Abstract
Background
Several arenaviruses cause severe hemorrhagic fever and aseptic meningitis in humans for which no licensed vaccines are available. A major obstacle for vaccine development is pathogen heterogeneity within the Arenaviridae family. Evidence in animal models and humans indicate that T cell and antibody-mediated immunity play important roles in controlling arenavirus infection and replication. Because CD4+ T cells are needed for optimal CD8+ T cell responses and to provide cognate help for B cells, knowledge of epitopes recognized by CD4+ T cells is critical to the development of an effective vaccine strategy against arenaviruses. Thus, the goal of the present study was to define and characterize CD4+ T cell responses from a broad repertoire of pathogenic arenaviruses (including lymphocytic choriomeningitis, Lassa, Guanarito, Junin, Machupo, Sabia, and Whitewater Arroyo viruses) and to provide determinants with the potential to be incorporated into a multivalent vaccine strategy.
Results
By inoculating HLA-DRB1*0101 transgenic mice with a panel of recombinant vaccinia viruses, each expressing a single arenavirus antigen, we identified 37 human HLA-DRB1*0101-restricted CD4+ T cell epitopes from the 7 antigenically distinct arenaviruses. We showed that the arenavirus-specific CD4+ T cell epitopes are capable of eliciting T cells with a propensity to provide help and protection through CD40L and polyfunctional cytokine expression. Importantly, we demonstrated that the set of identified CD4+ T cell epitopes provides broad, non-ethnically biased population coverage of all 7 arenavirus species targeted by our studies.
Conclusions
The identification of CD4+ T cell epitopes, with promiscuous binding properties, derived from 7 different arenavirus species will aid in the development of a T cell-based vaccine strategy with the potential to target a broad range of ethnicities within the general population and to protect against both Old and New World arenavirus infection.
Background
Several arenaviruses within the Arenaviridae family are rodent-borne human pathogens. Infection outcomes can range from subclinical disease to central nervous system damage [1], aseptic meningitis [2], congenital deformities [3,4], and severe hemorrhagic fever (reviewed in [5]). Mortality among patients with arenaviral hemorrhagic fever ranges from 15 to 30% [6,7]. Accordingly, arenavirus infections are considered a serious human public health problem. Despite the pathogenicity of arenaviruses, there are no licensed vaccines available and the live attenuated Junin virus (JUNV) vaccine, Candid #1, only has investigational new drug status in the U.S. [8]. Moreover, antiviral therapies are limited to the use of hyperimmune plasma [1] or the guanosine analogue ribavirin, which can lead to adverse side effects such as thrombocytosis, severe anemia, and birth defects [9,10]. Because of their pathogenicity and the lack of vaccines and antivirals to prevent and treat infection, arenaviruses are also regarded as a potential bioterrorism threat, and as such are classified as Class A pathogens. Thus, there is a need to develop novel prophylactic vaccination strategies to combat arenavirus infection.
Several studies have reported a beneficial role for both T cell and antibody-mediated immunity in countering arenavirus infections. Vaccine strategies aimed at generating a CD8+ T cell-mediated response confer protection against virus challenge in murine [11-14], guinea pig [15,16], and non-human primate [17] models of infection of two Old World arenaviruses, Lassa virus (LASV) and lymphocytic choriomeningitis virus (LCMV). In humans, cell-mediated immunity also seems to be critical for protection against LASV infection, as neutralizing antibodies appear several weeks or months after viral clearance [18,19], and treatment of infected patients with hyperimmune plasma does not protect against disease [20]. In contrast, antibody-mediated immunity seems to play an important role protecting against New World arenavirus infection, as administration of immune plasma at an early infection stage significantly reduces morbidity and mortality [21]. T cell responses might also be involved in countering New World arenavirus infection as, similar to LASV infection, neutralizing antibodies often appear several weeks after resolution of infection of the New World arenavirus, JUNV [22]. JUNV-specific T cell responses have also been detected in patients vaccinated with Candid #1 [8].
An important component in developing protective CD8+ T cell and antibody-mediated immunity is the generation of effective CD4+ T cell help. Several previous studies, conducted in murine models of LCMV infection, have demonstrated that virus-specific CD4+ T cells play an essential role in priming optimal CD8+ T cell responses in vivo. Infection of mice lacking CD4+ T cells (either by transient depletion or knock-out) with LCMV Armstrong led to the failure of LCMV-specific CD8+ T cells to expand upon antigen re-encounter, thus demonstrating that CD4+ T cell help is required for secondary memory CTL expansion during acute virus infection [23]. In mouse models of chronic LCMV infection, it has long been established that CD4+ T cell help is required to control viremia [24,25]. However, it was recently demonstrated that IL-21, produced by antigen-specific CD4+ T cells, is the critical helper factor needed to sustain effector CD8+ T cell activity and contain viremia during chronic LCMV infection [26-28]. In addition to providing help, in the case of LCMV infection, CD4+ T cells have been shown to have direct effector function mediated by cytokine secretion and cytolytic activity [29].
Given the importance of CD4+ T cells, a critical step in the development of a vaccine strategy against arenaviruses is the definition of arenavirus-specific CD4+ T cell epitopes with the capacity to induce help. However, one of the major obstacles in designing an arenavirus vaccine is the genetic diversity found amongst the different members of the Arenaviridae family, as well as the variability within a single arenavirus species. To overcome this challenge, it might be possible to combine antigens or epitopes derived from several arenavirus species in the same vaccine, and thus provide effective multivalent protection.
Thus, the goal of the present study was to identify HLA class II-restricted CD4+ T cell epitopes derived from arenaviruses associated with disease in humans that have the competency to provide help in a vaccination setting. We specifically targeted the 4 viral proteins (glycoprotein precursor (GPC), RNA-dependent RNA polymerase (L), nucleocapsid protein (NP), and zinc-finger binding protein (Z)) that are encoded by the 7 different species, including Guanarito virus (GTOV), JUNV, LASV, LCMV, Machupo virus (MACV), Sabia virus (SABV), and Whitewater Arroyo virus (WWAV). Because of the high degree of HLA class II polymorphism expressed in the human population, we focused on defining HLA-DRB1*0101-restricted CD4+ T cell epitopes, as these epitopes have demonstrated broad reactivity with other HLA-DR molecules [30], and thus have the potential to provide extensive population coverage across different ethnicities ([31] and J. Sidney and A. Sette, unpublished observations). Here, we tested whether it was possible to define HLA-DRB1*0101-restricted CD4+ T cell responses from the 7 arenaviruses of interest that provide broad population coverage, and demonstrate a polyfunctional phenotype with the potential to impart help.
Results
Identification of HLA-DRB1-restricted arenavirus CD4+ T cell epitopes
Recent studies have defined a large number of HLA class I-restricted CD8+ T cell epitopes derived from GTOV, JUNV, LASV, LCMV, MACV, SABV, and WWAV [11,12,14]. By contrast, very few human arenavirus-specific epitopes recognized by CD4+ T cells, and restricted by HLA class II, have been described. To identify arenavirus-specific CD4+ T cell epitopes, the common HLA-DR supertype specificity HLA-DRB1*0101 was selected, as HLA-DRB1*0101-restricted epitopes tend to also be broadly reactive with other HLA-DRB1, DRB3, DRB4, and DRB5 molecules, and thus have the potential to provide broad coverage across a highly diverse human population [31]. Accordingly, 3772 peptides, corresponding to 15-mer peptides overlapping by 10 amino acids and spanning the GPC, L, NP, and Z consensus protein sequences from the 7 target arenavirus species, were screened for their capacity to bind HLA-DRB1*0101. Using this approach, a total of 299 high affinity (50% inhibitory concentration; IC50 ≤ 50 nM) HLA-DRB1*0101 binding peptides were identified (Table 1 and data not shown).
