Abstract
A straight, non-sporulating, Gram-variable bacillus (HKU24T) was recovered from the blood culture of a patient with metastatic breast carcinoma. After repeated subculturing in BACTEC Plus Anaerobic/F blood culture broth, HKU24T grew on brucella agar as non-hemolytic, pinpoint colonies after 96 h of incubation at 37 °C in an anaerobic environment and aerobic environment with 5% CO2. Growth was enhanced with a streak of Staphylococcus aureus. HKU24T was non-motile and catalase-negative, but positive for alkaline phosphatase, β-glucosidase, and α-glucosidase. It hydrolyzed phenylphosphonate and reduced resazurin. 16S rRNA, groEL, gyrB, recA, and rpoB sequencing showed that HKU24T occupies a distinct phylogenetic position among the Leptotrichia species, being most closely related to Leptotrichia trevisanii. Using HKU24T groEL, gyrB, recA, and rpoB gene-specific primers, fragments of these genes were amplified from one of 20 oral specimens. Based on phenotypic and genotypic characteristics, we propose a new species, Leptotrichia hongkongensis sp. nov., to describe this bacterium.
Keywords: Leptotrichia hongkongensis, Bacterium, Blood culture, Anaerobic bacteria, Novel species
1. Introduction
Leptotrichia species are Gram-variable, non-sporulating, large, fusiform, non-motile bacilli. Since the first description in 1879 (Trevisan, 1879), Leptotrichia buccalis has been the only species in the genus Leptotrichia until 2001 (Tee et al., 2001). L. buccalis is part of the human normal oral flora and has been implicated as a cause of dental infections and bacteremia after tooth extractions (Crawford et al., 1974; Munson et al., 2004; Sutter, 1984). However, severe systemic infections associated with L. buccalis have been uncommon. Most of the serious cases occurred as bacteremia in patients with malignancies, with a significant proportion having neutropenia (Eribe and Olsen, 2008; Morgenstein et al., 1980; Reig et al., 1985; Schwartz et al., 1995; Ulstrup and Hartzen, 2006; Weinberger et al., 1991). Case reports of infective endocarditis associated with L. buccalis in patients with prosthetic heart valves or congenital heart defects have also been described (Caram et al., 2008; Duperval et al., 1984; Eribe and Olsen, 2008; Hammann et al., 1993). Since 2001, five additional species of Leptotrichia, including L. trevisanii, L. goodfellowii, L. hofstadii, L. shahii, and L. wadei, have been described (Eribe et al., 2004; Tee et al., 2001). All these five novel species have been isolated from human specimens, some from blood cultures and others from the oral cavity.
Recently, we isolated a bacterial strain, named HKU24T, from the blood culture of a patient with metastatic carcinoma of the breast. The strain exhibited distinct phenotypic characteristics that did not fit into patterns of any known bacterial species. Based on these observations, we hypothesized that HKU24T may represent a novel bacterial species. To test the hypothesis, we sequenced the 16S rRNA gene and four additional housekeeping genes (frequently used for bacterial identification and phylogenetic studies) of HKU24T and four closely-related Leptotrichia species identified by 16S rRNA gene sequencing. On the basis of both the phenotypic and genotypic characteristics, we proposed a new species, Leptotrichia hongkongensis sp. nov., to describe this bacterium.
2. Materials and methods
2.1. Patient and strains
Strain HKU24T was isolated from the blood culture of a patient with disseminated carcinoma of the breast. L. buccalis (HKU27) was isolated from the blood culture of a patient in Hong Kong (Woo et al., 2003b). L. shahii (CCUG 47503), L. wadei (CCUG 47505), and L. trevisanii (CCUG 49525) were obtained from the Culture Collection, University of Gőteborg (CCUG), Sweden.
2.2. Phenotypic characterization
Clinical specimens were collected and handled according to standard protocols (Murray et al., 2007). Blood cultures were performed with the BACTEC 9240 system with Plus Aerobic/F and Plus Anaerobic/F bottles (Becton Dickinson Microbiology Systems, Sparks, Md., USA). All suspect colonies were identified by standard conventional biochemical methods (Murray et al., 2007). All tests were performed in triplicate with freshly prepared media on separate occasions. In addition, the Vitek system (ANI and NH; bioMerieux Vitek, USA) was used for the identification of HKU24T. In vitro susceptibilities to penicillin, metronidazole, vancomycin, amoxicillin-clavulanate, and imipenem were determined using the E-test method.
2.3. Scanning electron microscopy (SEM)
Bacterial cells were washed twice using Milli-Q water. A suspension of the bacterium was settled onto a polycarbonate membrane (Nucleopore, USA) with a pore size of 5 µm for 5 min. The membrane was fixed in 2.5% (w/v) glutaraldehyde for 1 h and washed once in 0.1 mol/L sodium cacodylate buffer. Fixed material was dehydrated through a graded ethanol series from 30% to 90% in 20% steps, followed by two changes of absolute ethanol. Each of the stepwise changes was for 15 min. Dehydrated material in absolute ethanol was critical point-dried in a BAL-TEC CPD O30 Critical Point Drier using carbon dioxide as the drying agent. Critical point-dried material was mounted on to an aluminum stub and coated with palladium in BAL-TEC SCD 005 SEM coating system. Coated material was examined in Leica Cambridge Stereoscan 440 SEM operating at 12 kV and the specimen stage was tilted at zero degree (Woo et al., 2002c).
