Abstract
Proteasomes are large multi-catalytic protease complexes that are found in the cytosol and in the nucleus of eukaryotic cells with a central role in cellular protein turnover. The ubiquitin-proteasome system (UPS) is the predominant non-lysosomal protein degradation pathway that ensures the viability, proliferation and signaling of eukaryotic organisms. Overwhelming data exist implicating a critical role for the UPS in cerebral ischemic injury. and hypoxic trauma, and their associated oxidative, nitrosylative and energetic stress, underlie neurodegeneration following stroke, and evoke a discreet set of transcriptional events which have a complex and interdependent relationship with proteasomal function. Rapid elimination of denatured, misfolded and damaged proteins by the proteasome becomes a critical determinant of cell fate. Proof-of-principle has been obtained from animal models of cerebral ischemia, in which proteasome inhibitors reduce neuronal and astrocytic degeneration, cortical infarct volume, infarct neutrophil infiltration, and nuclear factor κB immunoreactivity. This neuroprotective efficacy has also been observed when proteasome inhibitors have been used 6 h after ischemic insult. Strategies aimed at effecting long- lasting changes in proteasomal function are not recommended, given the growing body of evidence implicating long-term proteasomal dysfunction in chronic neurodegenerative disease. These effects are likely due to the fact that the UPS is also essential for cellular growth, metabolism and repair, and untoward effects of proteasomal inhibition indicate that the development of short-lived proteasome inhibitors, or compounds which can spatially and temporally regulate the UPS, is a desirable clinical target. Studies in animal models indicate that the use of specific proteasome inhibitors may be beneficial in treating a host of acute neurological disorders, including ischemic stroke.
Keywords: Excitotoxicity, glutamate, hypoxia-inducible transcription factor 1α, inflammation, neurons, neurotoxicity, N-methyl-d-aspartate, nuclear factor κB, oxidative stress, proteasome, stroke, ubiquitin
Introduction
Ischemic neuronal cell death is a principal neuropathological feature of stroke, and constitutes a major source of morbidity and mortality, especially in the elderly. The widespread use of antihypertensive and antiplatelet drugs has reduced the age-adjusted incidence of this disorder, but little progress has been made in treatment [1]. The efficacy of treatments directed at revascularization, anticoagulation, inhibition of platelet function and thrombolysis is marginal or disputed. Moreover, neuroprotective strategies have predominantly been aimed at inhibiting ischemia-induced overactivation of glutamate receptors, ie, excitotoxicity, but these have not borne fruit [2••]. The therapeutic potential for developing small- molecule inhibitors for the treatment of stroke is significant, given that only a small area of the affected brain tissue, the ischemic core, is irreversibly damaged at the onset of stroke. A much larger volume of the brain tissue surrounding the ischemic core, known as the penumbra, has the potential to recover much of its function [3]. The pathophysiological mechanisms of brain injury following stroke include essential roles for inflammation, energetic dysfunction and protein destabilization [4], but targeting individual inflammatory mediators has proven difficult due to redundancy in function [5].
Several clues exist which can be used to guide the search for new therapeutic targets to limit stroke-induced cell death. Cerebral ischemia results in selective increased mRNA levels of genes involved in stress, inflammation, transcription and plasticity, as well as decreased mRNA levels of genes that control neurotransmitter function and ionic balance [6]. Ischemia activates synthesis of both potentially protective and potentially damaging proteins, including Bcl2 family members, heat shock protein (Hsp), inhibitors of apoptosis proteins (IAPs), hypoxia-inducible transcription factor (HIF)-1α, nuclear factor κB (NFκB), and a host of transcription factors which are all targets of 26S proteasome. The proteasome is an ATP-dependent protein degradation system, which degrades short-lived proteins under normal metabolic conditions, as well as bulk degrading long-lived proteins, partially digesting/processing some regulatory proteins and being involved in antigen presentation. Among the key regulatory proteins degraded by the 26S proteasome are the transcription factor c-Fos, M, S, and G1 phase-specific cyclins, cyclin dependent kinase inhibitors, p53 and a host of oncoproteins [7••,8,9].
Significant data exist to support the hypothesis that loss of protective and homeostatic proteins is a critical determinant of cell fate [10••]. This hypothesis is supported by the phenomenon of ischemic tolerance, in which mild ischemic injury increases resistance to subsequent stressors in a protein synthesis-dependent manner. The non-lethal activation of reactive oxygen species (ROS), and even caspases, is essential for neuroprotection [11]. The molecular chaperone Hsp70 is upregulated by the initial stressor, and it subsequently performs essential functions in protein refolding, as well as targeting denatured proteins to the proteasome for degradation. Inhibitors of proteasomal function rapidly increase Hsp70 expression and, like many other chaperones and chaperone-binding proteins, Hsp70 can block a host of acute and chronic conditions, including metabolic diseases, cancer, autoimmune disorders and neurodegeneration [12]. When protein refolding becomes untenable due to excessive denaturation, the majority of proteins become ubiquitinated and degraded by the proteasome. Rapid elimination of denatured, misfolded and damaged proteins by the ubiquitin-proteasome system (UPS) is a critical determinant of cell fate.
The degradation of cellular proteins is a highly complex, temporally controlled and tightly regulated process, and is essential for mitosis, energy and ion homeostasis, as well as revascularization and repair. Since a multitude of protein substrates are targeted and a myriad of processes are involved, it is not surprising that aberrations in the UPS are implicated in the pathogenesis of cerebral ischemia. The advent of powerful new genomic and proteomic technologies has substantially enhanced our understanding of the evolution of ischemic injury. Understanding the identity and interactions of proteins as a part of complex networks is the central problem in the post-genomic era. In this review we focus on the recent advances in our understanding of the UPS in cerebral ischemia, as well as on the novel approaches for stroke intervention that these studies have revealed.
Biology of the UPS
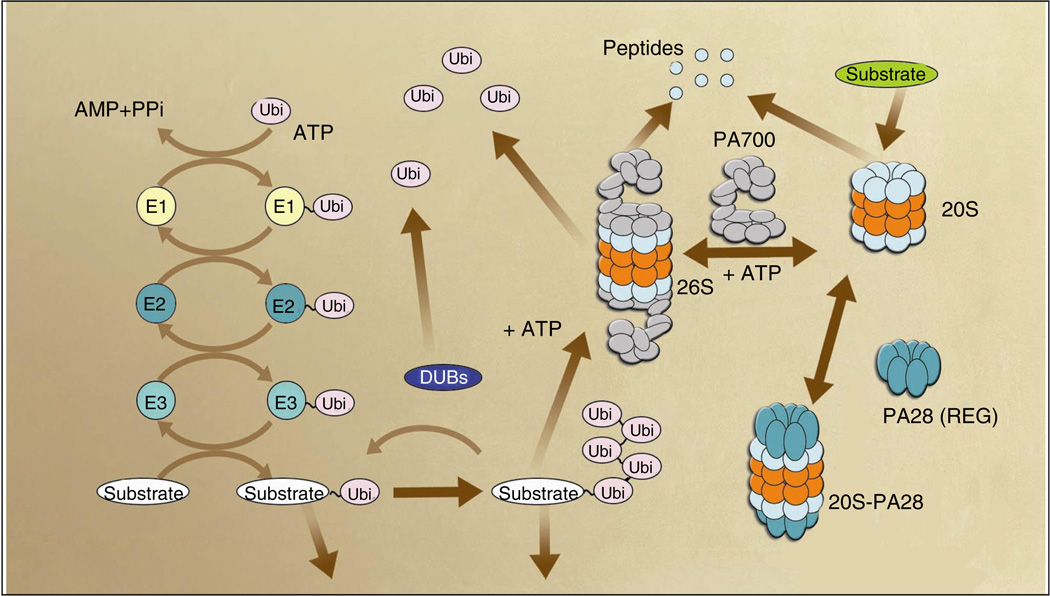
The UPS is essential for the non-lysosomal degradation of short-lived, mislocated, misfolded, mutant and damaged proteins in eukaryotic cells [13]. This pathway plays a major role in the breakdown of abnormal proteins that results from oxidative stress and mutations that may otherwise disrupt normal cellular homeostasis. There are two major steps in the protein degradation pathway. Firstly, substrates are polyubiquitinated in a process that is tightly controlled by both ubiquitinating and de-ubiquitinating enzymes, and secondly, polyubiquitinated substrates are selectively recognized and degraded by the 26S proteasome (Figure 1). Protein substrates are tagged by covalent attachment of multiple ubiquitin (Ub) molecules to generate the polyubiquitin chain that serves as a recognition marker for the proteasome. Ub is a well-conserved 76-amino acid polypeptide that controls many cellular processes and aids in protein and organelle trafficking, cell-cycle control, stress response, DNA repair, immune response, signal transduction, transcriptional regulation and endocytosis [14,15••]. The covalent attachment of Ub and Ub-like proteins has also emerged as a major means of post-translationally modifying macromolecules. At least ten different Ub-like modifications exist in mammals which can alter protein structure, half-life, enzymatic activity, interactions and subcellular localization [16–18]. These changes in mono-ubiquitination are particularly relevant for oncogenic protein transformation [19] and act as a signal in nuclear export for the tumor suppressor protein p53. Ub linkages via Lys63, rather than Lys48, activate the IκB kinase [20,21]. Receptor tyrosine kinases (RKTs) are downregulated in a proteasome-independent manner by ubiquitination, and overactivation of growth-factor sensitive RTKs has been linked to human neoplasias [22–25]. Targeted activation of proteins associated with cervical cancer, retinoblastoma and colon cancer can also be achieved by altering Ub ligases[26,27].
Figure 1. The Ub-proteasome pathway.