Table 1.
Summary of characteristics of arenavirus-derived HLA-DRB1*0101-restricted CD4+ T cell epitopes.
| Epitopea | Sequence | DRB1*0101 binding affinity (IC50 nM)b |
ELISPOT (net SFC/106 CD4+)c |
ICS (%IFN-γ+ CD4+)d |
|---|---|---|---|---|
| GTOV GPC131-145 | KGSPEFDWILGWTIK | 1.7 | 208 | 0.22 |
| GTOV L181-195 | DQEYHRLIHSLSKTS | 0.34 | 390 | 0.05 |
| GTOV L391-405 | RVLDILVARRLLLKK | 0.19 | 330 | 0.11 |
| GTOV L1826-1840 | IQLVFSSMINPLVIT | 0.23 | 163 | 0.04 |
| GTOV NP166-180 | KLNNQFGSMPALTIA | 0.12 | 122 | 0.05 |
| GTOV NP191-205 | NNVVQALTSLGLLYT | 0.29 | 97 | 0.05 |
| GTOV NP236-250 | ISGYNFSLSAAVKAG | 0.12 | 126 | 0.03 |
| GTOV NP541-555 | IPIQLLPNTLVFQAK | 0.25 | 142 | 0.11 |
| JUNV GPC46-60 | FFVFLALAGRSCTEE | 0.23 | 684 | 0.06 |
| JUNV L381-395 | VGQMLMLVNDRLLDI | 0.21 | 323 | 0.06 |
| JUNV L391-405 | RLLDILEAIKLIRKK | 0.48 | 333 | 0.08 |
| JUNV L411-425 | KWVQMCSRTLKNSHQ | 1.1 | 571 | 0.05 |
| JUNV L1491-1505 | MFIRNCARKVFNDIK | 2.0 | 425 | 0.11 |
| JUNV L1711-1725 | NKNFFWAVKPKAVRQ | 0.06 | 538 | 0.16 |
| LASV GPC236-250 | PSPIGYLGLLSQRTR | 0.14 | 497 | 0.07 |
| LASV GPC241-255 | YLGLLSQRTRDIYIS | 0.29 | 469 | 0.05 |
| LASV GPC476-490 | SCGLYKQPGVPVRWK | 1.5 | 273 | 0.04 |
| LCMV GPC421-435 | LRKDYIKRQGSTPLA | 4.8 | 268 | 0.16 |
| LCMV L256-270 | RNFQKVNPEGLIKEF | 5.6 | 217 | 0.07 |
| LCMV L946-960 | HLRKVILSEISFHLV | 2.1 | 533 | 0.07 |
| LCMV NP6-20 | EVKSFQWTQALRREL | 49 | 436 | 0.07 |
| LCMV NP521-535 | MDCIIFESASKARLP | 4.4 | 463 | 0.12 |
| MACV GPC96-110 | NSFYYMKGGVNTFLI | 0.21 | 365 | 0.08 |
| MACV GPC251-265 | SKTHLNFERSLKAFF | 1.7 | 427 | 0.12 |
| MACV GPC446-460 | ASLFLHLVGIPTHRH | 0.13 | 329 | 0.08 |
| MACV L391-405 | DRVLDILEAVKLIRK | 0.48 | 423 | 0.22 |
| MACV L636-650 | RYFLMAFANQIHHID | 0.24 | 269 | 0.06 |
| MACV L866-880 | DYLILKNLTGLVSAG | 0.21 | 322 | 0.13 |
| MACV L1491-1505 | TSFIRNCARKVFNDI | 0.11 | 400 | 0.07 |
| MACV L1711-1725 | NNQNFFWAVKPKVVR | 1.7 | 518 | 0.09 |
| MACV NP191-205 | NSVVQALTSLGLLYT | 0.78 | 176 | 0.06 |
| MACV Z21-35 | PSAEFRRTAPPSLYG | 2.0 | 313 | 0.16 |
| SABV GPC26-40 | VSLIAALKGMINLWK | 0.47 | 264 | 0.04 |
| SABV GPC436-450 | FTTTLFLHLVGFPTH | 0.92 | 1213 | 0.08 |
| SABV GPC441-455 | FLHLVGFPTHRHIRG | 0.09 | 823 | 0.05 |
| WWAV GPC46-60 | FIVFLLLAGRSCSYK | 1.2 | 701 | 0.10 |
| WWAV GPC386-400 | FRNQWLLESDHLISE | 1.6 | 517 | 0.39 |
a Peptide position within the GPC, L, NP, or Z protein of prototypic strains of GTOV, JUNV, LASV, LCMV, MACV, SABV, and WWAV.
b Each epitope was tested for binding to HLA-DRB1*0101.
c Peptide tested at 10 μg/ml in IFN-γ ELISPOT assays. Criteria for positivity are net SFC/106 cells ≥ 20, SI ≥ 1.4, and p-value ≤ 0.05 in two or more independent assays. Average IFN-γ responses are shown from two independent experiments.
d Peptide tested at 3 μg/ml in IFN-γ ICS assays. The epitope-specific responses were calculated by subtracting the total number of cells that scored positive for IFN-γ+ production in the absence of peptide. Criterion for positivity is SI ≥ 2.0 in two or more independent assays. Average IFN-γ responses are shown from two independent experiments.
Because human PBMC samples from arenavirus-exposed individuals were difficult to obtain, HLA-DRB1*0101 transgenic mice were used to identify human arenavirus epitopes [32]. To determine the in vivo antigenicity of the 299 peptides, HLA-DRB1*0101 transgenic mice were primed with a recombinant vaccinia virus (rVACV) expressing one of the arenavirus proteins and subsequently boosted with peptide pools emulsified in incomplete Freund's adjuvant (IFA) as described in the Methods. Mice were not infected with the native arenaviruses because the majority of arenaviruses studied require biosafety level-4 (BSL-4) containment. Inoculation of mice with the rVACV constructs still enabled the identification of endogenously processed CD4+ T cell epitopes. Eleven to 14 days after peptide immunization, purified splenic CD4+ T cells were screened for recognition of the arenavirus peptides through IFN-γ ELISPOT assays (Table 1 and data not shown).