2.4. DNA extraction
Bacterial DNA extraction was performed according to our previous publication (Woo et al., 2009). Briefly, 80 µl of NaOH (0.05 mol/L) was added to 20 µl of bacterial cells suspended in distilled water and the mixture was incubated at 60 °C for 45 min, followed by addition of 6 µl of Tris-HCl (pH 7.0), achieving a final pH of 8.0. The resultant mixture was diluted 100× and 5 µl of the diluted extract was used for polymerase chain reaction (PCR).
2.5. Sequencing of 16S rRNA, groEL, gyrB, recA, and rpoB genes
PCR amplification and DNA sequencing of the 16S rRNA, groEL, gyrB, recA, and rpoB genes of HKU24T, L. buccalis (HKU27), L. shahii (CCUG 47503), L. wadei (CCUG 47505), and L. trevisanii (CCUG 49525) were performed according to our previous publications on other anaerobic bacteria (Lau et al., 2004; Woo et al., 2002a; 2002b; 2003a; 2004a; 2007). Briefly, DNase I-treated distilled water and PCR master mix, which contains deoxynucleoside triphosphates (dNTPs), PCR buffer, and Taq polymerase, were used in all PCR reactions by adding 1.0 U of DNase I (Invitrogen, USA) to 40 µl of distilled water or PCR master mix, and then incubating the mixture at 25 °C for 15 min and subsequently at 95 °C for 10 min to inactivate the DNase I. The bacterial DNA extract and control were amplified with 0.5 µmol/L primers (Table 1). The PCR mixture (50 µl) contained bacterial DNA, PCR buffer (10 mmol/L Tris-HCl pH 8.3, 50 mmol/L KCl, 2 mmol/L MgCl2, and 0.01% (w/v) gelatin), 200 µmol/L of each dNTPs, and 1.0 U Taq polymerase (Applied Biosystems, Foster City, CA, USA). The mixtures were amplified in 40 cycles of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 2 min, and a final extension at 72 °C for 10 min in an automated thermal cycler (Applied Biosystems). DNase I-treated distilled water was used as the negative control. A total of 10 µl of each amplified product was electrophoresed in 1.5% (w/v) agarose gel, with a molecular size marker [PhiX174 DNA/BsuRI (HaeIII) Marker, Fermentas, Germany] in parallel. Electrophoresis in Tris-borate-ethylene diamine tetraacetic acid (EDTA) buffer was performed at 100 V for 1.5 h. The gel was stained with ethidium bromide (0.5 µg/ml) for 15 min, rinsed, and photographed under ultraviolet light illumination.
Table 1.
Primers used in this study
| Gene locus | Primers |
Amplicon size (bp) | |
| Forward | Reverse | ||
| 16S rRNA | LPW8385 5′-GAACGCTGACAGAATGCTTA-3′ | LPW8387 5′-CCAATCACTATCCACACCTTA-3′ | 1425 |
| groEL | LPW8389 5′-GTTGTGGAAGGNATGCARTTYGA-3′ | LPW8441 5′-CAGCTCCAACTTTTATTACAGCT-3′ | 555 |
| gyrB | LPW10271 5′-GGAAMWGAYRTAAGAGAAGG-3′ | LPW8399 5′-TTCATTTCTCCTAGNCCYTTRTA-3′ | 796 |
| recA | LPW8402 5′-GGTGCCGTTATGAAAYTNGGNGA-3′ | LPW10124 5′-GAACCAGGCTCCAGCTTT-3′ | 813 |
| rpoB | LPW8697 5′-AAATGGCACTTGAGCTGT-3′ | LPW8698 5′-CAATTCCAACAGTAATTCCA-3′ | 768 |
| groELa | LPW10130 5′-ATAATCGCTGAAGATGTG-3′ | LPW10132 5′-TAACTTCTCCACCTGTTAAT-3′ | 158 |
| gyrBa | LPW11369 5′-AAGCAACTTGAAATTCTATCTA-3′ | LPW11372 5′-ATATTGCCTGAAATCTTCTG-3′ | 271 |
| recAa | LPW11373 5′-CAGAAGGCTGGAGGAACG-3′ | LPW11376 5′-AGATTTTGAGATACTTCCTGTT-3′ | 308 |
| rpoBa | LPW11377 5′-ACCATCGAGTGGTAGACCA-3′ | LPW11380 5′-CTAATTCACCTTTACCAAAA-3′ | 285 |
Gene specific primers for the detection of L. hongkongensis from an oral specimen
The PCR product was gel-purified using the QIAquick PCR Purification Kit (QIAgen, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI Prism 3700 DNA Analyzer (Applied Biosystems), using the PCR primers. The sequences of the PCR products were compared with sequences of closely related species in GenBank by multiple sequence alignment using ClustalX 1.83 (Thompson et al., 1997).
2.6. Detection of Leptotrichia hongkongensis from oral specimens
Sterile polyester swabs were used to gently rub the oral mucosa and tooth surfaces of 20 healthy volunteers who were not on antibiotics in the past four weeks. Samples were suspended in 3 ml of saline followed by culturing in BACTEC Plus Anaerobic/F blood culture broth containing 40 µg/ml of metronidazole and 8 µg/ml of levofloxacin at 37 °C for 72 h. DNA was extracted from 1 ml of cultured bacterial cells using the method described above and the DNA extract further purified using QIAquick PCR Purification Kit (QIAgen).