Conjugation of Ub to the protein substrate proceeds via a three-step cascade mechanism. In the first step of ubiquitination, the Ub-activating enzyme, E1, activates Ub in an ATP-dependent manner to generate a high-energy thiol ester intermediate, E1-S~Ub. E2 enzymes, known as Ub-carrier proteins or Ub-conjugating enzymes, then transfer the activated Ub from E1 to the substrate that is specifically bound to a member of the Ub-protein ligase family, E3 via E2-S~Ub, an additional high energy thiol ester intermediate. E3s belong to the family of either HECT (homologous to E6-associated protein C-terminus) or RING (really interesting new gene) finger E3s and recognize protein substrates directly, or via active cysteine residues or through adapter subunits. E3s catalyze the last step in the conjugation process, namely the covalent attachment of Ub to the substrate. While there are 20 to 30 E2s, and presumably more than 100 E3s, there is only one E1 in mammals. Thus, E3s play a key role in the Ub-mediated proteolytic cascade by adding substrate, temporal, and even regional, specificity to the degradation cascade.
The second major step in the Ub-proteasome pathway is the degradation of polyubiquitinated proteins by the 26S proteasome, which are multisubunit proteases that only recognize modified substrates. The catalytic core of the 26S proteasome is the 20S proteasome which is composed of 28 subunits arranged in four heptameric stacked rings; two identical outer α~ rings and two identical inner β-rings, which form a cylindrical structure. The catalytic sites within the β-subunits are located inside the 20S particle. This region contains multiple subunits that possess peptidase activities: trypsin-like, chymotrypsin-like, caspase-like, peptidylglutamyl peptide hydrolase, branched chain amino acid-preferring, and small neutral amino acid-preferring. Most 20S complexes are capped by two 19S complexes, which provide the regulatory moiety of the 26S proteasome. The 19S particle contains at least 17 subunits, including ATPases, a de-ubiquitinating enzyme and polyubiquitin-binding subunits. The 19S regulatory particle (RP) can be further dissected into two multisubunit sub-complexes that are referred to as the lid and the base. The base, which generates a direct contact with the α-ring of the 20S complex, is made up of six homologous ATPases (Rpt1 to 6), together with three non-ATPase subunits (Rpn1, 2 and 10). The lid of the RP is made of eight non-ATPase subunits (Rpn 3 to 9) and is required for degradation of ubiquinated proteins. DUBs de-ubiquitinating enzymes,
In the Ub-proteasome degradation pathway, Ub is first activated by an activating enzyme and is then transferred to the conjugating enzyme. Subsequently, the conjugation is performed by a series of enzymes known as E1, E2 and E3. The E1 forms a high-energy thioester bond with Ub that is transferred to a reactive cysteine residue of the E2 enzyme. The final transfer of Ub to an ε-amino group of a reactive lysine residue of the substrate protein is brought about by E3, the ubiquitin ligase enzyme. The fate of ubiquitinated proteins depends in part on the length and linkage type of the Ub chain. In general, substrates with four or more Ub moieties linked via Lys29 or Lys48 are targeted for degradation by the 26S proteasome [28,29••] Alternate ubiquination of Lys63 is involved in regulation of endocytosis [30], mitochondrial inheritance [31], ribosome function [32], post-replicative DNA repair [33] and kinase activation [34]. Attachment of a single Ub moiety is essential to endocytosis of plasma membrane proteins, protein sorting and sub-nuclear trafficking [35]. Tagged proteins are degraded by the 26S proteasome, resulting in release of free and re-usable Ub. This process is mediated by Ub recycling enzymes, such as isopeptidases, also known as ubiquitin-specific proteases (UBPs) and de-ubiquitinating enzymes (DUBs) [29••]. These enzymes maintain sufficient free Ub levels for normal cell functioning and remove polyubiquitin chains that may interfere with the recognition of substrates by the proteasome [36].
The 26S proteasome is a multicomponent complex with a native molecular mass of approximately 2000 kDa (Figure 1). The proteasome is composed of a 20S catalytic core particle (CP), and a regulatory 19S regulatory particle (RP). The 19S RP recognizes ubiquitinated proteins and other potential substrates via two subunits referred to as Rpn10 and Rpt5 (S5a and S6' in mammalian cells, respectively) (Figure 2). The 19S RP also opens an orifice in the α-ring, allowing substrate entry into the proteolytic chamber. The 19S RP consists of approximately 17 subunits that include ATPases, a de-ubiquitinating enzyme, and polyubiquitin-binding subunits. ATPases provide energy for these processes as well as the assembly of the 26S proteasome and the injection of substrates into the catalytic chamber. The proteolytic core of this complex, the 20S proteasome, is composed of 28 subunits arranged in four heptameric, tightly stacked rings (α7, β7, β7 and α7) to form a cylindrical structure. The α-subunits make up the two outer rings and the β-subunits the two inner rings of the stack. The entrance to the active site of the complex is guarded by the α-subunits that allow access for the unfolded and extended polypeptides only. This core contains multiple peptidase activities, including chymotrypsin-like activity (ChTL, cleavage after hydrophobic side chains), caspase-like activity (CaL), postglutamyl peptidase activity (PGPH, cleavage after acidic side chains) and trypsin-like activity (TL, cleavage after basic side chains). Furthermore, two additional endopeptidase activities cleave bonds after branched-chain amino acids (BrAAP activity) and after small neutral amino acids (SNAAP activity) in mammalian proteasomes [37]. A new peptidase, tripeptidyl peptidase II (TPPII), that co-purifies with the 26S proteasome was identified [38,39]. TPPII is believed to participate in the degradation of extra-lysosomal polypeptides and may substitute for some metabolic functions of the proteasome, particularly in the absence of normal proteasome function [39,40]. The degradation products of the proteasome are short peptide fragments and amino acids that can be recycled to produce new proteins. These peptides are further degraded to amino acids by cytosolic amino- and carboxypeptidases [15••].
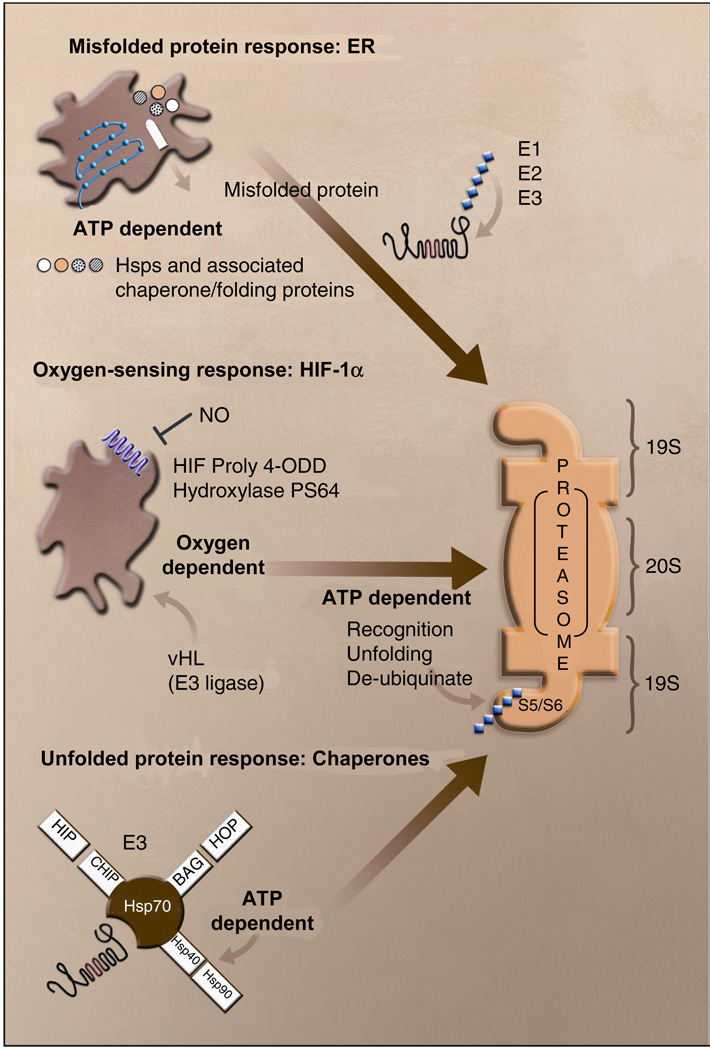
Figure 2. Cellular responses to ischemic stress.
Multiple energy and oxygen-dependent pathways converge on the proteasome in order to remove damaged proteins and activate transcription factors with roles in increasing oxygenation, vascularization and survival. Examples of these pathways are shown for the unfolded protein response in the endoplasmic reticulum (ER), the stabilization of hypoxin-reducible transcription factor (HIF)-1α and the chaperone triage system. Protein trafficking and assembly in the ER is responsive to multiple energetic and ionic perturbations, including calcium buffering, redox regulation, caspase activation and post-translational modification of proteins. These functions are dependent upon ATP content and redox-sensitive proteins, including the Sec ATPases (SEC). Misfolded proteins are targeted to the proteasome through ubiquitination (E1, E2 and E3 enzymes). Oxygen sensitivity is demonstrated in the HIF-1α subunit of the HIF transcription factor. Under normoxia, HIF hydroxylases modify proline residues within the oxygen-dependent domain (ODD) of HIF allowing for rapid ubiquination by the von Hippel–Lindau (vHL) E3 ligase. Anaerobic conditions block these modifications leading to enhanced HIF-1α expression and transcriptional activity. Finally, chaperones play an essential role in the stress response to ischemia. Some of the interactions of the stress-inducible chaperone Hsp70 are illustrated. Competition for Hsp70 binding leads to altered chaperone activity, in which the protein moves from a primary role in refolding, to one of targeting proteins to the proteasome to be degraded. Again, the function of this protein is dependent upon ATPase activity of interacting chaperones. BAG Bcl-2 binding athanogene, CHIP c-terminal Hsc70 (a constitutively expressed homolog of Hsp70)-interacting protein, HIP Hsc70-interacting protein, HOP Hsc70-Hsp90-organizing protein, Hsp heat shock protein, NO nitric oxide, PS 64
The UPS in cerebral ischemia
The UPS plays a complex and central role in cerebral ischemia/reperfusion injury [41•] Aberrant proteasomal function has been observed in experimental models of stroke [41•,42•,43••] and loss of proteasomal activity occurs within 10 min of global ischemia [44]. Even in these short periods, episodes of ischemia induce not only a transient decrease in proteasomal activity in the central nervous system (CNS), but also an accumulation of Ub proteins [45•] in multiple models of transient global, as well as focal, ischemia [46,47], and in in vitro models of neuronal hypoxia [48].