To demonstrate that the observed responses were CD4+ T cell-specific, we subsequently tested the ELISPOT positive peptides in IFN-γ intracellular cytokine staining (ICS) assays. These assays also provided a quantitative estimate of the frequency of responding arenavirus-specific CD4+ T cells. In total, 37 out of the 299 high affinity binding peptides were antigenic in both IFN-γ ELISPOT and ICS assays, with averaged responses ranging from 0.03 to 0.39% IFN-γ-producing CD4+ T cells. Figure 1 shows representative responses for each of the 37 CD4+ T cell epitopes that induced a positive IFN-γ response with a SI ≥ 2. Minor recognition of 4 out of 37 epitopes (GTOV NP236-250, GTOV L1826-1840, LCMV L946-960, and MACV GPC96-110) by CD8+ T cells was observed (data not shown), suggesting that these 15-mer peptides might also contain nested CD8+ T cell epitopes. The sequences of the 37 CD4+ T cell epitopes identified, along with the corresponding viral antigen, the HLA-DRB1*0101 binding affinity, and the averaged IFN-γ ELISPOT and ICS responses, are summarized in Table 1. An average of 5 epitopes (ranging from 2 to 10) were defined from each of the arenavirus species. The average binding affinity (IC50) of the 37 epitopes to HLA-DRB1*0101 was 0.72 nM, while the average IC50 of the remaining 262 non-epitopes was 0.96 nM. The difference between the two sets of peptides was not statistically significant, thus demonstrating that high binding affinity alone does not define a T cell epitope. Overall, these data demonstrate that the CD4+ T cell epitope set provides broad coverage of the 7 different arenaviruses of interest, suggesting their potential to be incorporated into a multivalent arenavirus vaccine strategy.
Figure 1.
Identification of antigenic arenavirus-derived CD4+ T cell epitopes in HLA-DRB1*0101 transgenic mice. Groups of HLA-DRB1*0101 transgenic mice were inoculated with a rVACV expressing a single arenavirus antigen, and 7 days later, immunized with an appropriate pool of GTOV, JUNV, LASV, LCMV, MACV, SABV, or WWAV peptides. Positive peptides in IFN-γ ELISPOT assays were tested in ICS assays using splenocytes from immunized HLA-DRB1*0101 mice as described in Materials and Methods. The numbers indicate the percent of CD4+ T cells producing IFN-γ following stimulation with 3 μg/ml of each of the listed peptides. A peptide was considered positive if the response was ≥ 2 SI above background in two experiments. Representative data from at least two independent experiments is shown.
Arenavirus-specific CD4+ T cells have a polyfunctional phenotype
Polyfunctional CD4+ T cell responses have been correlated with optimal protection against infection in vivo [33,34]. Because of the importance of IFN-γ, TNF-α, and IL-2 in mediating protection, we examined the frequency of IFN-γ, TNF-α, and IL-2 expressing CD4+ T cells elicited by a representative subset of the HLA-DRB1*0101-restricted epitopes derived from the GPC, L, and NP proteins of LCMV (LCMV GPC421-435, L946-960, or NP6-20, respectively). In these experiments, splenic CD4+ T cells were derived from HLA-DRB1*0101 transgenic mice that were primed with a rVACV expressing either the LCMV GPC, L, or NP and boosted with a single peptide emulsified in complete Freund's adjuvant (CFA). CFA was utilized in the peptide immunization mixture to expand antigen-specific CD4+ T cell subsets to detectable frequencies for the FACS analysis.
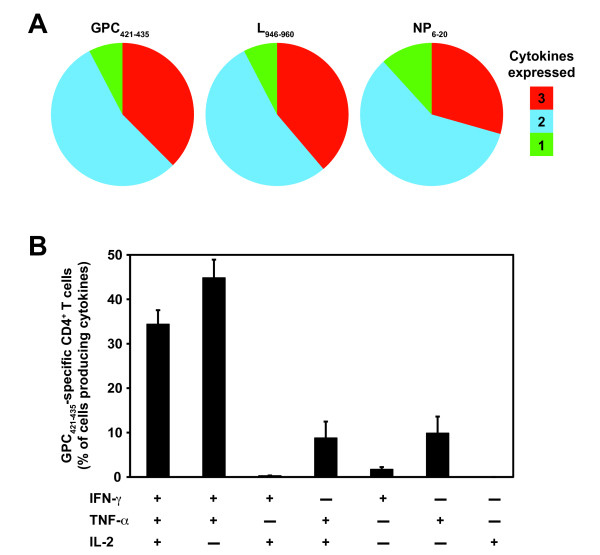
Utilizing multiparameter ICS assays, we quantified the fraction of the total cytokine response comprised of 3, any 2, or any 1 cytokine in response to in vitro stimulation with either the LCMV GPC421-435, L946-960, or NP6-20 epitope. We found that the different LCMV-specific epitopes induced similar cytokine expression patterns. Three cytokines were produced by 30 to 40% of cytokine-expressing CD4+ T cells, a little over half expressed any 2 cytokines, and ~10% produced any 1 cytokine (Figure 2A).
Figure 2.
Arenavirus-specific CD4+ T cells have a polyfunctional phenotype. Epitope-specific cytokine production (IFN-γ, TNF-α, and IL-2) from splenic CD4+ T cells of a group of three rVACV primed and peptide boosted HLA-DRB1*0101 transgenic mice was measured 11 to 14 days after peptide immunization. (A) The fraction of the total cytokine response comprising LCMV GPC421-435, L946-960, or NP6-20-specific CD4+ T cells expressing all 3 cytokines, any 2 cytokines, or any 1 cytokine. (B) The frequency of LCMV GPC421-435-specific CD4+ T cells expressing each of the seven possible combinations of IFN-γ, TNF-α, and IL-2. Results from one experiment are shown and are representative of at least two independent experiments. Error bars indicate SEM.
Next, we measured the frequency of CD4+ T cells expressing the different combinations of cytokines (Figure 2B, LCMV GPC421-435 shown as representative data). We detected 6 out of the 7 possible cytokine combinations, but the vast majority of CD4+ T cells were IFN-γ+TNF-α+IL-2+ and IFN-γ+TNF-α+. To a lesser extent, IFN-γ+IL-2+, TNF-α+IL-2+, single IFN-γ+, and single TNF-α+ producing CD4+ T cells were also detected. Although LCMV GPC421-435 is shown as representative data, the majority of CD4+ T cells restimulated with either the LCMV L946-960 or the NP6-20 peptide also were IFN-γ+TNF-α+IL-2+ and IFN-γ+TNF-α+ (Figure 2A and data not shown). Thus, these data show that the arenavirus-specific CD4+ T cell epitopes are capable of eliciting T cells primarily with a polyfunctional phenotype.
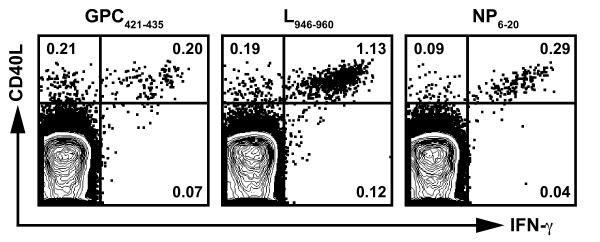
Interaction between CD40 and its ligand, CD40L (expressed by activated CD4+ T cells), is a key mediator in the priming of antigen-specific CD8+ T cells [35-37] and B cells [38]. Thus, to further characterize the LCMV-specific CD4+ T cells, we measured the frequency of cells expressing CD40L. In these experiments, splenic CD4+ T cells were derived from HLA-DRB1*0101 transgenic mice that were primed with a rVACV expressing either the LCMV GPC, L, or NP and boosted with the corresponding peptide emulsified in CFA. Following in vitro peptide restimulation, we found that the LCMV GPC421-435, L946-960, and NP6-20 epitopes induced robust expression of CD40L (Figure 3), indicating the competency of these CD4+ T cell subsets to provide help.
Figure 3.