Using L. hongkongensis gene specific primers (Table 1), a 135-bp fragment of the groEL gene, 269-bp fragment of the gyrB gene, 307-bp fragment of the recA gene, and 433-bp fragment of the rpoB gene of L. hongkongensis were amplified from the DNA extracts of the bacterial cells recovered from the oral specimens of the 20 healthy volunteers in four separate PCR reactions, using L. hongkongensis type strain HKU24T as the positive control and distilled water as the negative control. The PCR mixture (50 µl) contained bacterial DNA, PCR buffer (10 mmol/L Tris-HCl pH 8.3, 50 mmol/L KCl, 2 mmol/L MgCl2, and 0.01% (w/v) gelatin), 200 µmol/L of each dNTPs, and 1.0 U Taq polymerase (Applied Biosystems). The mixtures were amplified in 60 cycles of 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min, and a final extension at 72 °C for 10 min in an automated thermal cycler (Applied Biosystems). Agarose gel electrophoresis, purification of PCR products, and DNA sequencing were performed as described above.
2.7. Phylogenetic characterization
Phylogenetic tree construction was performed using the neighbor joining method with ClustalX 1.83 (Thompson et al., 1997). One thousand three hundred and eleven nucleotide positions of the 16S rRNA gene and 519, 692, 716, and 646 nucleotide positions of the groEL, gyrB, recA, and rpoB genes of HKU24T, respectively, were included in the analysis. A total of 158, 271, 308, and 244 nucleotide positions of the groEL, gyrB, recA, and rpoB genes, respectively, were included in the analysis of the sequences amplified from the oral specimen.
2.8. Nucleotide sequence accession numbers
The 16S rRNA, groEL, gyrB, recA, and rpoB gene sequences of L. hongkongensis (HKU24T), L. buccalis (HKU27), L. shahii (CCUG 47503), L. wadei (CCUG 47505), and L. trevisanii (CCUG 49525) have been lodged within the GenBank sequence database under accession numbers EU919515 and GU086169 to GU086196.
3. Results
3.1. Patient
A 66-year-old Chinese woman was admitted because of fever for three days. The patient had carcinoma of the left breast five years ago, having been treated with modified radical mastectomy, chemotherapy, and hormonal therapy. After defaulting further hormonal therapy for two years, the patient developed metastatic lesions in the lungs, pleura, lymph nodes, and multiple ribs one year ago, and was treated with multiple courses of chemotherapy. She also had histories of bilateral retinal detachment and cataract 20 years ago and colonic polyp. Four days before the scheduled 10th course of chemotherapy, she developed fever. On admission, her body temperature was 38.9 °C. Examination did not reveal any localizing signs. Total white cell count was 7.2×109 L−1 (neutrophil count of 5.5×109 L−1, lymphocyte count 0.8×109 L−1). Liver and renal function tests were normal. Blood culture was performed and empirical intravenous amoxicillin-clavulanate was commenced.
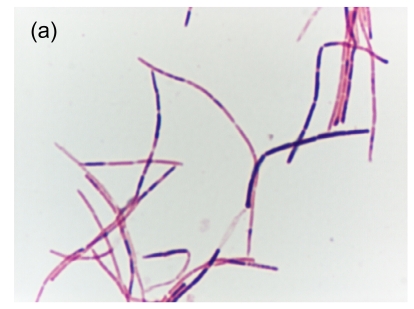
On Day 2 post-incubation, the anaerobic blood culture bottle turned positive with a non-sporulating Gram-variable bacillus (strain HKU24T) (Fig. 1a). The fever gradually subsided after three days of amoxicillin-clavulanate and antibiotics were continued for a total of 14 d. There was no relapse of the bacteremia up to the time of writing, one year after discharge.
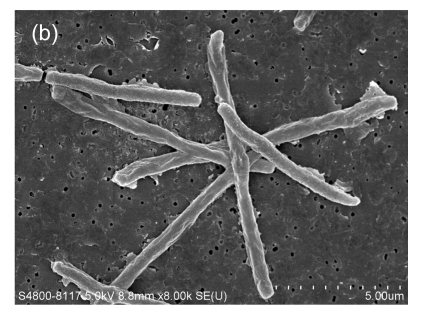
Fig. 1.
Gram smear and scanning electron micrograph of HKU24T. (a) Gram smear showing a straight, non-sporulating, Gram-variable bacillus; (b) Scanning electron micrograph showing that the bacterium is aflagellate and straight
Cells vary in length from 5.86 to 11.94 µm and diameter from 0.50 to 0.74 µm (mean=8.92 µm×0.62 µm, n=20)
3.2. Phenotypic characteristics
Strain HKU24T is a straight, non-sporulating, Gram-variable bacillus (Fig. 1a). On primary subculture, it does not grow on sheep blood agar, chocolate agar, or brucella agar. After repeated subculturing in BACTEC Plus Anaerobic/F blood culture broth, it grows on brucella agar as non-hemolytic, pinpoint colonies after 96 h of incubation at 37 °C in anaerobic environment, as well as the aerobic environment with 5% CO2. Growth is enhanced with a streak of Staphylococcus aureus. It is non-motile. It dose not produce catalase. The Vitek system (ANI and NH) showed that it was positive for alkaline phosphatase, β-glucosidase, and α-glucosidase. It hydrolyzed phenylphosphonate and reduced resazurin (Table 2). The minimal inhibitory concentrations of penicillin, metronidazole, vancomycin, amoxicillin-clavulanate, levofloxacin, and imipenem for the isolate were 0.006, 8, 0.5, 0.016, >32, and 0.012 µg/ml, respectively.
Table 2.