The temporal window of proteasomal inhibition is essential to determining fate, since long-term inhibition of the proteasome is a potent neurotoxic stimuli [49•]. However, mild proteasomal perturbation is a highly effective mechanism with which to induce neuroprotective protein expression, block deleterious effects associated with inflammation and enhance energetic status. These observations support the therapeutic potential of delivering small-molecule therapies, which can spatially, and/or temporally, restrict proteasomal degradation and enhance cell survival.
High-resolution confocal microscopy has demonstrated that protein aggregates are found surrounding nuclei and along dendrites in post-ischemic neurons [50•]. These aggregates contain ubiquitinated proteins, which can also be found in the neuronal soma, dendrites and axons. In the post-ischemic phase, ubiquitinated proteins are associated with intracellular membranous structures [50•,51]. Ub proteins in neuronal lysosomal vesicles and in late endosome-like organelles in the ischemic area may result from an attempt to eliminate accumulating Ub proteins by autophagy [50•,51]. Biochemical characterization of Ub-immunoreactive material is an area of active research. As in many chronic neurodegenerative diseases, Hsps co-localize with Ub proteins in the inclusions [52,53]. Expression of Hsp70 is increased following ischemia in vivo and in vitro [53–55], and in ischemic stroke [52]. The Hsps are highly conserved, abundantly expressed proteins with diverse functions, including the assembly of multiprotein complexes, transportation of nascent polypeptides and regulation of protein folding [56]. Hsp70 is the major inducible Hsp found in cells [57], and both it and its constitutively expressed homolog Hsc70, interact with many of the same binding partners and client proteins. In addition to aiding in protein refolding, the Hsp70 family can sequester activated caspases and other cell death proteins [58•,59,60].
Ischemic injury, ROS generation and injuries that induce protein denaturation increase Hsp70 protein expression [57], and overexpression of Hsp70 protects against glutamate toxicity, ischemia and oxidative injury [61,62]. This protein functions as a part of a multiprotein complex and association with different binding partners can dramatically alter its function. For instance, the E3 Ub chain formation protein C-terminal Hsc70 interacting protein (CHIP) competes with Hsc70-Hsp90-organizing protein for c-terminal binding to Hsp70. Similarly, Bcl-2 binding athanogene (BAG)-1 competes with Hsc70-interacting protein for N-terminal binding. Formation of BAG-1/Hsp70/CHIP complexes is thought to redirect Hsp activity away from protein refolding and towards ubiquitination and proteasomal degradation (Figure 2) [63]
The Ub ligase activity and link to proteasomal function of CHIP can be critical in mediating protein refolding and degradation in other degenerative conditions, including familial Parkinson's disease (PD) caused by mutations in the Parkin gene, as well as in the cystic-fibrosis transmembrane-conductance regulator which controls chloride-ion channel function, and the protein tau, which is altered in individuals with a number of neurodegenerative diseases including Alzheimer's disease [64–66]. Proteasome activity seems to be correlated with the stability of the neurodegenerative disease-associated proteins and their fragments, which are responsible for the generation of disease pathology. For PD, it is well known how the UPS malfunction occurs. Firstly, mutations of an E3 Ub ligase, Parkin, in PD abrogates its enzymatic activity for ubiquitinating substances, including unfolded Pael receptor (Pael-R), and is toxic for neurons [64,67] and, secondly, the loss of Ub C-terminal hydrolase L1 (UCHL1) activity by mutations increases susceptibility to the illness [68]. Furthermore, the accumulation of Ub conjugates seems to be uncommon in neuronal death; the Ub conjugates are accumulated by β-amyloid protein in primary cortical neuron cell cultures and proteasome inhibition potentiates β-amyloid-induced neuronal death [69], although treatment with proteasome inhibitors effectively reduces neuronal and astrocytic degeneration during the ischemic stress produced by stroke [43••].
It has been reported that proteasome inhibition is sufficient to induce both the formation of cellular aggresomes composed of aggregation-prone proteins and neuronal death [70,71], but it remains to be determined whether protein aggregation causes neuronal degeneration or is a rescuing mechanism in the cell. The six identified members of the BAG family of proteins are generally considered to confer resistance to cell death. These proteins have a variety of binding partners and are essential for cell division, death and differentiation [72]. Recently, the role of BAG5 has been revealed as more complex than originally thought, since it promotes neurodegeneration in dopaminergic cells by sequestering Parkin via the inhibition of Hsp70 [73]. The complexity of each of these protein-protein interactions for decisions of refolding, stress signaling and cell fate in chronic neurodegerative diseases speaks of the need to develop subtle and physiologically relevant means with which to regulate protein stability, particularly in the CNS.
With regard to hypoxic signaling, Tanaka et al [74] have shown that protein disulfide isomerase (PDI), an enzyme that also catalyzes protein-folding reactions, is also upregulated in response to CNS hypoxia. Like Hsp70, PDI is intimately linked with the proteasome. It shuttles proteins for proteasomal degradation via interactions with the Ub-like protein ubiquitin. Moreover, enhancing PDI function confers cytoprotection following stroke.
One of the most rapid changes induced by hypoxia is an increase in the expression of HIF-1α. The HIF-1 complex is composed of two protein subunits, HIF-1β/ARNT (aryl hydrocarbon receptor nuclear translocator), which is constitutively expressed, and HIF-1α, which is rapidly ubiquinated and degraded to near negligible levels in normoxic cells. HIF-1α expression is increased by hypoxia, resulting in enhanced expression of genes associated with oxygen binding, glycolysis, glucose transport, vasodilation and angiogenesis [75]. The rapid turnover of HIF-1α in normal cells is mediated by hydroxylation on two proline residues within a conserved oxygen-dependent degradation domain. In the presence of oxygen, these prolyl residues are enzymatically hydroxylated, which promotes interaction with the von Hippel–Lindau (vHL) tumor suppressor protein, leading to ubiquitylation by vHL-associated E3 ligase and subsequent proteasomal destruction. As a consequence of gene mutations, individuals with vHL disease are susceptible to retinal angiomas, hemangioblastoma lesions of the brain and spinal cord, as well as tumors in other areas, including the pancreas, kidney and adrenal glands [76]. These tissues are generally highly vascularized tissues and overproduce peptides and hormones, such as erythropoietin and vascular endothelial growth factor, which are involved in angiogenesis and hypoxia sensing. The hydroxylation of HIF-1α is mediated by three to four HIF prolyl hydroxylases (HIF PHDs) (Figure 2) which are substantially altered by stress and are subject to proteasomal degradation. Siah E3 Ub ligases are induced by hypoxia, and their binding to HIF PHDs removes the impetus to degrade HIF, resulting in HIF complex stabilization, nuclear translocation and transcription. The importance of this feedback loop has recently been demonstrated in Siah knockout cells, wherein HIF-1α expression does not increase as oxygen levels drop [77]. Thus, the function and stability of HIF are intimately linked to proteasomal function. Not surprisingly, proteasome inhibitors produce dramatic increases in HIF-1α [78], and also accelerated revascularization and red blood cell maturation to enhance oxygen binding and delivery in vitro [79]. However, the mechanism by which hypoxia inhibits vHL-mediated ubiquitination of HIF-1α is currently unknown.
Given the linkage between energetic status and protein aggregation, it is tempting to speculate that the loss of E3 Ub ligases, such as Siah, into proteinacious aggregates formed during ischemia and chronic neurodegenerative diseases, compromises the ability of neurons to mount an appropriate stress response through HIF and other transcriptional elements [80].
UPS response to changes in cellular homeostasis
As outlined in Figure 1, ubiquitination and degradation processes are heavily dependent on ATP. Loss of ATP resulting from oxygen deprivation has profound and immediate consequences on a host of cellular functions, including proteasomal function [42•,81,82]; 1 h transient focal cerebral ischemia induces marked depletion of the E3 ligase parkin, but does not affect levels of E2. Parkin upregulation can protect cells from injury induced by endoplasmic reticulum (ER) stress, which suggests that parkin depletion may increase the sensitivity of neurons to stimuli managed by the ER, including the unfolded protein response and calcium dysregulation which are induced by loss of cellular ATP levels [83]. In the ischemic core, ATP-and Ub-dependent degradation mediated by the 26S proteasome is impaired, while the ATP- and Ub-independent degradation mediated by 20S proteasome is unimpeded. This is likely due to the dissociation of 26S proteasomes into 20S proteasomes and PA700 caps, which occurs under stress. While the 26S proteasome activity recovers in many regions following reperfusion, particularly vulnerable areas, including the CA1 region of the hippocampus, PA700 and 20S proteasomes do not fully reassociate - a problem which may underlie the delayed neuronal cell death in these regions [42•].
Another consequence of oxygen deprivation is the increase in cellular acidosis. While the molecular site of action of changes in pH has not been localized within the proteasome, it is clear that loss of proteasome activity occurs as pH levels decrease. The 20S subunit contains activity of five different proteases, including those which resemble trypsin, chymotrypsin and caspases, all of which are less effective with decreasing pH [84]. Proteasomes may also be altered by direct transient denaturation induced by pH, and indirectly by enhanced formation of free radical formation (via iron delocalization and the Fenton reaction) [85], or more specifically by altering catalytic activity of the proteasomal complex, as well as by altering the action of Ub-protein-ligase complexes [86,87].