Arenavirus-specific CD4+ T cells express CD40L. Splenic CD4+ T cells from rVACV primed and peptide boosted HLA-DRB1*0101 transgenic mice were stimulated with the indicated peptides (LCMV GPC421-435, L946-960, or NP6-20), and stained for intracellular IFN-γ and CD40L. Plots are gated on CD4+ T cells and the numbers indicate the frequency of cells expressing IFN-γ and CD40L. Results are representative of at least two independent experiments.
Arenavirus CD4+ T cell epitopes degenerately bind to additional HLA-DR molecules
Previous work has established that epitopes with the capacity to bind with high affinity to HLA-DRB1*0101 tend to also be broadly reactive with other HLA-DRB1, DRB3, DRB4, and DRB5 molecules, and thus have the ability to provide extensive population coverage across different ethnicities ([30,31] and J. Sidney and A. Sette, unpublished observations). As the HLA-DR supertype consists of a group of molecules that share largely overlapping peptide binding specificities, we tested the binding affinity of the 37 arenavirus-specific CD4+ T cell epitopes to 14 common HLA-DRB1, DRB3, DRB4, and DRB5 molecules, as described in Materials and Methods. The number of allelic molecules bound is summarized in Table 2. The 37 different arenavirus-specific CD4+ T cell epitopes bound, on average, 8 different HLA-DR molecules (range 2 to 13). These data demonstrate that the HLA-DRB1*0101-restricted epitopes extensively bind to several HLA-DR molecules, suggesting that these epitopes have the capacity to provide relatively broad coverage across different ethnic populations.
Table 2.
Number of HLA-DRB1, DRB3, DRB4, and DRB5 molecules bound by the arenavirus-specific CD4+ T cell epitopes.
| Epitopea | No. DRB1 molecules boundb | No. DRB3/4/5 molecules boundc | Total No. DR molecules bound |
|---|---|---|---|
| GTOV GPC131-145 | 4 | 1 | 5 |
| GTOV L181-195 | 3 | 1 | 4 |
| GTOV L391-405 | 9 | 1 | 10 |
| GTOV L1826-1840 | 8 | 2 | 10 |
| GTOV NP166-180 | 8 | 1 | 9 |
| GTOV NP191-205 | 8 | 2 | 10 |
| GTOV NP236-250 | 10 | 1 | 11 |
| GTOV NP541-555 | 9 | 2 | 11 |
| JUNV GPC46-60 | 6 | 1 | 7 |
| JUNV L381-395 | 10 | 3 | 13 |
| JUNV L391-405 | 8 | 1 | 9 |
| JUNV L411-425 | 6 | 1 | 7 |
| JUNV L1491-1505 | 8 | 2 | 10 |
| JUNV L1711-1725 | 8 | 1 | 9 |
| LASV GPC236-250 | 7 | 2 | 9 |
| LASV GPC241-255 | 7 | 1 | 8 |
| LASV GPC476-490 | 1 | 1 | 2 |
| LCMV GPC421-435 | 6 | 2 | 8 |
| LCMV L256-270 | 3 | 1 | 4 |
| LCMV L946-960 | 8 | 3 | 11 |
| LCMV NP6-20 | 7 | 2 | 9 |
| LCMV NP521-535 | 5 | 2 | 7 |
| MACV GPC96-110 | 3 | 1 | 4 |
| MACV GPC251-265 | 8 | 2 | 10 |
| MACV GPC446-460 | 2 | 1 | 3 |
| MACV L391-405 | 7 | 1 | 8 |
| MACV L636-650 | 10 | 3 | 13 |
| MACV L866-880 | 7 | 2 | 9 |
| MACV L1491-1505 | 9 | 3 | 12 |
| MACV L1711-1725 | 8 | 1 | 9 |
| MACV NP191-205 | 8 | 2 | 10 |
| MACV Z21-35 | 8 | 2 | 10 |
| SABV GPC26-40 | 7 | 2 | 9 |
| SABV GPC436-450 | 5 | 1 | 6 |
| SABV GPC441-455 | 7 | 1 | 8 |
| WWAV GPC46-60 | 5 | 1 | 6 |
| WWAV GPC386-400 | 6 | 2 | 8 |
a Peptide position within the GPC, L, NP, or Z protein of prototypic strains of GTOV, JUNV, LASV, LCMV, MACV, SABV, and WWAV.
b Each epitope was tested for binding to 14 common HLA molecules within the DR supertype. The number of HLA molecules bound corresponds to the epitope having an IC50 ≤ 200 nM to the specific HLA molecule.
c The number of DRB3*0101, DRB4*0101, and DRB5*0101 molecules bound by the arenavirus epitopes with an IC50 ≤ 200 nM.
Broadly reactive HLA-DR-restricted arenavirus CD4+ T cell epitopes provide widespread population coverage
Next, based on the capacity of each CD4+ T cell epitope to bind different common HLA-DR molecules, we estimated the breadth of population coverage afforded by this epitope set. Here, we defined coverage as the extent or degree to which different ethnic populations worldwide recognize the CD4+ T cell epitope set. A high degree of coverage suggests that the epitope set might also be able to provide broad protection against a variety of arenaviruses. To estimate population coverage, we calculated the theoretical coverage in various populations afforded by the CD4+ T cell epitopes for each of the 7 arenaviruses using the Population Coverage Calculation Tool available through the Immune Epitope Database and Analysis Resource [39]. Because allele frequency data for the HLA-DRB3, DRB4, and DRB5 loci are not available, these calculations are based on the HLA-DRB1 allele frequencies alone, and thus likely represent conservative estimates. For the present analysis, biologically relevant binding was defined as an IC50 ≤ 200 nM.
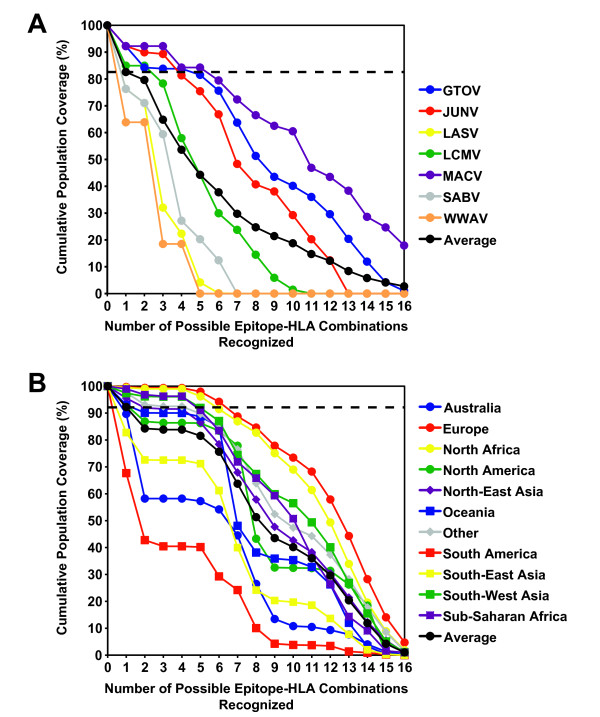
The total coverage of the general population provided by the corresponding set of epitopes for each of the 7 different arenaviruses is shown in Figure 4A. In this case, coverage is shown as an average across 11 different population groups characterized by the Database of MHC [40]. The epitopes identified from GTOV, JUNV, and MACV provided the broadest population coverage, with 92.3% of the overall population recognizing at least one or more epitope(s), while WWAV epitopes provided the least amount of coverage (63.9%). The remaining viruses, LASV, LCMV, and SABV provided 76.3%, 85.0%, and 76.3% population coverage, respectively. When averaged over the 7 different viruses, the defined epitopes provided population coverage of 82.6% of the entire population. It is important to note that coverage typically entailed recognition of multiple HLA-epitope combinations. For example, from Figure 4A, it can be seen that in an average population, about 35% of individuals can be expected to recognize 6 or more HLA-epitope combinations. Thus, this analysis suggests that the defined epitopes provide not only wide-ranging coverage, but also an appreciable degree of redundant coverage, which may be important in the context of RNA viruses with high mutation rates.