Biochemical profiles of the blood culture isolate HKU24T by Vitek ANI and NH systems
| Biochemical reactions/enzymes/substrates | Vitek ANI | Vitek NH |
| Arginine dehydrogenase | − | |
| Ornithine decarboxylase | − | |
| Alkaline phosphatase | + | |
| Phosphate choline | − | − |
| Urease | − | − |
| Penicillinase | − | |
| Phenylphosphonate | + | |
| Reduction of triphenyltetrazolium | − | − |
| Reduction of resazurin | + | |
| Oxidation/fermentation of: | ||
| Arabinose | − | |
| Glucose | − | − |
| Raffinose | − | |
| Trehalose | − | |
| Xylose | − | |
| α-arabinosidase | − | |
| α-fucosidase | − | |
| β-fucosidase | − | |
| α-galactosidase | − | |
| β-galactosidase | − | − |
| α-glucosidase | + | |
| β-glucosidase | + | |
| β-glucuronidase | − | |
| α-mannosidase | − | |
| β-lactosidase | − | |
| β-xylosidase | − | |
| N-acetyl-glucosaminidase | − | |
| Alanine arylamidase | − | − |
| Benzoyl-arginine arylamidase | − | |
| Gamma-glutamyl-arylamidase | − | − |
| Leucine arylamidase | − | |
| Lysine arylamidase | − | − |
| Proline arylamidase | − | − |
| Glycine arylamidase | − |
3.3. Scanning electron microscopy
A scanning electron micrograph of HKU24T is shown in Fig. 1b. Bacterial cells were aflagellate straight bacilli.
3.4. Molecular and phylogenetic characterization by 16S rRNA gene sequencing
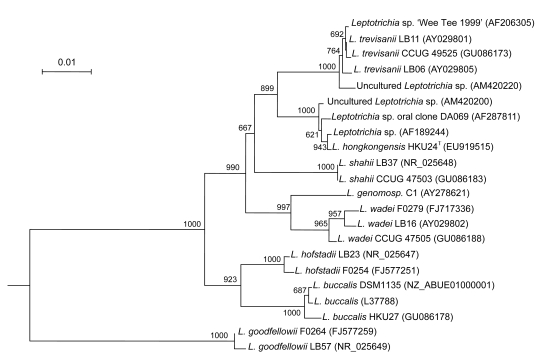
Sequencing of the 16S rRNA gene of HKU24T showed that there were two (0.15%) base differences between the 16S rRNA gene sequence of HKU24T and that of an unnamed bacterium isolated in 1999 (AF189244), five (0.38%) between the 16S rRNA gene sequence of HKU24T and that of Leptotrichia species oral clone DA 069 (AF287811), six (0.45%) between the 16S rRNA gene sequence of HKU24T and that of an uncultured Leptotrichia species oral clone 502H05 (AM420200), 34 (2.59%) between the 16S rRNA gene sequence of HKU24T and that of L. trevisanii (AY029801), 46 (3.51%) between the 16S rRNA gene sequence of HKU24T and that of L. shahii (NR_025648), 59 (4.50%) between the 16S rRNA gene sequence of HKU24T and that of L. wadei (AY029802), 70 (5.33%) between the 16S rRNA gene sequence of HKU24T and that of L. hofstadii (AY029803), 83 (6.33%) between the 16S rRNA gene sequence of HKU24T and that of L. buccalis (L37788.1), and 141 (10.76%) base differences between the 16S rRNA gene sequence of HKU24T and that of L. goodfellowii (AY029807) (Fig. 2).
Fig. 2.
Phylogenetic relationship of 16S rRNA between the L. hongkongensis HKU24T and other Leptotrichia species. The tree was inferred from 16S rRNA data by the neighbor-joining method and rooted using the 16S rRNA gene sequence of Fusobacterium nucleatum (FJ471645). Bootstrap values were calculated from 1000 trees. The scale bar indicates the estimated number of substitutions per 100 bases. Names and accession numbers are given as cited in the GenBank database
3.5. Phylogenetic characterization by groEL, gyrB, recA, and rpoB gene sequencing
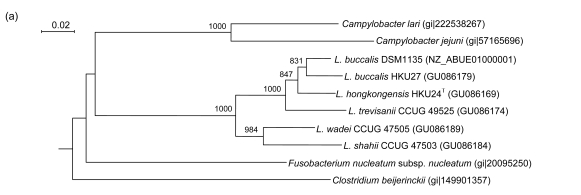
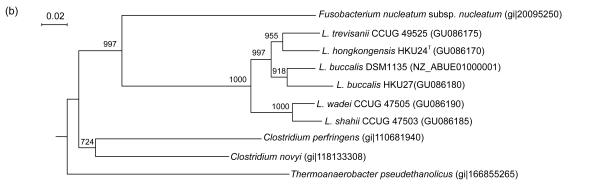
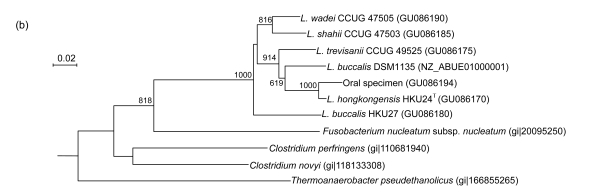
PCR of the partial groEL genes of HKU24T, L. shahii (CCUG 47503), L. wadei (CCUG 47505), L. trevisanii (CCUG 49525), and L. buccalis (HKU27) showed bands at about 550 bp. There were 22 (4.24%) base differences between the groEL gene of HKU24T and that of L. buccalis (DSM1135), 26 (5.01%) between the groEL gene of HKU24T and that of L. buccalis (HKU27), 35 (6.74%) between the groEL gene of HKU24T and that of L. trevisanii (CCUG 49525), 56 (10.79%) between the groEL gene of HKU24T and that of L. wadei (CCUG 47505), and 68 (13.10%) between the groEL gene of HKU24T and that of L. shahii (CCUG 47503) (Fig. 3a).