UPS and excitotoxic cell death
The loss of ATP that occurs in ischemic injury results in an inability to remove glutamate from the synaptic cleft by ATP-dependent transport, reversal of the glutamate transporters and excessive stimulation of N-methyl-d-aspartate (NMDA) and other glutamate receptors [88,89]. The overstimulation of NMDA receptors leads to calcium influx [90], which is associated with enhanced cytotoxic activation of nitric oxide synthase [91], calpain [92], phospholipase A2 [93] and mitogen-activated protein kinases [94]. NMDA-induced calcium entry can also uncouple respiration from ATP synthesis and result in production of free radicals, thus enhancing this cytotoxic cascade [88,89,95]. Chronic proteasome inhibition is deleterious to cells since it severely impedes mitochondrial calcium buffering, and results in loss of mitochondrial membrane potential (Δψm), formation of dense mitochondrial deposition and cytochrome C release into the cytosol, with secondary dilation of the rough ER, formation of cytoplasmic vacuoles and activation of caspase 3 [96–99].
In the context of stroke-induced injury in the infracted core, the early phase of glutamate-induced necrosis apparently does not require proteasome activation [100]. However, in the region most therapeutic agents are designed to rescue, a complex interaction feedback loop exists between glutamate-mediated excitation and proteasomal degradation, in that stimulation of glutamate receptors by NMDA application leads to ubiquitination of scaffolding protein PSD-95 via the E3 Ub ligase Mdm2 and its proteasomal degradation [101–104]. This process requires calcium-dependent dephosphorylation of protein kinase A substrates [105,106]. PSD-95 degradation likely has important ramifications for synaptic transmission, stability and plasticity, particularly given that PSD-95 is also associated with glutamatergic α-amino-3-hydroxymethyl-4-isoxazolyl-propionic acid receptors, which are endocytosed in an NMDA receptor-dependent manner.
Moreover, Arundine and Tymianski [107] have shown that an HIV-Tat fusion protein containing C terminal NMDA residues, designed to interfer with PSD-95/receptor interactions, blocks excitotoxic cell death in vitro, as well as blocking ischemia in vivo. This suggests that proteasomal degradation of PSD-95 is essential for dampening excitotoxic insults, and that global proteasome inhibition would have long lasting effects on synaptic efficacy, as well as possibly leading to sustained glutamatergic transmission and enhanced neurotoxicity under pathophysiological conditions. The observation that NMDA receptor stimulation upregulates expression of the p112 proteasome subunit of PSD-95 suggests that glutamate receptor antagonists may exacerbate cell death induced by exposure to proteasome inhibitors [108]. In summary, these observations suggest that an important reciprocal relationship exists between the proteasome and synaptic activity. Following cerebral ischemia-reperfusion injury, there is a time-dependent decrease in proteasome activity, which is associated with oxidative stress [100]. ROS modify several proteasome subunits (α1, α2 and α4) and impair proteasome activity [109,110]. 20S proteasomes can degrade mildly oxidized proteins without previous ubiquitination, but they are unable to degrade extensively oxidized proteins [111,112]. Moreover, oxidative damage enhances the effects of proteasome inhibition, leading to protein aggregation and cell death [113]. The oxidation of disulfide bonds within transcription factors and binding proteins, as well as the ionic dysfunction associated with energetic failure, results in free transition metals that oxidize the 20S proteasome. Trypsin-like (TL) activity gradually increases, while chymotrypsin-like (ChTL) and peptidylglutamyl-peptide hydrolase (PGPH) activities are substantially inhibited [85].
Proteasome interaction with apoptogenic proteins
Caspases are a family of highly conserved cysteine proteases that are required for programmed cell death [114], and they cleave substrates, including structural proteins, DNA repair and cleavage enzymes and kinases, as well as proteins which inhibit apoptosis [115] Activation of caspases 1, 3, 8, 9 and 11 and release of cytochrome C have all been observed in cerebral ischemia [116]. Moreover, inhibition of caspases reduces tissue damage and improves neurological outcome [117,118] even when delivered following damage [119,120]. During apoptosis, caspases cleave subunits of the 19S regulatory complex of the proteasome, including S6' (Rpt5) and S5a (Rpn10), whose role is to recognize polyubiquitinated substrates, as well as S1 (Rpn2), which cooperates with other subunits to hold together the lid and base. These cleavage events inhibit the proteasomal degradation of Ub-dependent and -independent cellular substrates, including pro-apoptotic molecules such as Smac, caspases themselves, p53 and other pro-apoptotic molecules. Caspase cleavage of the proteasome is thought to enhance the execution of the apoptotic program by providing a feedforward amplification loop [121••]. The existence of this positive feedback loop in caspase-mediated proteasome inactivation suggests that blocking 19S proteasome is vital for the effective function of the apoptotic machinery. Another consequence of this proteolysis is the partial dissociation of the 19S regulatory complex from the 20S proteasome. Uncoupling of the regulatory particle from the catalytic core particle may diminish proteasome-mediated ATP consumption and preserve cellular energy for certain ATP-sensitive steps of apoptosis.
UPS and gene-mediated effects on nuclear factor κB
Nuclear factor (NF)κB is central to the regulation and expression of the stress response to inflammation and oxidative stress [122]. The 26S proteasome can activate members of the NFκB/Rel transcription factor family through the proteolytic removal of its endogenous inhibitor, IκB [123]. NFκB is normally cytoplasmically bound to IκBs, and the best-characterized member of this family, IkBα, binds the p50/p65 heterodimer of NFκB in the cytoplasm. Following stressful stimuli, two serine residues of IκBα are specifically phosphorylated, serving as a potent ubiquitination signal to stimulate the degradation of IκBα by proteasome. NFκB is released from IκBα, revealing a nuclear localization signal [124]. Nuclear trafficking and promoter binding upregulates a host of genes critical to determining cell fate following stress [125].
Preclinical trials are aimed at blocking NFκB activity using gene therapy, peptides, small-molecules and proteasome inhibitors, while other neuroprotective compounds (eg, antioxidants, natural products, salicylates and non-steroidal anti-inflammatory drugs) can also block NFκB [126]. Several studies have now demonstrated that free-radical damage following ischemia is at least partially mediated by NFκB. NFκB is activated in the core and penumbra 1 day after 90-min temporary focal ischemia of the cortex, and the proteasome inhibitor MLN-519 attenuates damage measured 24 h after 2-h ischemia [127], as well as inducing inflammatory cytokines and cell adhesion molecules [127–129].
Pharmacological inhibition of proteasome: The proteasome inhibitors
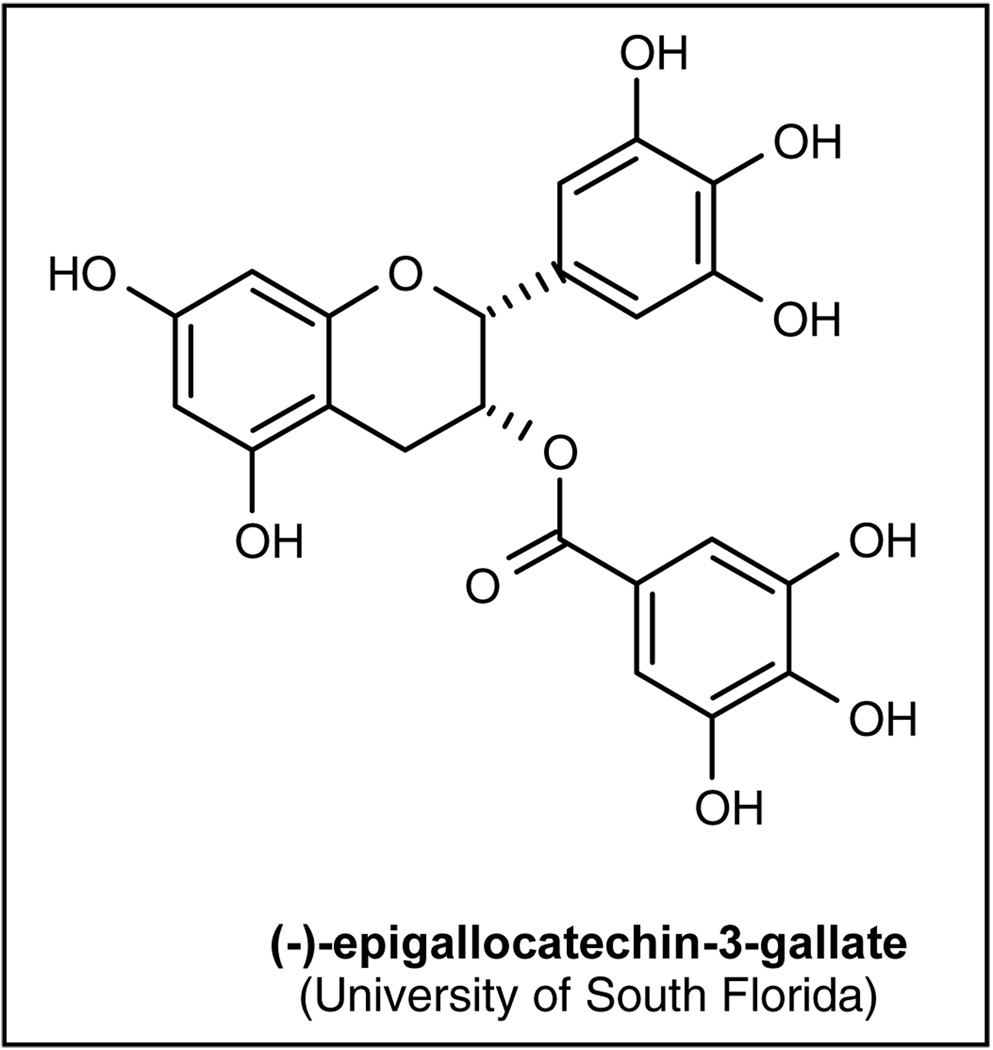
Several natural and synthetic compounds that act as proteasome inhibitors with different chemical characteristics have been reported, and several reviews detail their properties and mechanisms of action [41•,43••,130,131•]. These compounds either block the active sites of the 20S proteasome core particle non-covalently by specific hydrogen binding with the main-chain atoms of the protein and without modifying the nucleophilic Thr1 residue [132], or by targeting all individual active subunits with comparable affinity (peptide vinyl sulfones) [133]. The tripeptide aldehyde compounds are reversible inhibitors of ChTL, PGPH and TL activities of the proteasome. Another class of compounds, vinyl sulfones, act as suicide substrates for the active site nucleophiles. Lactacystin, from a third group of compounds, is a covalent inhibitor of the ChTL and TL activities of the proteasome, and its action is thought to be due to the action of its β-lactone form, which is produced on incubation in the aqueous medium. Some of these inhibitors also have a significant inhibitory effect on the activity of TPPII. Several new inhibitors have been reported in later years. Eponemycin and epoximycin were originally described as anticancer agents produced by a Streptomyces strain, but were later found to be proteasome inhibitors. They covalently modify the catalytically active N-terminal Thr residues of β5 and β2 subunits, which results in a strong inhibition of the ChTL activity and a much weaker inhibition of the PGPH and TL activities [134,135]. Gliotoxin, a fungal metabolite responsible for various toxic effects accompanying Aspergillus fumigatus and Candida albicans infections, is also an effective inhibitor of the proteasome [136]. Several other antibiotics that inhibit the different activities of the proteasome have also been described. Tannic acid and ester bond-containing tea polyphenols, such as (−)-epigallocatechin-3-gallate (EGCG; University of South Florida, Figure 3), which are commonly present in green tea, inhibit the ChTL activity of the proteasome, causing cell-cycle block and apoptosis [137,138], and various drugs used in clinical practice interact with the UPS. The inhibitor of the HIV protease, ritonavir, is also a potent inhibitor of the ChTL activity of the proteasome [139] Lovastatin, an inhibitor of the hydroxymethyl glutaryl-coenzyme A reductase, used commonly to lower cholesterol plasma levels also partially inhibits some proteasome activities, while doxorubicin binds to the proteasome without inhibiting it [140]. Vinblastine inhibits proteasome activity in vitro and causes accumulation of ubiquitinated proteins in vivo, as well as exerting its effects on microtubules [141].
