity of this detection scheme. The data points in Figure 5 lead to a directly observed detection limit of 0.06 nM for mercuric ion, which is more than two orders of magnitude below the EPA limit. This high sensitivity can be attributed to the following factors: 1) The IR laser used to excite the photon upconverting nanoparticles does not directly excite SYBR Green I. Nor does the IR laser excite any possible fluorescent impurities in the sample. Thus the background signal is extremely low. 2) There is an approximate 11-fold increase in the quantum yield of SYBR Green I when binding to double-stranded DNA (~0.8), as compared to single-stranded DNA (Zipper et al., 2004). This would minimize the interference from the free SYBR Green I present in the solution or those bound to single-stranded DNA. 3) The narrow emission from the NaYF4:Yb3+,Tm3+ nanoparticles allows one to monitor signals at a wavelength where there is little overlap between the emissions of the photon upconverting nanoparticles (donor) and SYBR Green I (acceptor). It is worthwhile to point out that the NaYF4:Yb3+,Tm3+ nanoparticles are extremely stable against photobleaching, which allows more intense laser to be used to increase the signal level. This is an advantage over the fluorescence-based detection schemes, where photobleaching hinders the use of too powerful an excitation source.
Figure 5.
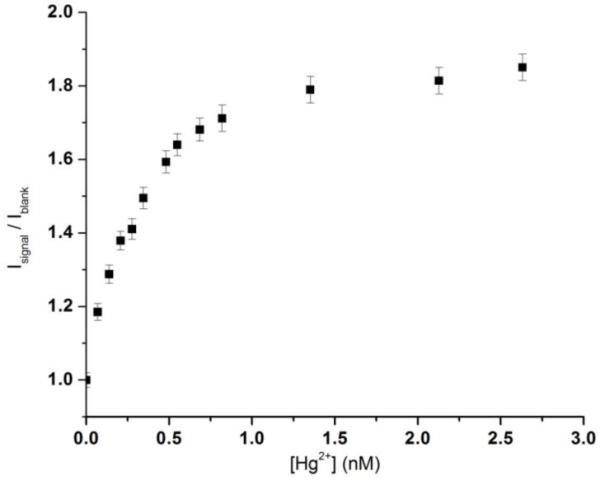
Isignal/Iblank vs. [Hg ion] in buffer solution.
The selectivity of this detection scheme for mercuric ion is also investigated. It has been reported in the literature that PDCA would mask many potentially interfering metal ions and help improve the detection specificity toward mercuric ions (Li et al., 2009; Darbha et al., 2007). We adopt the same strategy and add an excess amount of PDCA in the assay mixture. A number of common divalent metal ions, Ba2+, Cd2+, Cu2+, Ni2+, Pb2+, Zn2+, are individually added into the mixture of SYBR Green I, PDCA, and the DNA-functionalized NaYF4:Yb3+,Tm3+ nanoparticles. As shown in Figure 6, the Isignal/Iblank values for these potentially interfering metal ions at different concentrations are fluctuating around one, essentially representing the noise of the measurements. They are significantly below the value of Isignal/Iblank at 0.06 nM of mercuric ion. The high specificity most likely results from a few factors: 1) Strong T-Hg2+-T interaction mediates the formation of hairpin structure of DNA. 2) The presence of PDCA masks the potentially interfering metal ions. 3) Elevated operating temperature reduces the possibility of forming hairpin structure of DNA through other routes than Hg2+ mediation.
Figure 6.
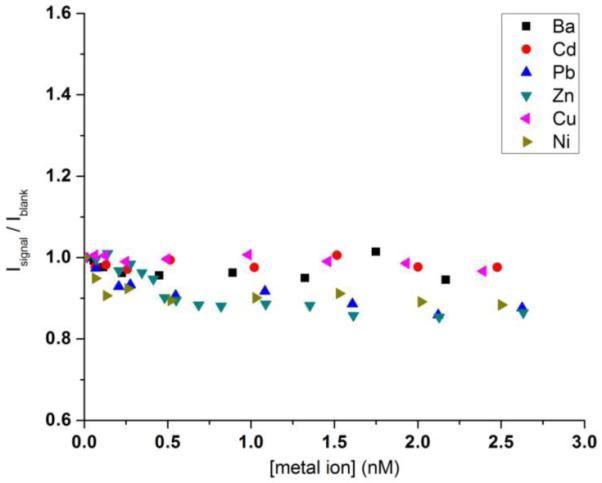
Isignal/Iblank vs. [other metal ion] in buffer solution.
4. Conclusions
In conclusion, we have proposed and demonstrated a new scheme for mercuric ions detection in aqueous media, using a single-stranded DNA as Hg2+-capturing element, photon upconverting particles as energy donor and an appropriate intercalating dye as energy acceptor. By using NaYF4:Yb3+,Tm3+ nanoparticles as donor and SYBR green I as acceptor, we can quantitatively detect the presence of mercuric ions with a directly observed detection limit of 0.06 nM. The presence of many common metal ions (Ba2+, Cd2+, Cu2+, Ni2+, Pb2+, Zn2+, Na+, K+) does not interfere with the detection of mercuric ion. These results show that such label-free design displays high sensitivity and specificity. Further effort to expand the method to detect non-ionic form of mercury, such as methyl mercury, is currently underway.