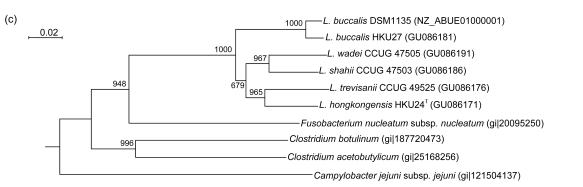
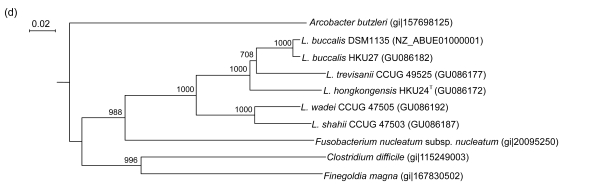
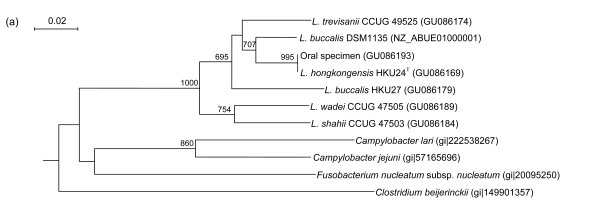
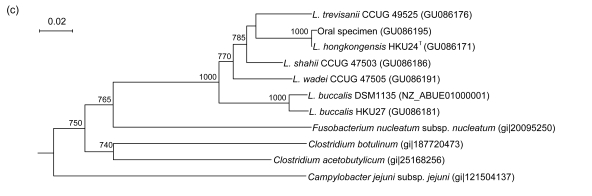
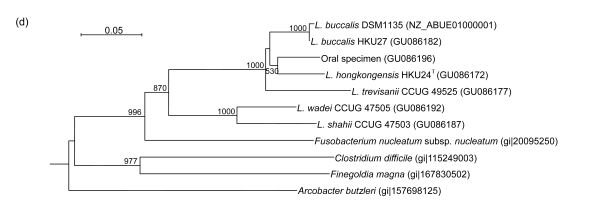
Fig. 3.
Phylogenetic relationships of groEL (a), gyrB (b), recA (c), and rpoB (d) genes between the L. hongkongensis HKU24T and other Leptotrichia species. The trees were constructed by the neighbor-joining method and bootstrap values were calculated from 1000 trees. The corresponding nucleotide sequences of Escherichia coli (NC_013364) were used as outgroups. The scale bar indicates the estimated number of substitutions per 50 bases. All names and accession numbers are given as cited in the GenBank database
PCR of the partial gyrB genes of HKU24T, L. shahii (CCUG 47503), L. wadei (CCUG 47505), L. trevisanii (CCUG 49525), and L. buccalis (HKU27) showed bands at about 800 bp. There were 40 (5.78%) base differences between the gyrB gene of HKU24T and that of L. trevisanii (CCUG 49525), 47 (6.79%) between the gyrB gene of HKU24T and that of L. buccalis (DSM1135), 62 (8.96%) between the gyrB gene of HKU24T and that of L. buccalis (HKU27), 74 (10.69%) between the gyrB gene of HKU24T and that of L. wadei (CCUG 47505), and 79 (11.42%) between the gyrB gene of HKU24T and that of L. shahii (CCUG 47503) (Fig. 3b).
PCR of the partial recA genes of HKU24T, L. shahii (CCUG 47503), L. wadei (CCUG 47505), L. trevisanii (CCUG 49525), and L. buccalis (HKU27) showed bands at about 800 bp. There were 56 (7.82%) base differences between the recA gene of HKU24T and that of L. trevisanii (CCUG 49525), 67 (9.36%) between the recA gene of HKU24T and that of L. shahii (CCUG 47503), 75 (10.47%) between the recA gene of HKU24T and that of L. wadei (CCUG 47505), and 80 (11.17%) between the recA gene of HKU24T and those of L. buccalis (HKU27) and L. buccalis (DSM1135) (Fig. 3c).
PCR of the partial rpoB genes of HKU24T, L. shahii (CCUG 47503), L. wadei (CCUG 47505), L. trevisanii (CCUG 49525), and L. buccalis (HKU27) showed bands at about 750 bp. There were 59 (9.13%) base differences between the rpoB gene of HKU24T and that of L. buccalis (DSM1135), 60 (9.29%) between the rpoB gene of HKU24T and that of L. buccalis (HKU27), 75 (11.61%) between the rpoB gene of HKU24T and that of L. trevisanii (CCUG 49525), 118 (18.27%) between the rpoB gene of HKU24T and that of L. wadei (CCUG 47505), and 130 (20.12%) between the rpoB gene of HKU24T and that of L. shahii (CCUG 47503) (Fig. 3d).
3.6. Detection of Leptotrichia hongkongensis from oral specimens
Using L. hongkongensis gene specific primers, the 158-bp, 271-bp, 308-bp, and 285-bp fragments of the groEL, gyrB, recA, and rpoB genes, respectively, could be amplified from the DNA extracted from bacterial cells recovered from one of the 20 oral specimens from healthy volunteers. Sequencing of the purified PCR products showed that there were 0, 7, 1, and 18 base differences between the 158-bp, 271-bp, 308-bp, and 244-bp fragments of the groEL, gyrB, recA, and rpoB genes, respectively, amplified from the DNA extracted from bacterial cells recovered from the oral specimen and the corresponding region of L. hongkongensis strain HKU24T (Fig. 4).