Figure 3.
The structure of (−)-epigallocatechin-3-gallate
Effects of proteasome inhibitors in animal models of focal ischemia and reperfusion injury
While the specificity of all proteasome inhibitors is not yet known, several of these compounds have been tested in stroke models (Table 1) [127,129,142–144]. One of the most well understood drugs is MLN-519, which consistently reduces cerebral infarct volume after middle cerebral artery occlusion in a dose-dependent manner with a therapeutic window of up to 6 h after onset of ischemia. MLN-519 also exhibits a neuroprotective effect following focal brain ischemia with a 50 to 60% reduction of infarct volume and decreased leukocyte infiltration, as well as prevention of NFκB activation following reperfusion [127,129]. When MLN-519 was used in combination with tissue plasminogen activator (tPA) in an embolic stroke model, it decreased infarct volume and improved neurological outcome 1 week after the ischemic episode, as well as eliminating hemorrhage associated with tPA treatment, even when administered 6 h after vessel occlusion [145]. These neuroprotective effects were also replicated in models of cerebral hemorrhage [146•]
Table 1.
Animal models of cerebral ischemia and proteasome inhibitor treatment.
| Treatment | Year | Model of ischemia/strain | Results | Comments | Reference |
|---|---|---|---|---|---|
| CVT-634 | 2000 | 90 min MCAO/inbred SH rats. | Smaller infarct of 13 ± 2% (p < 0.01) and 2 ± 2% (p < 0.001) of hemispheric volume at 1 day and 7 days. |
Regional cerebral blood flows were not affected. |
[144] |
| MLN-519 | 2000 | Temporary MCAO/Sprague- Dawley, rats |
Neuroprotection approached 60%. Neutrophil infiltration at 24 h was significantly decreased (63 to 70%, p < 0.05). |
The neuroprotective effect is in part caused by a reduction in the leukocyte inflammatory response. |
[127] |
| MLN-519 | 2001 | Cardio-embolic stroke model/ Wistar, rats. |
Combination treatment with tPA even at 6 h significantly (p < 0.05) reduced infarct volume, improved neurological recovery, and did not increase the incidence of hemorrhagic transformation. |
Combination treatment extends the neuroprotective effect to at least 6 h after embolization. |
[145] |
| MLN-519 | 2003 | Temporary MCAO/Sprague- Dawley, rats. |
Treatment up to 6 h after MCAO (4 h after reperfusion) reduced neuronal and astrocytic degeneration, decreased cortical infarct volume (− 48%) and increased neurologic recovery (+51%), with a reduced neutrophil infiltration (−38%) and a decrease in activated NFκB immunoreactivity (−45%). |
These effects were related to > 80% reduction in blood proteasome levels. |
[129] |
| MLN-519 | 2003 | Temporary MCAO/Sprague- Dawley rats. |
The most striking effects of intravenous treatment were associated with reductions in ICAM-1 expression at 3 h followed by reductions in E- selectin (12 to 72 h). Less dramatic reductions were observed in IL-1β (3 to 24 h) and TNFα (24 h) with no apparent effects on IL-6 and VCAM-1 mRNA levels |
Immunohistochemical analysis revealed that the genes most dramatically affected had highest expression in endothelial cells and leukocytes. |
[142] |
| MLN-519 | 2004 | Temporary MCAO/Sprague- Dawley rats. |
Core infarct volume in vehicle-treated rats was reduced with delayed treatments of 6, 8 or 10 h after injury (p < 0.05) and was associated with reductions in neuronal and axonal degeneration. |
Neuroprotective treatment provides an extended treatment window of up to 10 h after ischemia/reperfusion brain injury. |
[143] |
ICAM intercellular adhesion molecule, IL interleukin, MCAO middle cerebral artery occlusion, NFκb nuclear factor κB, SH standard housing, TNF tumor necrosis factor, VCAM vascular cell adhesion molecule.
The hypothesis of the dual role of the UPS in stroke
The proteasome clearly represents a central target for the processing and metabolism of proteins, having critical roles in excitability, outgrowth, neuroprotection, metabolism and repair and an understanding of its role is now beginning. Given this complexity, no single role for the UPS in cerebral ischemia is likely to emerge. Indeed, considerable controversy exists over the role of the UPS in neuronal ischemic cell death, and this may result from differences in experimental approaches and endpoints. However, in many animal models, proteasome inhibitors exert a surprisingly consistent neuroprotective effect. The evidence presented in this review suggests that UPS may be involved in the acute neurodestructive phase that occurs immediately after stroke, but that after this period it assumes its normal physiological functions, which include promotion of neuronal survival (the yin-yang effect). Time- and dose-dependent proteasome inhibition will likely promote neuronal survival following stroke by helping neurons evoke chaperones, dampen inflammation and promote revascularization and energetic repletion. However, neurons subjected to prolonged and/or complete proteasome inhibition are likely to be severely damaged, as a result of inability to resume requisite communication, protein turnover and protein trafficking. This hypothesis is supported by the observation that the temporal window of decreased proteasome activity is coincident with the therapeutic window during which proteasome inhibitors promote neuronal survival following stroke.
The post-ischemic impairment of proteasome activity likely also leads to accumulation of Ub conjugates, contributing to loss of neuronal function. While neurons can withstand relatively long periods with intracellular accumulations of ubiquitinated proteins such as found in neurodegenerative disorders [147–150], they are highly sensitive to damage elicited by an inflammatory response. Therefore proteasome inhibitors are of interest in stroke medicine, since they are able to prevent inflammation, NFκB activation and cell death if administered in a temporally restricted manner [144,151].
The problem of defining the temporal window during which proteasome inhibitors are efficacious is particularly important, given that protracted use of these compounds results in neuronal cell death [90,100,152,153]. In addition to the temporal constraints, important spatial considerations must also be addressed in terms of the means to deliver a proteasome inhibitory cocktail, given that the neurotransmitter profile and metabolic activity of cells alter the efficacy of proteasome inhibitors [154•]. Hypoxic endothelia showed a >10-fold increase in sensitivity to inhibitors of proteasome activation [155].
The neurochemical and transcriptional uniqueness of neurons is also particularly important for eliciting proteasome-dependent inflammation pathways. While cell culture studies have shown that activation of NFκB in neurons protects them against excitotoxic and metabolic insults [7••] data from mice lacking the p50 subunit of NFκB suggest that NFκB activation enhances ischemic neuronal death. However, these effects differ between cell types in that activation of NFκB in microglia promotes ischemic neuronal degeneration, whereas activation of NFκB in neurons is neuroprotective [7••].
An increasing number of patients have been receiving treatment with proteasome inhibitors without serious adverse reactions. Velcade has now been launched for the treatment of multiple myeloma and is in phase II trials for other malignancies [156]. A phase I trial of MLN-519 also showed no widespread adverse effects at doses which corresponded to the desired therapeutic values [157], although development of this drug has now been discontinued [158].
The most serious possible complication of inhibiting proteasomes may involve the central nervous system, since 'aggresome' formation has been evoked as a model for neurodegenerative disorders [13,159] Short-term treatment with proteasome inhibitors has important therapeutic potential for increasing survival following stroke. As the roles of the ubiquination machinery, the factors which dictate substrate specificity and the interactions of ischemia-induced biochemical changes on discreet regions of the proteasome become known, we will likely be capable of designing improved therapeutic agents with which to selectively target components of the UPS.
Conclusions
Researchers and pharmaceutical companies are now poised to develop highly relevant biochemical and cell-based high-throughput screening assays with which to identify small-molecule inhibitors of the proteasome. Cell-based screens using non-invasive bioluminescence imaging of 26S proteasome function is a particularly intriguing strategy to use in order to meet these goals. Thus, transgenic mice have been developed, which have green fluorescent protein-tagged proteasome substrates that can be followed over time to determine the regional and temporal effectiveness of proteasome inhibitors [160••]. There are currently more than 20 clinical trials underway in the US and Canada targeting the proteasome. Non-invasive methods to image protein degradation in humans is not yet possible, making it difficult to assess whether proteasome inhibitors reached their targets and their stability in vivo.