Figure 4.
The identified arenavirus-specific CD4+ T cell epitopes provide broad population coverage. The theoretical population coverage was calculated based on the binding affinity data for each HLA-DRB1-restricted epitope and the reported frequencies of each HLA-DRB1 allele in different ethnic populations. Biologically relevant binding was defined as an IC50 ≤ 200 nM. (A) The average population coverage for each of the 7 arenaviruses. The horizontal dashed line indicates that 82.6% of the population, on average, recognizes one or more arenavirus epitope. (B) For GTOV, shown as representative data, the number of possible peptide-HLA allele combinations as a function of the fraction of each ethnic population (%) is shown. The horizontal dashed line represents the fraction of individuals (92.3%) that recognize one or more GTOV peptide in an average population.
The coverage provided by the epitope panel was also reasonably balanced throughout the major ethnic groups. As representative data, Figure 4B shows the coverage afforded by the GTOV-specific CD4+ T cell epitopes across several major population groups. As shown, the GTOV epitopes provide coverage ranging from about 67.7% of South Americans, to 99.7% for Europeans, with an average coverage across all ethnic populations of 92.3%. The depth of coverage is exemplified by the fact that, on average, over 80% of the individuals in the general population would be expected to present 5 or more HLA-epitope combinations. These data provide a proof of concept validation that a CD4+ T cell epitope set can be defined, affording broad coverage across different pathogenic arenavirus species and multiple HLA class II molecules.
Discussion
Prior to this study, little was known about human arenavirus-specific CD4+ T cells. To our knowledge, only 6 human CD4+ T cell epitopes derived from a single arenavirus species, LASV, had been previously described [41,42]. These earlier studies relied on identifying epitopes from T cell clones generated from LASV antibody (Ab) positive individuals. Because PBMC from arenavirus-immune donors were not available, we utilized HLA-DRB1*0101 transgenic mice to carry out an extensive screen of 299 arenavirus-derived peptides that bound with high affinity to HLA-DRB1*0101. Furthermore, we bypassed the requirement for BSL-4 containment for most arenavirus species considered by developing a panel of 24 rVACV vectors that expressed the different arenavirus antigens of interest. Utilizing this strategy, we identified 37 different CD4+ T cell epitopes from 7 pathogenic arenavirus species. Two of these epitopes (GTOV NP191-205 and MACV NP191-205) overlapped with the orthologous LASV NP190-202 epitope defined with T cell clones from LASV-immune individuals [41], suggesting that our approach identified CD4+ T cell epitopes relevant for humans. In the future, we plan to examine CD4+ T cell responses in arenavirus-immune human donors in order to assess the degree of overlap between arenavirus-specific responses recognized in HLA transgenic mice and humans. However, it has been demonstrated with other infectious pathogens, such as Mycobacterium tuberculosis, that HLA class II transgenic mice recognize the same pathogen-derived T cell epitopes as humans [43,44].
Given that CD4+ T cells are essential for maintaining effective CTL responses during an arenavirus infection [23], knowledge of human CD4+ T cell epitopes is a crucial step in the development of an arenavirus vaccine that induces effective T cell-mediated immunity. As it is not likely that separate vaccines for each arenavirus species will be generated, one strategy might be the development of a multivalent vaccine that targets multiple species within the Arenaviridae family. This approach has proven itself in the case of the currently licensed vaccines against Streptococcus pneumoniae (i.e. Pneumovax 23), which contains capsular polysaccharide antigens from 23 of the most prevalent pneumococcal serotypes [45], and human papillomavirus (i.e. Gardasil), which consists of capsid proteins derived from 4 different serotypes prominently associated with disease [46].
Herein, we defined a set of CD4+ T cell epitopes from 7 different arenaviruses, with promiscuous binding characteristics. Together, these epitopes afforded ~83% coverage of the general population, on average, and thus, could be incorporated into a multivalent vaccine targeting multiple arenavirus species and ethnicities. For our analyses, we have excluded coverage at the HLA-DRB3, DRB4, and DRB5 loci, and also used a conservative threshold of 200 nM to define biologically relevant binding. Previously, we have shown that over 90% of known HLA-DR epitopes have an affinity < 1000 nM [30]. Thus, our coverage may represent a low estimate.
In similar studies, we described the identification of a collection of human class I-restricted CD8+ T cell epitopes derived from the same 7 arenaviruses that afforded 60% coverage of the general population [11,14,47]. When immunized as a peptide cocktail, the CD8+ T cell epitopes protected HLA transgenic mice against challenge with rVACVs expressing either Old or New World arenavirus GPC [14]. It is tempting to speculate that combining arenavirus-specific CD4+ and CD8+ T cell epitopes into a single multivalent vaccine might enhance the protective capacity of the CD8+ T cell response. Furthermore, we showed previously that cross-reactive T cell recognition of orthologous peptides derived from different arenavirus species further increased the coverage afforded by the CD8+ T cell epitopes. Likewise, we defined several CD4+ T cell epitopes that shared orthologous sequences between two or more arenavirus species (i.e. JUNV/WWAV GPC46-60, GTOV/JUNV/MACV L391-405, JUNV/MACV L1491-1505, JUNV/MACV L1711-1725, and GTOV/MACV NP191-205), suggesting the potential for CD4+ T cell cross-reactivity in vivo, and thus even greater virus and population coverage.
Three of the CD4+ T cell responses identified in this study, (MACV Z21-35, JUNV GPC46-60, and WWAV GPC46-60), were directed against regions that contained nested human CD8+ T cell epitopes. Our previous study defined a HLA-A*1101-restricted epitope, MACV Z27-36, and a HLA-A*0201-restricted epitope, WWAV GPC42-50 [14]. Overlapping murine CD4+ and CD8+ T cell epitopes have been described in both H-2b and H-2d mice infected with LCMV [48-50], and influenza virus infection of H-2b mice [51]. Thus, it seems that epitope sharing between both murine and human CD4+ and CD8+ T cells might be a general phenomenon amongst viruses with small proteomes. These overlapping epitopic regions could be of particular importance when designing a multivalent vaccine strategy that targets both arenavirus-specific CD4+ and CD8+ T cell responses.