Fig. 4.
Phylogenetic relationships of groEL (a), gyrB (b), recA (c), and rpoB (d) genes between the L. hongkongensis amplified from the oral specimen of the healthy volunteer in the present study and other Leptotrichia species. The trees were constructed by the neighbor-joining method and bootstrap values were calculated from 1000 trees. The corresponding nucleotide sequences of Escherichia coli (NC_013364) were used as outgroups. The scale bar indicates the estimated number of substitutions per 20 or 50 bases as indicated. All names and accession numbers are given as cited in the GenBank database
4. Discussion
We describe a case of Leptotrichia bacteremia in a non-neutropenic patient with widely disseminated carcinoma of the breast and propose a novel species, named L. hongkongensis, in the Leptotrichia genus. The clinical significance of the bacterium was evident by its isolation from blood culture as pure growth, the patient’s systemic inflammatory response to the bacteremia, and the prompt response to amoxicillin-clavulanate treatment. The clinical setting was also similar to those in other cases of Leptotrichia infection, in which the bacterium was isolated from the blood culture of an immunocompromised patient with malignancy (Eribe and Olsen, 2008; Morgenstein et al., 1980; Reig et al., 1985; Schwartz et al., 1995; Ulstrup and Hartzen, 2006; Weinberger et al., 1991). Phenotypic characterization showed that the characteristics of HKU24T were similar to those of a bacterium isolated from the blood culture of a neutropenic 50-year-old male patient with acute myeloid leukemia on chemotherapy, also a typical clinical setting of Leptotrichia infection (Patel et al., 1999). These two strains and other species in the genus Leptotrichia were all non-sporulating, non-motile Gram-variable bacilli. On the other hand, unlike other Leptotrichia species, these two strains were catalase-negative and exhibited fastidious growth which was enhanced with a streak of S. aureus. 16S rRNA gene sequence analysis showed that there were only two to six base differences between the sequence of HKU24T and that bacterium, and two other oral clones (AF287811 and AM420200), but more than 2.5% difference compared to those of all other bacteria (Fig. 2) (Paster et al., 2001). Due to their unique phenotypic characteristics, the high nucleotide identity of their 16S rRNA gene sequences, and the significant difference from other Leptotrichia species, these two strains should constitute a novel Leptotrichia species. Interestingly, the strain recovered previously utilized glucose and sucrose (Patel et al., 1999), but this was not observed in HKU24T using both the ANI and NH panels of the Vitek system. Therefore, basic phenotypic tests and 16S rRNA gene sequencing should be used for identification of L. hongkongensis. Such variation in biochemical profiles is not uncommonly observed in other bacteria, such as viridans streptococci, where biochemical tests are relatively less useful for their identification (Woo et al., 2004b; 2004c). Collection of additional strains and performing phenotypic tests would reveal the predominant biochemical profile of the bacterium.
Based on sequence analysis of five gene loci, HKU24T is most closely related to L. trevisanii. In order to determine its phylogenetic position, we sequenced four additional housekeeping genes of HKU24T and the four most closely related Leptotrichia species shown by 16S rRNA gene sequence analysis. Results showed that HKU24T is most closely related to L. trevisanii for the gyrB and recA genes, in additional to the 16S rRNA gene, with high bootstraps values of 955 and 965, respectively (Fig. 3). Furthermore, HKU24T is also quite closely related to L. trevisanii for the groEL and rpoB genes. Interestingly, HKU24T is mostly closely related to L. buccalis for the groEL gene, with a high bootstrap value of 831, and is also quite closely related to L. buccalis for the gyrB and rpoB genes (Fig. 3). This is in contrast to its relatively distant relationship with L. buccalis in the 16S rRNA and recA genes (Figs. 2 and 3).
The oral cavity is the natural reservoir of L. hongkongensis. L. buccalis is a member of the human oral flora (Duperval et al., 1984; Weinberger et al., 1991). It causes infective endocarditis as a result of transient bacteremia due to dental procedures and neutropenic bacteremia as a result of mucositis (Caram et al., 2008; Crawford et al., 1974; Duperval et al., 1984; Eribe and Olsen, 2008; Hammann et al., 1993; Morgenstein et al., 1980; Munson et al., 2004; Reig et al., 1985; Schwartz et al., 1995; Sutter, 1984; Ulstrup and Hartzen, 2006; Weinberger et al., 1991). Among the five recently described novel Leptotrichia species, L. hofstadii, L. shahii, and L. wadei have been recovered in the human oral cavity (Eribe et al., 2004). As for L. hongkongensis, 16S rRNA gene sequence analysis showed that it is clustered with two oral clones (AF287811 and AM420200). Therefore, we used a molecular method, using multiple gene targets, to detect the presence of L. hongkongensis in oral specimens of healthy volunteers (Woo et al., 2008). Since the type strain of L. hongkongensis is resistant to metronidazole and levofloxacin, we used BACTEC Plus Anaerobic/F blood culture broth with these two antibiotics for selection and enrichment of L. hongkongensis in the oral specimens, as no L. hongkongensis was detected if antibiotics were not used in the enrichment procedure (data not shown). Since no L. hongkongensis specific primers could be designed for the 16S rRNA gene, we used L. hongkongensis specific primers for the other four housekeeping genes that we used for phylogenetic analysis to screen for L. hongkongensis in oral specimens. All four genes could be amplified from the oral specimen collected from the same healthy volunteer, and sequencing and phylogenetic analysis confirmed that they were all L. hongkongensis. These showed that similar to most other Leptotrichia species, the oral cavity is the natural reservoir of L. hongkongensis.