Supplementary Material
Acknowledgement
This work was partially supported by Grant Number RR-016480 from NCRR of NIH and the Water Innovation Fund of the State of New Mexico.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auzel FE. Chem. Rev. 2004;104:139. doi: 10.1021/cr020357g. [DOI] [PubMed] [Google Scholar]
- Boyer JC, Manseau MP, Murray JI, van Veggel FCJM. Langmuir. 2010;26:1157. doi: 10.1021/la902260j. [DOI] [PubMed] [Google Scholar]
- Butler OT, Cook JM, Harrington CF, Hill SJ, Rieuwerts J, Miles DLJ. Atomic Spectrometry Update. Environmental Analysis. Anal. At. Spectrum. 2006;21:217. [Google Scholar]
- Che Y, Yang X, Zang L. Chem. Comm. 2008;12:1413. doi: 10.1039/b719384j. [DOI] [PubMed] [Google Scholar]
- Chiang C-K, Huang C-C, Liu C-W, Chang H-T. Anal. Chem. 2008;80:3716. doi: 10.1021/ac800142k. [DOI] [PubMed] [Google Scholar]
- Corstjens PLAM, Zuiderwijk M, Nilsson M, Feindt H, Niedbala RS, Tanke HJ. Anal. Biochem. 2003;312:191. doi: 10.1016/s0003-2697(02)00505-5. [DOI] [PubMed] [Google Scholar]
- Darbha GK, Ray A, Ray PC. ACS Nano. 2007;1(3):208. doi: 10.1021/nn7001954. [DOI] [PubMed] [Google Scholar]
- Fong B, Mei W, Siu TS, Lee J, Sai K, Tam SJ. Anal. Toxicol. 2007;31:281. doi: 10.1093/jat/31.5.281. [DOI] [PubMed] [Google Scholar]
- Hilderbrand SC, Shao F, Salthouse C, Mahmood U, Weissleder R. Chem. Comm. 2009:4188. doi: 10.1039/b905927j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylander LD, Goodsite ME. Sci. Total Environ. 2006;368:352. doi: 10.1016/j.scitotenv.2005.11.029. [DOI] [PubMed] [Google Scholar]
- Kim IB, Bunz UHF. J. Am. Chem. Soc. 2006;128:2818. doi: 10.1021/ja058431a. [DOI] [PubMed] [Google Scholar]
- Kumar M, Guo Y, Zhang P. Biosens. Bioelectron. 2009;24:1522. doi: 10.1016/j.bios.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Zhang P. Langmuir. 2009;25(11):6024. doi: 10.1021/la900936p. [DOI] [PubMed] [Google Scholar]
- Lee JS, Han MS, Mirkin CA. Angew. Chem., Int. Ed. 2007;46:4093. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- Li L, Dong S, Wang E. Anal. Chem. 2009;81(6):2144. doi: 10.1021/ac900188y. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Togashi H, Tashiro M, Yamaguchi H, Oda S, Kudo M, Tanaka Y, Kondo Y, Sawa R, Fujimoto T, Machinami T, Ono A. J. Am. Chem. Soc. 2006;128:2172. doi: 10.1021/ja056354d. [DOI] [PubMed] [Google Scholar]
- Naccache R, Vetrone F, Mahalingam V, Cuccia LA, Capobianco JA. Chem. Mater. 2009;21:717. [Google Scholar]
- Nolan EM, Lippard S. Chem. Rev. 2008;108:3443. doi: 10.1021/cr068000q. [DOI] [PubMed] [Google Scholar]
- Ono A, Togashi H. Angew. Chem. Int. Ed. 2004;43:4300. doi: 10.1002/anie.200454172. [DOI] [PubMed] [Google Scholar]
- U.S. Government Printing Office; Washington, DC: 2005. U.S. EPA. EPA-452/R-05-003. [Google Scholar]
- van de Rijke F, Zijlmans H, Li S, Vail T, Raap AK, Niedbala RS, Tanke HJ. Nat. Biotechnol. 2001;19:273. doi: 10.1038/85734. [DOI] [PubMed] [Google Scholar]
- Walsh MK, Wang X, Weimer BC. J. Biochem. Biophys. Methods. 2001;47(3):221. doi: 10.1016/s0165-022x(00)00146-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Tian B, Lu J, Wang J, Luo D, MacDonald D. Electroanalysis. 1998;10(6):399. [Google Scholar]
- Wang J, Liu B. Chem. Comm. 2008:4759. doi: 10.1039/b806885b. [DOI] [PubMed] [Google Scholar]
- Wang LY, Yan RX, Huo ZG, Wang L, Zeng JH, Bao J, Wang X, Peng Q, Li YD. Angew. Chem. Int. Ed. 2005;44:6054. doi: 10.1002/anie.200501907. [DOI] [PubMed] [Google Scholar]
- Yi GS, Chow GM. Adv. Funct. Mater. 2006;16:2324. [Google Scholar]
- Zahir F, Rizwi SJ, Haq SK, Khan RH. Environ. Toxicol. Pharmacol. 2005;20:351. doi: 10.1016/j.etap.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Zhang P, Rogelj S, Nguyen K, Wheeler D. J. Am. Chem. Soc. 2006;128:12410. doi: 10.1021/ja0644024. [DOI] [PubMed] [Google Scholar]
- Zhang P, Steelant W, Kumar M, Scholfield M. J. Am. Chem. Soc. 2007;129:4526. doi: 10.1021/ja0700707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Wang QC, Zhang XW, Zheng DM, Zhang ZS, Zhang SQ. Sci. Total Environ. 2007;387:96. doi: 10.1016/j.scitotenv.2007.07.044. [DOI] [PubMed] [Google Scholar]
- Zhu XJ, Fu ST, Wong WK, Guo HP, Wong WY. Angew. Chem., Int. Ed. 2006;45:3150. doi: 10.1002/anie.200600248. [DOI] [PubMed] [Google Scholar]
- Zipper H, Brunner H, Bernhagen J, Vitzthum F. Nucl. Acids Res. 2004;32:e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