The preclinical profile of even the current group of nonselective proteasome inhibitors is superior to many previously investigated compounds, and the observed protection afforded by proteasome inhibitors has been replicated in a variety of model systems. While these compounds are promising agents for the treatment of acute ischemic injury, there is an immediate need for preclinical research on the pharmacokinetics, safety profile and toxicity of these compounds, prior to entering more rigorous tests of clinical efficacy.
Acknowledgements
Both authors have contributed equally to the preparation of the manuscript. They would like to thank Kylie Beck of the Vanderbilt Kennedy Center for her exceptional graphical contribution to the figures. This research was supported in part by NICHD Grant P30HD15052 and NIH grant DK48831.
References
- 1.Caso V, Hacke W. The very acute stroke treatment: Fibrinolysis and after. Clin Exp Hypertens. 2002;24(7–8):595–602. doi: 10.1081/ceh-120015335. [DOI] [PubMed] [Google Scholar]
- 2. Ikonomidou C, Turski L. Why did NMDA receptor antagonists fail clinical trials for stroke and traumatic brain injury? Lancet Neurol. 2002;1(6):383–386. doi: 10.1016/s1474-4422(02)00164-3.. •• This paper is a well argued critical revision of NMDA antagonists in stroke medicine and outlines the reasons for their failure.
- 3.Dirnagl U, Iadecola C, Moskowitz MA. Pathobiology of ischaemic stroke: An integrated view. Trends Neurosci. 1999;22(9):391–397. doi: 10.1016/s0166-2236(99)01401-0. [DOI] [PubMed] [Google Scholar]
- 4.del Zoppo G, Ginis I, Hallenbeck JM, Iadecola C, Wang X, Feuerstein GZ. Inflammation and stroke: Putative role for cytokines, adhesion molecules and iNOS in brain response to ischemia. Brain Pathol. 2000;10(1):95–112. doi: 10.1111/j.1750-3639.2000.tb00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanimirovic D, Satoh K. Inflammatory mediators of cerebral endothelium: A role in ischemic brain inflammation. Brain Pathol. 2000;10(1):113–126. doi: 10.1111/j.1750-3639.2000.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghavendra Rao VL, Bowen KK, Dhodda VK, Song G, Franklin JL, Gavva NR, Dempsey RJ. Gene expression analysis of spontaneously hypertensive rat cerebral cortex following transient focal cerebral ischemia. J Neurochem. 2002;83(5):1072–1086. doi: 10.1046/j.1471-4159.2002.01208.x. [DOI] [PubMed] [Google Scholar]
- 7. Mattson MP, Camandola S. NF-κB in neuronal plasticity and neurodegenerative disorders. J Clin Invest. 2001;107(3):247–254. doi: 10.1172/JCI11916.. •• This is an introductory review of the role of NFκB in the physiology and pathology of the CNS.
- 8.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288(5467):874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci USA. 2001;98(15):8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stenzel-Poore MP, Stevens SL, Xiong Z, Lessov NS, Harrington CA, Mori M, Meller R, Rosenzweig HL, Tobar E, Shaw TE, Chu X, Simon RP. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: Similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362(9389):1028–1037. doi: 10.1016/S0140-6736(03)14412-1.. •• This paper describes a new approach to the molecular mechanisms of neuroprotection. A mouse model of neuroprotection in stroke was used and gene expression profiling was conducted to identify potential neuroprotective genes and their associated pathways.
- 11.McLaughlin B, Hartnett KA, Erhardt JA, Legos JJ, White RF, Barone FC, Aizenman E. Caspase 3 activation is essential for neuroprotection in preconditioning. Proc Natl Acad Sci USA. 2003;100(2):715–720. doi: 10.1073/pnas.0232966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golab J, Bauer TM, Daniel V, Naujokat C. Role of the ubiquitin-proteasome pathway in the diagnosis of human diseases. Clin Chim Acta. 2004;340(1–2):27–40. doi: 10.1016/j.cccn.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: A cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29(1):15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 14.Fang S, Lorick KL, Jensen JP, Weissman AM. RING finger ubiquitin protein ligases: Implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin Cancer Biol. 2003;13(1):5–14. doi: 10.1016/s1044-579x(02)00095-0. [DOI] [PubMed] [Google Scholar]
- 15. Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001.. •• This paper is a well-written review of the UPS from two authorities in the field.
- 16.Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci. 2003;28(11):598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 18.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 19.Huang DT, Walden H, Duda D, Schulman BA. Ubiquitin-like protein activation. Oncogene. 2004;23(11):1958–1971. doi: 10.1038/sj.onc.1207393. [DOI] [PubMed] [Google Scholar]
- 20.Brooks CL, Li M, Gu W. Monoubiquitination: The signal for p53 nuclear export? Cell Cycle. 2004;3(4):436–438. [PubMed] [Google Scholar]
- 21.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 22.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 23.Shtiegman K, Yarden Y. The role of ubiquitylation in signalling by growth factors: Implications to cancer. Semin Cancer Biol. 2003;13(1):29–40. doi: 10.1016/s1044-579x(02)00097-4. [DOI] [PubMed] [Google Scholar]
- 24.Marmor MD, Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23(11):2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- 25.Bache KG, Slagsvold T, Stenmark H. Defective downregulation of receptor tyrosine kinases in cancer. EMBO J. 2004;23(14):2707–2712. doi: 10.1038/sj.emboj.7600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cong F, Zhang J, Pao W, Zhou P, Varmus H. A protein knockdown strategy to study the function of β-catenin in tumorigenesis. BMC Mol Biol. 2003;4(1):10. doi: 10.1186/1471-2199-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Zheng N, Zhou P. Exploring the functional complexity of cellular proteins by protein knockout. Proc Natl Acad Sci USA. 2003;100(24):14127–14132. doi: 10.1073/pnas.2233012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickart CM. Back to the future with ubiquitin. Cell. 2004;116(2):181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 29. Ciechanover A. The ubiquitin - proteasome pathway: On protein death and cell life. EMBO J. 1998;17(24):7151–7160. doi: 10.1093/emboj/17.24.7151.. •• This is a classic paper describing the UPS.
- 30.van Kerkhof P, Alves dos Santos CM, Sachse M, Klumperman J, Bu G, Strous GJ. Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol Biol Cell. 2001;12(8):2556–2566. doi: 10.1091/mbc.12.8.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisk HA, Yaffe MP. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J Cell Biol. 1999;145(6):1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102(1):67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 33.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15(3):1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 35.Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106(5):527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- 36.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 37.Orlowski M, Cardozo C, Michaud C. Evidence for the presence of five distinct proteolytic components in the pituitary multicatalytic proteinase complex. Properties of two components cleaving bonds on the carboxyl side of branched chain and small neutral amino acids. Biochemistry. 1993;32(6):1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 38.Kierszenbaum AL. The 26S proteasome: Ubiquitin-mediated proteolysis in the tunnel. Mol Reprod Dev. 2000;57(2):109–110. doi: 10.1002/1098-2795(200010)57:2<109::AID-MRD1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Bury M, Mlynarczuk I, Pleban E, Hoser G, Kawiak J, Wojcik C. Effects of an inhibitor of tripeptidyl peptidase II (Ala-Ala-Phe-chloromethylketone) and its combination with an inhibitor of the chymotrypsin-like activity of the proteasome (PSI) on apoptosis, cell cycle and proteasome activity in U937 cells. Folia Histochem Cytobiol. 2001;39(2):131–132. [PubMed] [Google Scholar]
- 40.Wang EW, Kessler BM, Borodovsky A, Cravatt BF, Bogyo M, Ploegh HL, Glas R. Integration of the ubiquitin-proteasome pathway with a cytosolic oligopeptidase activity. Proc Natl Acad Sci USA. 2000;97(18):9990–9995. doi: 10.1073/pnas.180328897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Napoli M, Papa F. The proteasome system and proteasome inhibitors in stroke: Controlling the inflammatory response. Curr Opin Invest Drugs. 2003;4(11):1333–1342.. • This is the first review article to describe the role of the UPS in cerebral ischemia.
- 42. Asai A, Tanahashi N, Qiu JH, Saito N, Chi S, Kawahara N, Tanaka K, Kirino T. Selective proteasomal dysfunction in the hippocampal CA1 region after transient forebrain ischemia. J Cereb Blood Flow Metab. 2002;22(6):705–710. doi: 10.1097/00004647-200206000-00009.. • This paper reports that the irreversible loss of proteasome function underlies the delayed neuronal death induced by transient forebrain ischemia in the hippocampal CA1 region.
- 43. Wojcik C, Di Napoli M. Ubiquitin-proteasome system and proteasome inhibition: New strategies in stroke therapy. Stroke. 2004;35(6):1506–1518. doi: 10.1161/01.STR.0000126891.93919.4e.. •• This is an introductory and updated review article on the role of UPS and protesome inhibitors in stroke.
- 44.Kim CH, Kim JH, Hsu CY, Ahn YS. Zinc is required in pyrrolidine dithiocarbamate inhibition of NF-κB activation. FEBS Lett. 1999;449(1):28–32. doi: 10.1016/s0014-5793(99)00390-7. [DOI] [PubMed] [Google Scholar]
- 45. Kamikubo T, Hayashi T. Changes in proteasome activity following transient ischemia. Neurochem Int. 1996;28(2):209–212. doi: 10.1016/0197-0186(95)00071-2.. • This paper details how proteasome activity might not be irreversibly impaired after transient ischemia. However, transient inhibition of ATP-dependent conversion of 20S to 26S proteasomes in vitro must be one of the causes of the accumulation of the ubiquitin-protein conjugates in the early reperfusion period.