Leptotrichia hongkongensis sp. nov. is described as follows: Leptotrichia hongkongensis (hong.kong. en’sis. N.L. fem. adj. hongkongensis of Hong Kong, where the type strain was isolated). Cells are straight, non-sporulating, Gram-variable bacilli. On primary subculture, it does not grow on sheep blood agar, chocolate agar, or brucella agar. After repeated subculturing in BACTEC Plus Anaerobic/F blood culture broth, it grows on brucella agar as non-hemolytic, pinpoint colonies after 96 h of incubation at 37 °C in anaerobic environment, as well as the aerobic environment with 5% CO2. Growth is enhanced with a streak of S. aureus. It is non-motile. It dose not produce catalase. It is positive for alkaline phosphatase, β-glucosidase, and α-glucosidase. It hydrolyzes phenylphosphonate and reduces resazurin. The organism was isolated from the blood culture of a patient with disseminated carcinoma of the breast. The type strain of L. hongkongensis is strain HKU24T.
5. Conclusions
L. hongkongensis is a novel Leptotrichia species isolated from the blood culture of a patient with metastatic breast carcinoma. Phylogenetically, L. hongkongensis is most closely related to L. trevisanii. The oral cavity is the natural reservoir of L. hongkongensis.
Footnotes
Project partly supported by the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for Department of Health of the Hong Kong Special Administrative Region of China, the Research Grant Council Grant, the University Development Fund, the Outstanding Young Researcher Award, and the Committee for Research and Conference Grant, The University of Hong Kong, China
References
- 1.Caram LB, Linefsky JP, Read KM, Murdoch DR, Lalani T, Woods CW, Reller LB, Kanj SS, Premru MM, Ryan S, et al. Leptotrichia endocarditis: report of two cases from the International Collaboration on Endocarditis (ICE) database and review of previous cases. European Journal of Clinical Microbiology & Infectious Diseases. 2008;27(2):139–143. doi: 10.1007/s10096-007-0406-1. [DOI] [PubMed] [Google Scholar]
- 2.Crawford JJ, Sconyers JR, Moriarty JD, King RC, West JF. Bacteremia after tooth extractions studied with the aid of prereduced anaerobically sterilized culture media. Applied Microbiology. 1974;27(5):927–932. doi: 10.1128/am.27.5.927-932.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duperval R, Béland S, Marcoux JA. Infective endocarditis due to Leptotrichia buccalis: a case report. Canadian Medical Association Journal. 1984;130(4):422–424. [PMC free article] [PubMed] [Google Scholar]
- 4.Eribe ER, Olsen I. Leptotrichia species in human infections. Anaerobe. 2008;14(3):131–137. doi: 10.1016/j.anaerobe.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Eribe ER, Paster BJ, Caugant DA, Dewhirst FE, Stromberg VK, Lacy GH, Olsen I. Genetic diversity of Leptotrichia and description of Leptotrichia goodfellowii sp. nov., Leptotrichia hofstadii sp. nov., Leptotrichia shahii sp. nov. and Leptotrichia wadei sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2004;54(Pt. 2):583–592. doi: 10.1099/ijs.0.02819-0. [DOI] [PubMed] [Google Scholar]
- 6.Hammann R, Iwand A, Brachmann J, Keller K, Werner A. Endocarditis caused by a Leptotrichia buccalis-like bacterium in a patient with a prosthetic aortic valve. European Journal of Clinical Microbiology and Infectious Diseases. 1993;12(4):280–282. doi: 10.1007/BF01967258. [DOI] [PubMed] [Google Scholar]
- 7.Lau SK, Woo PC, Woo GK, Fung AM, Wong MK, Chan KM, Tam DM, Yuen KY. Eggerthella hongkongensis sp. nov. and Eggerthella sinensis sp. nov., two novel Eggerthella species, account for half of the cases of Eggerthella bacteremia. Diagnostic Microbiology and Infectious Disease. 2004;49(4):255–263. doi: 10.1016/j.diagmicrobio.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Morgenstein AA, Citron DM, Orisek B, Finegold SM. Serious infection with Leptotrichia buccalis. Report of a case and review of the literature. The American Journal of Medicine. 1980;69(5):782–785. doi: 10.1016/0002-9343(80)90452-0. [DOI] [PubMed] [Google Scholar]
- 9.Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. Journal of Clinical Microbiology. 2004;42(7):3023–3029. doi: 10.1128/JCM.42.7.3023-3029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray PR, Baron EJ, Jorgensen JH, et al. Manual of Clinical Microbiology. 9th Ed. Washington D.C.: American Society for Microbiology; 2007. [Google Scholar]