The ability of T cell-based vaccine candidates to induce protective immunity against infection has largely been associated with the capacity of antigen-specific CD4+ and CD8+ T cells to produce multiple effector functions simultaneously (reviewed in [52]). A recent study evaluating the protective efficacy of vaccine formulations against Leishmania major found that vaccine-elicited protection was best correlated with the concurrent release of IFN-γ, TNF-α, and IL-2 by antigen-specific CD4+ T cells [33]. Similarly, several studies have demonstrated a strong association between the maintenance of highly polyfunctional T cell responses and non-progressive HIV infection [53-55]. Here, we showed that a substantial percentage of the LCMV-specific CD4+ T cells produced a polyfunctional response, characterized by simultaneous release of IFN-γ, TNF-α, and IL-2, following peptide stimulation. Finally, we demonstrated that the LCMV-specific CD4+ T cells expressed CD40L following peptide stimulation, indicating their propensity to provide CD8+ T cell help [35-37]. Taken together, these data suggest that the identified CD4+ T cell epitopes might help induce a protective cell-mediated immune response in a vaccination setting.
Conclusions
In conclusion, the identification of CD4+ and CD8+ T cell epitopes from 7 different arenavirus species might lead to the development of a T cell-based vaccine strategy protecting against Old and New World arenavirus infection responsible for hemorrhagic fever and aseptic meningitis in humans. Promiscuous epitopes with the capacity to bind to multiple alleles within a HLA supertype are of particular relevance in generating a vaccine that targets a broad range of ethnicities within the general population. The validation of both CD4+ and CD8+ T cell epitopes in arenavirus-immune individuals, and their formulation in a multivalent construct would also be the logical next steps in the further exploration of this concept.
Methods
Peptide synthesis
Peptides were synthesized as crude material by Pepscan Systems (Lelystad, The Netherlands). Candidate epitopes were resynthesized by A and A Labs (San Diego, CA) and purified to 95% or greater homogeneity by reverse-phase HPLC. The IEDB submission identification number for HLA-DRB1*0101-restricted arenavirus-specific CD4+ T cell epitopes is 1000404.
MHC peptide-binding assay
Quantitative assays to measure the binding affinity of peptides to purified HLA-DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0404, DRB1*0405, DRB1*0701, DRB1*0802, DRB1*0901, DRB1*1101, DRB1*1302, DRB1*1501, DRB3*0101, DRB4*0101, and DRB5*0101 molecules were based on the inhibition of binding of a radiolabeled standard peptide, and were performed essentially as described elsewhere [31,56]. Briefly, after a 2-day incubation, binding of the radiolabeled peptide to the corresponding MHC class II molecule was determined by capturing MHC/peptide complexes on Greiner Lumitrac 600 microplates (Greiner Bio-One, Monroe, NC) coated with the L243 Ab (anti-DRA), and measuring bound cpm using the Topcount microscintillation counter (Packard Instrument). The concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50) was then calculated. Peptides were typically tested at 6 different concentrations covering a 100,000-fold dose range, and in 3 or more independent assays. Under the conditions utilized, where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the KD values.
Mice
HLA-DRB1*0101 transgenic mice on a FVB/N background [32] were crossed with C57BL/6J Aβ° mice to generate animals on a C57BL/6J background with no endogenous mouse MHC class II. HLA-DRB1*0101 Aβ° transgenic mice (referred to as HLA-DRB1*0101) retained the expression of mouse MHC class I. Mice were bred and maintained in the animal facilities at the La Jolla Institute for Allergy and Immunology (La Jolla, CA). All mouse studies followed guidelines set by the National Institutes of Health and the Institutional Animal Care and Use Committee-approved animal protocols (Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC# 000840) and Office of Laboratory Animal Welfare (OLAW# A3779-01)).
Viruses and immunizations
rVACV were generated as previously described [14], and are available from the Biodefense and Emerging Infections Research Resources Repository (BEI Resources; http://www.beiresources.org; BEIR NR-15486-NR-15509) [57]. In total, 24 different rVACV were constructed, each expressing a single arenavirus protein, either the GPC, L, NP, or Z protein. rVACV expressing the JUNV Z, SABV L, WWAV L, and WWAV Z were not generated, and therefore, peptides derived from these viral proteins were not tested for antigenicity in mice. Groups of three HLA-DRB1*0101 transgenic mice were injected i.p. with 107 PFU rVACV, as this infectious dose has been routinely used in previous studies [11,14,47]. Seven days after rVACV inoculation, mice were peptide-immunized s.c. at the base of the tail with a pool of 4 to 6 peptides (15 μg/peptide), or 15 μg of a single peptide in PBS emulsified in IFA or CFA, respectively. The immunized peptides corresponded to the arenavirus protein expressed by the rVACV used to inoculate the mice. On day 11 to 14 post-peptide immunization, mice were sacrificed and splenic CD4+ T cells were analyzed by ex vivo ELISPOT and ICS assays for cytokine production.
IFN-γ ELISPOT assay
The mouse IFN-γ ELISPOT assay was performed as previously described [58]. In brief, 2 × 105 splenic CD4+ T cells purified by anti-CD4 magnetic beads [Miltenyi Biotec, Auburn, CA] from immunized HLA-DRB1*0101 transgenic mice were cultured with 105 peptide-pulsed CD11c+ dendritic cells (DC). Anti-CD11c magnetic beads (Miltenyi Biotec) were used to purify CD11c+ DCs from HLA-DRB1*0101 transgenic mice immunized s.c. 12 days prior with 106 B16 cells expressing fms-like tyrosine kinase 3 ligand. For peptide pulsing, DCs were incubated with 10 μg/ml peptide for at least 1 h at 37°C, followed by 3 washes to remove excess peptide. Each assay was performed in triplicate wells. After a 20 h incubation at 37°C, plates were developed, and responses calculated as described [58]. Criteria for positivity were net spot-forming cells (SFC)/106 cells ≥ 20, stimulation index (SI) ≥ 1.4, and p-value ≤ 0.05 using a Student's t test in at least 2 out of 3 experiments.
Multiparameter ICS assay
Splenocytes from immunized HLA-DRB1*0101 transgenic mice were cultured in the presence of 3 μg/ml of arenavirus peptide and 1 μl/ml Golgiplug (containing Brefeldin A; BD Biosciences, San Diego, CA) in complete RPMI medium. After 5-6 h, cells were harvested and stained for cell surface antigens CD4 and CD8. After washing, cells were fixed, permeabilized, and stained for IFN-γ, TNF-α, IL-2, and CD40L using a Cytofix/Cytoperm kit, according to manufacturer's directions (BD Biosciences). Approximately 500,000 viable lymphocytes per sample were acquired on a BD LSR II flow cytometer. The frequency of CD4+ T cells responding to each arenavirus peptide was quantified by determining the total number of gated CD4+ and cytokine+ cells using FlowJo software (Tree Star, San Carlos, CA).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MFK, MM, JG, and CO performed the cellular immunoassays. JB constructed and validated the rVACV. HHB generated the arenavirus protein consensus sequences. JS carried out the population coverage analysis. MJB and AS conceived the study. BP, SC, DMA, MJB, and HG aided with data analyses. MFK, JB, and AS wrote the manuscript. All authors participated in discussions, and reviewed and approved the final manuscript version.
Contributor Information
Maya F Kotturi, Email: maya@liai.org.
Jason Botten, Email: Jason.Botten@uvm.edu.
Matt Maybeno, Email: mmaybeno@liai.org.
John Sidney, Email: jsidney@liai.org.
Jean Glenn, Email: jglenn@liai.org.
Huynh-Hoa Bui, Email: hbui@isisph.com.
Carla Oseroff, Email: coseroff@liai.org.
Shane Crotty, Email: shane@liai.org.