- 46.Gubellini P, Bisso GM, Ciofi-Luzzatto A, Fortuna S, Lorenzini P, Michalek H, Scarsella G. Ubiquitin-mediated stress response in a rat model of brain transient ischemia/hypoxia. Neurochem Res. 1997;22(1):93–100. doi: 10.1023/a:1027389623767. [DOI] [PubMed] [Google Scholar]
- 47.Vannucci SJ, Mummery R, Hawkes RB, Rider CC, Beesley PW. Hypoxia-ischemia induces a rapid elevation of ubiquitin conjugate levels and ubiquitin immunoreactivity in the immature rat brain. J Cereb Blood Flow Metab. 1998;18(4):376–385. doi: 10.1097/00004647-199804000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Magnusson K, Wieloch T. Impairment of protein ubiquitination may cause delayed neuronal death. Neurosci Lett. 1989;96(3):264–270. doi: 10.1016/0304-3940(89)90389-3. [DOI] [PubMed] [Google Scholar]
- 49. Ding Q, Bruce-Keller AJ, Chen Q, Keller JN. Analysis of gene expression in neural cells subject to chronic proteasome inhibition. Free Radic Biol Med. 2004;36(4):445–455. doi: 10.1016/j.freeradbiomed.2003.10.025.. a. • This paper describes how, despite the fact that chronic low-level proteasome inhibition alters the expression of a limited number of genes, it also significantly alters the expression of genes that are highly relevant in aging and neurodegenerative disorders.
- 50. Hu BR, Janelidze S, Ginsberg MD, Busto R, Perez-Pinzon M, Sick TJ, Siesjo BK, Liu CL. Protein aggregation after focal brain ischemia and reperfusion. J Cereb Blood Flow Metab. 2001;21(7):865–875. doi: 10.1097/00004647-200107000-00012.. a. • This paper details how proteins are severely aggregated in hippocampal neurons vulnerable to transient brain ischemia. The accumulation of protein aggregates could cause ischemic neuronal death.
- 51.Hu BR, Martone ME, Jones YZ, Liu CL. Protein aggregation after transient cerebral ischemia. J Neurosci. 2000;20(9):3191–3199. doi: 10.1523/JNEUROSCI.20-09-03191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tytell M, Brown WR, Moody DM, Challa VR. Immunohistochemical assessment of constitutive and inducible heat-shock protein 70 and ubiquitin in human cerebellum and caudate nucleus. Mol Chem Neuropathol. 1998;35(1–3):97–117. doi: 10.1007/BF02815118. [DOI] [PubMed] [Google Scholar]
- 53.Noga M, Hayashi T, Tanaka J. Gene expressions of ubiquitin and Hsp70 following focal ischaemia in rat brain. Neuroreport. 1997;8(5):1239–1241. doi: 10.1097/00001756-199703240-00036. [DOI] [PubMed] [Google Scholar]
- 54.Hara T, Mies G, Hata R, Hossmann KA. Gene expressions after thrombolytic treatment of middle cerebral artery clot embolism in mice. Stroke. 2001;32(8):1912–1919. doi: 10.1161/01.str.32.8.1912. [DOI] [PubMed] [Google Scholar]
- 55.Nowak TS, Jr, Bond U, Schlesinger MJ. Heat shock RNA levels in brain and other tissues after hyperthermia and transient ischemia. J Neurochem. 1990;54(2):451–458. doi: 10.1111/j.1471-4159.1990.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 56.Welch WJ. Heat shock proteins functioning as molecular chaperones: Their roles in normal and stressed cells. Philos Trans Soc Lond Biol. 1993;339(1289):327–333. doi: 10.1098/rstb.1993.0031. [DOI] [PubMed] [Google Scholar]
- 57.Yenari MA, Giffard RG, Sapolsky RM, Steinberg GK. The neuroprotective potential of heat shock protein 70 (Hsp70) Mol Med Today. 1999;5(12):525–531. doi: 10.1016/s1357-4310(99)01599-3. [DOI] [PubMed] [Google Scholar]
- 58. Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, Tailor P, Morimoto RI, Cohen GM, Green DR. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol. 2000;2(8):469–475. doi: 10.1038/35019501.. a. • This is the first in a series of papers to re-evaluate the molecular mechanisms of protection afforded by chaperones.
- 59.Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat Cell Biol. 2000;2(8):476–483. doi: 10.1038/35019510. [DOI] [PubMed] [Google Scholar]
- 60.Lee JE, Yenari MA, Sun GH, Xu L, Emond MR, Cheng D, Steinberg GK, Giffard RG. Differential neuroprotection from human heat shock protein 70 overexpression in in vitro and in vivo models of ischemia and ischemia-like conditions. Exp Neurol. 2001;170(1):129–139. doi: 10.1006/exnr.2000.7614. [DOI] [PubMed] [Google Scholar]
- 61.Xu L, Lee JE, Giffard RG, Overexpression of bcl-2. bcl-xL or Hsp70 in murine cortical astrocytes reduces injury of co-cultured neurons. Neurosci Lett. 1999;277(3):193–197. doi: 10.1016/s0304-3940(99)00882-4. [DOI] [PubMed] [Google Scholar]
- 62.Hoehn B, Ringer TM, Xu L, Giffard RG, Sapolsky RM, Steinberg GK, Yenari MA. Overexpression of Hsp72 after induction of experimental stroke protects neurons from ischemic damage. J Cereb Blood Flow Metab. 2001;21(11):1303–1309. doi: 10.1097/00004647-200111000-00006. [DOI] [PubMed] [Google Scholar]
- 63.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27(7):368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 64.Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10(1):55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- 65.Shimura H, Schwartz D, Gygi SP, Kosik KS. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem. 2004;279(6):4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 66.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3(1):100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 67.Imai Y, Soda M, Murakami T, Shoji M, Abe K, Takahashi R. A product of the human gene adjacent to parkin is a component of Lewy bodies and suppresses Pael receptor-induced cell death. J Biol Chem. 2003;278(51):51901–51910. doi: 10.1074/jbc.M309655200. [DOI] [PubMed] [Google Scholar]
- 68.Maraganore DM, de Andrade M, Lesnick TG, Farrer MJ, Bower JH, Hardy JA, Rocca WA. Complex interactions in Parkinson's disease: A two-phased approach. Mov Disord. 2003;18(6):631–636. doi: 10.1002/mds.10431. [DOI] [PubMed] [Google Scholar]
- 69.Song S, Kim SY, Hong YM, Jo DG, Lee JY, Shim SM, Chung CW, Seo SJ, Yoo YJ, Koh JY, Lee MC, et al. Essential role of E2-25K/Hip-2 in mediating amyloid-β neurotoxicity. Mol Cell. 2003;12(3):553–563. doi: 10.1016/j.molcel.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 70.McNaught KS, Mytilineou C, Jnobaptiste R, Yabut J, Shashidharan P, Jennert P, Olanow CW. Impairment of the ubiquitin-proteasome system causes dopaminergic cell death and inclusion body formation in ventral mesencephalic cultures. J Neurochem. 2002;81(2):301–306. doi: 10.1046/j.1471-4159.2002.00821.x. [DOI] [PubMed] [Google Scholar]
- 71.Zhang HG, Wang J, Yang X, Hsu HC, Mountz JD. Regulation of apoptosis proteins in cancer cells by ubiquitin. Oncogene. 2004;23(11):2009–2015. doi: 10.1038/sj.onc.1207373. [DOI] [PubMed] [Google Scholar]
- 72.Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3(10):237–241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 73.Kalia SK, Lee S, Smith PD, Liu L, Crocker SJ, Thorarinsdottir TE, Glover JR, Fon EA, Park DS, Lozano AM. BAG5 inhibits parkin and enhances dopaminergic neuron degeneration. Neuron. 2004;44(6):931–945. doi: 10.1016/j.neuron.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 74.Tanaka S, Uehara T, Nomura Y. Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J Biol Chem. 2000;275(14):10388–10393. doi: 10.1074/jbc.275.14.10388. [DOI] [PubMed] [Google Scholar]
- 75.Schmidt-Kastner R, Zhang BT, Webster K, Kietzmann T, Zhao W, Busto R, Ginsberg MD. Hypoxia-inducible factor-1 (HIF-1) in experimental brain ischemia. Scientific World J. 2002;2:123–124. doi: 10.1100/tsw.2002.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richard S, Campello C, Taillandier L, Parker F, Resche F. Haemangioblastoma of the central nervous system in von Hippel-Lindau disease. French VHL Study Group. J Intern Med. 1998;243(6):547–553. doi: 10.1046/j.1365-2796.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 77.Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia. Cell. 2004;117(7):941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 78.Kallio PJ, Wilson WJ, O'Brien S, Makino Y, Poellinger L. Regulation of the hypoxia-inducible transcription factor 1a by the ubiquitin-proteasome pathway. J Biol Chem. 1999;274(10):6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 79.Li J, Post M, Volk R, Gao Y, Li M, Metais C, Sato K, Tsai J, Aird W, Rosenberg RD, Hampton TG, et al. PR39, a peptide regulator of angiogenesis. Nat Med. 2000;6(1):49–55. doi: 10.1038/71527. [DOI] [PubMed] [Google Scholar]
- 80.Soucek T, Cumming R, Dargusch R, Maher P, Schubert D. The regulation of glucose metabolism by HIF-1 mediates a neuroprotective response to amyloid β peptide. Neuron. 2003;39(1):43–56. doi: 10.1016/s0896-6273(03)00367-2. [DOI] [PubMed] [Google Scholar]
- 81.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416(6882):763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 82.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7(3):627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 83.Mengesdorf T, Jensen PH, Mies G, Aufenberg C, Paschen W. Down-regulation of parkin protein in transient focal cerebral ischemia: A link between stroke and degenerative disease? Proc Natl Acad Sci USA. 2002;99(23):15042–15047. doi: 10.1073/pnas.232588799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hampton MB, Fadeel B, Orrenius S. Redox regulation of the caspases during apoptosis. Ann NY Acad Sci. 1998;854:328–335. doi: 10.1111/j.1749-6632.1998.tb09913.x. [DOI] [PubMed] [Google Scholar]