- 11.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. Journal of Bacteriology. 2001;183(12):3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel JB, Clarridge J, Schuster MS, Waddington M, Osborne J, Nachamkin I. Bacteremia caused by a novel isolate resembling Leptotrichia species in a neutropenic patient. Journal of Clinical Microbiology. 1999;37(6):2064–2067. doi: 10.1128/jcm.37.6.2064-2067.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reig M, Baquero F, García-Campello M, Loza E. Leptotrichia buccalis bacteremia in neutropenic children. Journal of Clinical Microbiology. 1985;22(2):320–321. doi: 10.1128/jcm.22.2.320-321.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz DN, Schable B, Tenover FC, Miller RA. Leptotrichia buccalis bacteremia in patients treated in a single bone marrow transplant unit. Clinical Infectious Diseases. 1995;20(4):762–767. doi: 10.1093/clinids/20.4.762. [DOI] [PubMed] [Google Scholar]
- 15.Sutter VL. Anaerobes as normal oral flora. Reviews of Infectious Diseases. 1984;6(Suppl. 1):S62–S66. doi: 10.1093/clinids/6.supplement_1.s62. [DOI] [PubMed] [Google Scholar]
- 16.Tee W, Midolo P, Janssen PH, Kerr T, Dyall-Smith ML. Bacteremia due to Leptotrichia trevisanii sp. nov. European Journal of Clinical Microbiology and Infectious Diseases. 2001;20(11):765–769. doi: 10.1007/s100960100618. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trevisan V. Prime linee d'introduzione allo studio dei Batterj italiani. Rend Reale Ist Lombardo Science. 1879;12:133–151. (in Italian) [Google Scholar]
- 19.Ulstrup AK, Hartzen SH. Leptotrichia buccalis: a rare cause of bacteraemia in non-neutropenic patients. Scandinavian Journal of Infectious Diseases. 2006;38(8):712–716. doi: 10.1080/00365540500452465. [DOI] [PubMed] [Google Scholar]
- 20.Weinberger M, Wu T, Rubin M, Gill VJ, Pizzo PA. Leptotrichia buccalis bacteremia in patients with cancer: report of four cases and review. Reviews of Infectious Diseases. 1991;13(2):201–206. doi: 10.1093/clinids/13.2.201. [DOI] [PubMed] [Google Scholar]
- 21.Woo PC, Fung AM, Lau SK, Hon E, Yuen KY. Diagnosis of pelvic actinomycosis by 16S ribosomal RNA gene sequencing and its clinical significance. Diagnostic Microbiology and Infectious Disease. 2002;43(2):113–118. doi: 10.1016/S0732-8893(02)00375-9. [DOI] [PubMed] [Google Scholar]
- 22.Woo PC, Fung AM, Lau SK, Yuen KY. Identification by 16S rRNA gene sequencing of Lactobacillus salivarius bacteremic cholecystitis. Journal of Clinical Microbiology. 2002;40(1):265–267. doi: 10.1128/JCM.40.1.265-267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo PC, Tam DM, Leung KW, Lau SK, Teng JL, Wong MK, Yuen KY. Streptococcus sinensis sp. nov., a novel species isolated from a patient with infective endocarditis. Journal of Clinical Microbiology. 2002;40(3):805–810. doi: 10.1128/JCM.40.3.805-810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo PC, Fung AM, Lau SK, Teng JL, Wong BH, Wong MK, Hon E, Tang GW, Yuen KY. Actinomyces hongkongensis sp. nov. a novel Actinomyces species isolated from a patient with pelvic actinomycosis. Systematic and Applied Microbiology. 2003;26(4):518–522. doi: 10.1078/072320203770865819. [DOI] [PubMed] [Google Scholar]
- 25.Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, Leung KW, Tam DM, Que TL, Yuen KY. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. Journal of Clinical Microbiology. 2003;41(5):1996–2001. doi: 10.1128/JCM.41.5.1996-2001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo PC, Lau SK, Woo GK, Fung AM, Yiu VP, Yuen KY. Bacteremia due to Clostridium hathewayi in a patient with acute appendicitis. Journal of Clinical Microbiology. 2004;42(12):5947–5949. doi: 10.1128/JCM.42.12.5947-5949.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo PC, Teng JL, Leung KW, Lau SK, Tse H, Wong BH, Yuen KY. Streptococcus sinensis may react with Lancefield group F antiserum. Journal of Medical Microbiology. 2004;53(Pt. 11):1083–1088. doi: 10.1099/jmm.0.45745-0. [DOI] [PubMed] [Google Scholar]
- 28.Woo PC, Tse H, Chan KM, Lau SK, Fung AM, Yip KT, Tam DM, Ng KH, Que TL, Yuen KY. Streptococcus milleri endocarditis caused by Streptococcus anginosus . Diagnostic Microbiology and Infectious Disease. 2004;48(2):81–88. doi: 10.1016/j.diagmicrobio.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Woo PC, Lau SK, Lin AW, Curreem SO, Fung AM, Yuen KY. Surgical site abscess caused by Lactobacillus fermentum identified by 16S ribosomal RNA gene sequencing. Diagnostic Microbiology and Infectious Disease. 2007;58(2):251–254. doi: 10.1016/j.diagmicrobio.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Woo PC, Teng JL, Tsang SN, Tse CW, Lau SK, Yuen KY. Oral cavity as natural reservoir for Streptococcus sinensis . Clinical Microbiology and Infection. 2008;14(11):1075–1079. doi: 10.1111/j.1469-0691.2008.02083.x. [DOI] [PubMed] [Google Scholar]
- 31.Woo PC, Teng JL, Tsang AK, Tse H, Tsang VY, Chan KM, Lee EK, Chan JK, Ma SS, Tam DM, et al. Development of a multi-locus sequence typing scheme for Laribacter hongkongensis, a novel bacterium associated with freshwater fish-borne gastroenteritis and traveler’s diarrhea. BMC Microbiology. 2009;9(1):21. doi: 10.1186/1471-2180-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]