Bjoern Peters, Email: bpeters@liai.org.
Howard Grey, Email: hgrey@liai.org.
Daniel M Altmann, Email: d.altmann@csc.mrc.ac.uk.
Michael J Buchmeier, Email: m.buchmeier@uci.edu.
Alessandro Sette, Email: alex@liai.org.
Acknowledgements
We thank Louis Huynh, Carrie Moore, Sandy Ngo, and Amiyah Steen for performing the MHC binding assays, and Josie Babin and Justine Swann for assistance with the cellular assays. We thank Joey Ting and Danh Do for help in constructing the rVACVs. We are also grateful to Ravi Kolla for helpful discussions. This work was supported by the National Institutes of Health Grant NIH-AI-50840 (MJB) and Contract NO1-AI-40023 (AS). MJB, JB, and BP were also supported in part by funds from the Pacific Southwest RCE AI065359. La Jolla Institute for Allergy and Immunology & Kyowa Hakko Kirin California publication number 1232.
References
- Buchmeier MJ, de la Torre JC, Peters CJ. In: Fields Virology. Knipe DM, Howley PM, editor. Vol. 2. Lippincott Williams & Wilkins, a Wolters Klumer Business; 2007. Arenaviridae: the viruses and their replication; pp. 1791–1828. [Google Scholar]
- Barton LL, Hyndman NJ. Lymphocytic choriomeningitis virus: reemerging central nervous system pathogen. Pediatrics. 2000;105:E35. doi: 10.1542/peds.105.3.e35. [DOI] [PubMed] [Google Scholar]
- Barton LL, Mets MB, Beauchamp CL. Lymphocytic choriomeningitis virus: emerging fetal teratogen. Am J Obstet Gynecol. 2002;187:1715–1716. doi: 10.1067/mob.2002.126297. [DOI] [PubMed] [Google Scholar]
- Bonthius DJ, Wright R, Tseng B, Barton L, Marco E, Karacay B, Larsen PD. Congenital lymphocytic choriomeningitis virus infection: spectrum of disease. Ann Neurol. 2007;62:347–355. doi: 10.1002/ana.21161. [DOI] [PubMed] [Google Scholar]
- Gonzalez JP, Emonet S, de Lamballerie X, Charrel R. Arenaviruses. Curr Top Microbiol Immunol. 2007;315:253–288. doi: 10.1007/978-3-540-70962-6_11. full_text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiztegui JI. Clinical and epidemiological patterns of Argentine haemorrhagic fever. Bull World Health Organ. 1975;52:567–575. [PMC free article] [PubMed] [Google Scholar]
- McCormick JB, Webb PA, Krebs JW, Johnson KM, Smith ES. A prospective study of the epidemiology and ecology of Lassa fever. J Infect Dis. 1987;155:437–444. doi: 10.1093/infdis/155.3.437. [DOI] [PubMed] [Google Scholar]
- Maiztegui JI, McKee KT Jr, Barrera Oro JG, Harrison LH, Gibbs PH, Feuillade MR, Enria DA, Briggiler AM, Levis SC, Ambrosio AM, Halsey NA, Peters CJ. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J Infect Dis. 1998;177:277–283. doi: 10.1086/514211. [DOI] [PubMed] [Google Scholar]
- Enria DA, Briggiler AM, Levis S, Vallejos D, Maiztegui JI, Canonico PG. Tolerance and antiviral effect of ribavirin in patients with Argentine hemorrhagic fever. Antiviral Res. 1987;7:353–359. doi: 10.1016/0166-3542(87)90017-9. [DOI] [PubMed] [Google Scholar]
- McKee KT Jr, Huggins JW, Trahan CJ, Mahlandt BG. Ribavirin prophylaxis and therapy for experimental argentine hemorrhagic fever. Antimicrob Agents Chemother. 1988;32:1304–1309. doi: 10.1128/aac.32.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botten J, Alexander J, Pasquetto V, Sidney J, Barrowman P, Ting J, Peters B, Southwood S, Stewart B, Rodriguez-Carreno MP, Mothe B, Whitton JL, Sette A, Buchmeier MJ. Identification of protective Lassa virus epitopes that are restricted by HLA-A2. J Virol. 2006;80:8351–8361. doi: 10.1128/JVI.00896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botten J, Whitton JL, Barrowman P, Sidney J, Whitmire JK, Alexander J, Ting JP, Bui HH, Sette A, Buchmeier MJ. HLA-A2-Restricted Protection against Lethal Lymphocytic Choriomeningitis. J Virol. 2007;81:2307–2317. doi: 10.1128/JVI.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi MF, Peters B, Buendia-Laysa F Jr, Sidney J, Oseroff C, Botten J, Grey H, Buchmeier MJ, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi MF, Botten J, Sidney J, Bui HH, Giancola L, Maybeno M, Babin J, Oseroff C, Pasquetto V, Greenbaum JA, Peters B, Ting J, Do D, Vang L, Alexander J, Grey H, Buchmeier MJ, Sette A. A multivalent and cross-protective vaccine strategy against arenaviruses associated with human disease. PLoS Pathog. 2009;5:e1000695. doi: 10.1371/journal.ppat.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auperin DD, Esposito JJ, Lange JV, Bauer SP, Knight J, Sasso DR, McCormick JB. Construction of a recombinant vaccinia virus expressing the Lassa virus glycoprotein gene and protection of guinea pigs from a lethal Lassa virus infection. Virus Res. 1988;9:233–248. doi: 10.1016/0168-1702(88)90033-0. [DOI] [PubMed] [Google Scholar]
- Morrison HG, Bauer SP, Lange JV, Esposito JJ, McCormick JB, Auperin DD. Protection of guinea pigs from Lassa fever by vaccinia virus recombinants expressing the nucleoprotein or the envelope glycoproteins of Lassa virus. Virology. 1989;171:179–188. doi: 10.1016/0042-6822(89)90525-4. [DOI] [PubMed] [Google Scholar]
- Fisher-Hoch SP, Hutwagner L, Brown B, McCormick JB. Effective vaccine for lassa fever. J Virol. 2000;74:6777–6783. doi: 10.1128/JVI.74.15.6777-6783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg JC. Current progress towards vaccines for arenavirus-caused diseases. Vaccine. 1992;10:89–95. doi: 10.1016/0264-410X(92)90022-C. [DOI] [PubMed] [Google Scholar]
- Fisher-Hoch SP, McCormick JB. Towards a human Lassa fever vaccine. Rev Med Virol. 2001;11:331–341. doi: 10.1002/rmv.329. [DOI] [PubMed] [Google Scholar]
- McCormick JB. Clinical, epidemiologic, and therapeutic aspects of Lassa fever. Med Microbiol Immunol. 1986;175:153–155. doi: 10.1007/BF02122438. [DOI] [PubMed] [Google Scholar]
- Maiztegui JI, Fernandez NJ, de Damilano AJ. Efficacy of immune plasma in treatment of Argentine haemorrhagic fever and association between treatment and a late neurological syndrome. Lancet. 1979;2:1216–1217. doi: 10.1016/S0140-6736(79)92335-3. [DOI] [PubMed] [Google Scholar]
- McKee KT Jr, Oro JG, Kuehne AI, Spisso JA, Mahlandt BG. Safety and immunogenicity of a live-attenuated Junin (Argentine hemorrhagic fever) vaccine in rhesus macaques. Am J Trop Med Hyg. 1993;48:403–411. doi: 10.4269/ajtmh.1993.48.403. [DOI] [PubMed] [Google Scholar]
- Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- Battegay M, Moskophidis D, Rahemtulla A, Hengartner H, Mak TW, Zinkernagel RM. Enhanced establishment of a virus carrier state in adult CD4+ T-cell-deficient mice. J Virol. 1994;68:4700–4704. doi: 10.1128/jvi.68.7.4700-4704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaesser H, Sauer K, Brooks DG. IL-21 is required to control chronic viral infection. Science. 2009;324:1569–1572. doi: 10.1126/science.1174182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324:1572–1576. doi: 10.1126/science.1175194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich A, Kisielow J, Schmitz I, Freigang S, Shamshiev AT, Weber J, Marsland BJ, Oxenius A, Kopf M. IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science. 2009;324:1576–1580. doi: 10.1126/science.1172815. [DOI] [PubMed] [Google Scholar]
- Jellison ER, Kim SK, Welsh RM. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J Immunol. 2005;174:614–618. doi: 10.4049/jimmunol.174.2.614. [DOI] [PubMed] [Google Scholar]
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- Calvo-Calle JM, Strug I, Nastke MD, Baker SP, Stern LJ. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3:1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann DM, Douek DC, Frater AJ, Hetherington CM, Inoko H, Elliott JI. The T cell response of HLA-DR transgenic mice to human myelin basic protein and other antigens in the presence and absence of human CD4. J Exp Med. 1995;181:867–875. doi: 10.1084/jem.181.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah PA, Patel DT, de Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- Ciuffreda D, Comte D, Cavassini M, Giostra E, Buhler L, Perruchoud M, Heim MH, Battegay M, Genne D, Mulhaupt B, Malinverni R, Oneta C, Bernasconi E, Monnat M, Cerny A, Chuard C, Borovicka J, Mentha G, Pascual M, Gonvers JJ, Pantaleo G, Dutoit V. Polyfunctional HCV-specific T-cell responses are associated with effective control of HCV replication. Eur J Immunol. 2008;38:2665–2677. doi: 10.1002/eji.200838336. [DOI] [PubMed] [Google Scholar]
- Bennett SR, Carbone FR, Karamalis F, Flavell RA, Miller JF, Heath WR. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature. 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- Ridge JP, Di Rosa F, Matzinger P. A conditioned dendritic cell can be a temporal bridge between a CD4+ T-helper and a T-killer cell. Nature. 1998;393:474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- Schoenberger SP, Toes RE, Voort EI van der, Offringa R, Melief CJ. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Singe RM, Mack SJ, Lancaster A, Nelson MP, Erlich H, Frenandez-Vina M, Thomson G. In: Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference; Seattle, WA. Hansen JA, editor. IHWG Press; 2007. Single Locus Polymorphism of Classical HLA Genes; pp. 653–704. [Google Scholar]
- ter Meulen J, Badusche M, Kuhnt K, Doetze A, Satoguina J, Marti T, Loeliger C, Koulemou K, Koivogui L, Schmitz H, Fleischer B, Hoerauf A. Characterization of human CD4(+) T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J Virol. 2000;74:2186–2192. doi: 10.1128/JVI.74.5.2186-2192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulen J, Badusche M, Satoguina J, Strecker T, Lenz O, Loeliger C, Sakho M, Koulemou K, Koivogui L, Hoerauf A. Old and New World arenaviruses share a highly conserved epitope in the fusion domain of the glycoprotein 2, which is recognized by Lassa virus-specific human CD4+ T-cell clones. Virology. 2004;321:134–143. doi: 10.1016/j.virol.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Geluk A, Taneja V, van Meijgaarden KE, Zanelli E, Abou-Zeid C, Thole JE, de Vries RR, David CS, Ottenhoff TH. Identification of HLA class II-restricted determinants of Mycobacterium tuberculosis-derived proteins by using HLA-transgenic, class II-deficient mice. Proc Natl Acad Sci USA. 1998;95:10797–10802. doi: 10.1073/pnas.95.18.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalam AK, Rajagopalan G, Taneja V, David CS. HLA class II transgenic mice mimic human inflammatory diseases. Adv Immunol. 2008;97:65–147. doi: 10.1016/S0065-2776(08)00002-3. full_text. [DOI] [PubMed] [Google Scholar]
- Bogaert D, Hermans PW, Adrian PV, Rumke HC, de Groot R. Pneumococcal vaccines: an update on current strategies. Vaccine. 2004;22:2209–2220. doi: 10.1016/j.vaccine.2003.11.038. [DOI] [PubMed] [Google Scholar]
- Bryan JT. Developing an HPV vaccine to prevent cervical cancer and genital warts. Vaccine. 2007;25:3001–3006. doi: 10.1016/j.vaccine.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Botten JW, Kotturi MF. Adaptive immunity to Lymphocyctic choriomeningitis virus: new insights into antigenic determinants. Future Virology. 2007;2:495–508. doi: 10.2217/17460794.2.5.495. [DOI] [Google Scholar]
- Homann D, Lewicki H, Brooks D, Eberlein J, Mallet-Designe V, Teyton L, Oldstone MB. Mapping and restriction of a dominant viral CD4+ T cell core epitope by both MHC class I and MHC class II. Virology. 2007;363:113–123. doi: 10.1016/j.virol.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe BR, Stewart BS, Oseroff C, Bui HH, Stogiera S, Garcia Z, Dow C, Rodriguez-Carreno MP, Kotturi M, Pasquetto V, Botten J, Crotty S, Janssen E, Buchmeier MJ, Sette A. Chronic lymphocytic choriomeningitis virus infection actively down-regulates CD4+ T cell responses directed against a broad range of epitopes. J Immunol. 2007;179:1058–1067. doi: 10.4049/jimmunol.179.2.1058. [DOI] [PubMed] [Google Scholar]
- Dow C, Oseroff C, Peters B, Nance-Sotelo C, Sidney J, Buchmeier M, Sette A, Mothe BR. Lymphocytic choriomeningitis virus infection yields overlapping CD4+ and CD8+ T-cell responses. J Virol. 2008;82:11734–11741. doi: 10.1128/JVI.00435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe SR, Miller SC, Brown DM, Adams PS, Dutton RW, Harmsen AG, Lund FE, Randall TD, Swain SL, Woodland DL. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine. 2006;24:457–467. doi: 10.1016/j.vaccine.2005.07.096. [DOI] [PubMed] [Google Scholar]
- Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, de Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall MG, Precopio ML, Ambrozak DA, Jaye A, McMichael AJ, Whittle HC, Roederer M, Rowland-Jones SL, Koup RA. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur J Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Oseroff C, Guercio MFD, Sette A, Grey H. In: Current Protocols in Immunology I. Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, editor. New York: John Wiley & Sons; 1998. Measurement of MHC/peptide interactions by gel filtration; pp. 18.13.11–18.13.19. [Google Scholar]
- Biodefense and Emerging Infections Research Resources Repository. http://www.beiresources.org
- Moutaftsi M, Bui HH, Peters B, Sidney J, Salek-Ardakani S, Oseroff C, Pasquetto V, Crotty S, Croft M, Lefkowitz EJ, Grey H, Sette A. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]