- 85.Amici M, Forti K, Nobili C, Lupidi G, Angeletti M, Fioretti E, Eleuteri AM. Effect of neurotoxic metal ions on the proteolytic activities of the 20S proteasome from bovine brain. J Biol Inorg Chem. 2002;7(7–8):750–756. doi: 10.1007/s00775-002-0352-4. [DOI] [PubMed] [Google Scholar]
- 86.Fruh K, Gossen M, Wang K, Bujard H, Peterson PA, Yang Y. Displacement of housekeeping proteasome subunits by MHC-encoded LMPs: A newly discovered mechanism for modulating the multicatalytic proteinase complex. EMBO J. 1994;13(14):3236–3244. doi: 10.1002/j.1460-2075.1994.tb06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9(6):788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 88.Peng TI, Jou MJ, Sheu SS, Greenamyre JT. Visualization of NMDA receptor-induced mitochondrial calcium accumulation in striatal neurons. Exp Neurol. 1998;149(1):1–12. doi: 10.1006/exnr.1997.6599. [DOI] [PubMed] [Google Scholar]
- 89.Dugan LL, Sensi SL, Canzoniero LM, Handran SD, Rothman SM, Lin TS, Goldberg MP, Choi DW. Mitochondrial production of reactive oxygen species in cortical neurons following exposure to N-methyl-d-aspartate. J Neurosci. 1995;15(10):6377–6388. doi: 10.1523/JNEUROSCI.15-10-06377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mattson MP. Excitotoxic and excitoprotective mechanisms: Abundant targets for the prevention and treatment of neurodegenerative disorders. Neuromolecular Med. 2003;3(2):65–94. doi: 10.1385/NMM:3:2:65. [DOI] [PubMed] [Google Scholar]
- 91.Dawson VL, Dawson TM, London ED, Bredt DS, Snyder SH. Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures. Proc Natl Acad Sci USA. 1991;88(14):6368–6371. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Siman R, Noszek JC, Kegerise C. Calpain I activation is specifically related to excitatory amino acid induction of hippocampal damage. J Neurosci. 1989;9(5):1579–1590. doi: 10.1523/JNEUROSCI.09-05-01579.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dumuis A, Sebben M, Haynes L, Pin JP, Bockaert J. NMDA receptors activate the arachidonic acid cascade system in striatal neurons. Nature. 1988;336(6194):68–70. doi: 10.1038/336068a0. [DOI] [PubMed] [Google Scholar]
- 94.Stanciu M, Wang Y, Kentor R, Burke N, Watkins S, Kress G, Reynolds I, Klann E, Angiolieri MR, Johnson JW, DeFranco DB. Persistent activation of ERK contributes to glutamate-induced oxidative toxicity in a neuronal cell line and primary cortical neuron cultures. J Biol Chem. 2000;275(16):12200–12206. doi: 10.1074/jbc.275.16.12200. [DOI] [PubMed] [Google Scholar]
- 95.Reynolds IJ, Hastings TG. Glutamate induces the production of reactive oxygen species in cultured forebrain neurons following NMDA receptor activation. J Neurosci. 1995;15(1):3318–3327. doi: 10.1523/JNEUROSCI.15-05-03318.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wagenknecht B, Hermisson M, Groscurth P, Liston P, Krammer PH, Weller M. Proteasome inhibitor-induced apoptosis of glioma cells involves the processing of multiple caspases and cytochrome C release. J Neurochem. 2000;75(6):2288–2297. doi: 10.1046/j.1471-4159.2000.0752288.x. [DOI] [PubMed] [Google Scholar]
- 97.Ling YH, Liebes L, Zou Y, Perez-Soler R. Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. J Biol Chem. 2003;278(36):33714–33723. doi: 10.1074/jbc.M302559200. [DOI] [PubMed] [Google Scholar]
- 98.Ding Q, Keller JN. Proteasomes and proteasome inhibition in the central nervous system. Free Radic Biol Med. 2001;31(5):574–584. doi: 10.1016/s0891-5849(01)00635-9. [DOI] [PubMed] [Google Scholar]
- 99.Qiu JH, Asai A, Chi S, Saito N, Hamada H, Kirino T. Proteasome inhibitors induce cytochrome c-caspase-3-like protease-mediated apoptosis in cultured cortical neurons. J Neurosci. 2000;20(1):259–265. doi: 10.1523/JNEUROSCI.20-01-00259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bobba A, Canu N, Atlante A, Petragallo V, Calissano P, Marra E. Proteasome inhibitors prevent cytochrome C release during apoptosis but not in excitotoxic death of cerebellar granule neurons. FEBS Lett. 2002;515(1–3):8–12. doi: 10.1016/s0014-5793(02)02231-7. [DOI] [PubMed] [Google Scholar]
- 101.Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40(3):595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6(3):231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 103.Ehrlich I, Malinow R. Postsynaptic density 95 controls AMPA receptor incorporation during long-term potentiation and experience-driven synaptic plasticity. J Neurosci. 2004;24(4):916–927. doi: 10.1523/JNEUROSCI.4733-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu CL, Martone ME, Hu BR. Protein ubiquitination in postsynaptic densities after transient cerebral ischemia. J Cereb Blood Flow Metab. 2004;24(11):1219–1225. doi: 10.1097/01.WCB.0000136706.77918.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cohen NA, Brenman JE, Snyder SH, Bredt DS. Binding of the inward rectifier K+ channel Kir 2.3 to PSD-95 is regulated by protein kinase A phosphorylation. Neuron. 1996;17(4):759–767. doi: 10.1016/s0896-6273(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 106.Suen PC, Wu K, Xu JL, Lin SY, Levine ES, Black IB. NMDA receptor subunits in the postsynaptic density of rat brain: Expression and phosphorylation by endogenous protein kinases. Brain Res Mol Brain Res. 1998;59(2):215–228. doi: 10.1016/s0169-328x(98)00157-0. [DOI] [PubMed] [Google Scholar]
- 107.Arundine M, Tymianski M. Molecular mechanisms of calcium-dependent neurodegeneration in excitotoxicity. Cell Calcium. 2003;34(4–5):325–337. doi: 10.1016/s0143-4160(03)00141-6. [DOI] [PubMed] [Google Scholar]
- 108.Snider BJ, Tee LY, Canzoniero LM, Babcock DJ, Choi DW. NMDA antagonists exacerbate neuronal death caused by proteasome inhibition in cultured cortical and striatal neurons. Eur J Neurosci. 2002;15(3):419–428. doi: 10.1046/j.0953-816x.2001.01867.x. [DOI] [PubMed] [Google Scholar]
- 109.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276(32):30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 110.Carrard G, Bulteau AL, Petropoulos I, Friguet B. Impairment of proteasome structure and function in aging. Int J Biochem Cell Biol. 2002;34(11):1461–1474. doi: 10.1016/s1357-2725(02)00085-7. [DOI] [PubMed] [Google Scholar]
- 111.Grune T, Merker K, Sandig G, Davies KJ. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun. 2003;305(3):709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 112.Shringarpure R, Grune T, Mehlhase J, Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem. 2003;278(1):311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- 113.Demasi M, Davies KJ. Proteasome inhibitors induce intracellular protein aggregation and cell death by an oxygen-dependent mechanism. FEBS Lett. 2003;542(1–3):89–94. doi: 10.1016/s0014-5793(03)00353-3. [DOI] [PubMed] [Google Scholar]
- 114.Thornberry NA, Lazebnik Y. Caspases: Enemies within. Science. 1998;281(5381):1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 115.Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: A comprehensive update of caspase substrates. Cell Death Differ. 2003;10(1):76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Benchoua A, Guegan C, Couriaud C, Hosseini H, Sampaio N, Morin D, Onteniente B. Specific caspase pathways are activated in the two stages of cerebral infarction. J Neurosci. 2001;21(18):7127–7134. doi: 10.1523/JNEUROSCI.21-18-07127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Friedlander RM, Gagliardini V, Hara H, Fink KB, Li W, MacDonald G, Fishman MC, Greenberg AH, Moskowitz MA, Yuan J. Expression of a dominant negative mutant of interleukin-1β converting enzyme in transgenic mice prevents neuronal cell death induced by trophic factor withdrawal and ischemic brain injury. J Exp Med. 1997;185(5):933–940. doi: 10.1084/jem.185.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Trapp T, Korhonen L, Besselmann M, Martinez R, Mercer EA, Lindholm D. Transgenic mice overexpressing XIAP in neurons show better outcome after transient cerebral ischemia. Mol Cell Neurosci. 2003;23(2):302–313. doi: 10.1016/s1044-7431(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 119.Hara H, Fink K, Endres M, Friedlander RM, Gagliardini V, Yuan J, Moskowitz MA. Attenuation of transient focal cerebral ischemic injury in transgenic mice expressing a mutant ICE inhibitory protein. J Cereb Blood Flow Metab. 1997;17(4):370–375. doi: 10.1097/00004647-199704000-00002. [DOI] [PubMed] [Google Scholar]
- 120.Fink K, Zhu J, Namura S, Shimizu-Sasamata M, Endres M, Ma J, Dalkara T, Yuan J, Moskowitz MA. Prolonged therapeutic window for ischemic brain damage caused by delayed caspase activation. J Cereb Blood Flow Metab. 1998;18(10):1071–1076. doi: 10.1097/00004647-199810000-00003. [DOI] [PubMed] [Google Scholar]
- 121. Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM. Caspase activation inhibits proteasome function during apoptosis. Mol Cell. 2004;14(1):81–93. doi: 10.1016/s1097-2765(04)00156-x.. •• This paper describes the first demonstration that the induction of apoptosis may regulate the activity of the proteasome. During apoptosis, caspase activation results in the cleavage of three specific subunits of the 19S regulatory complex of the proteasome. This caspase-mediated cleavage inhibits the proteasomal degradation of ubiquitin-dependent and -independent cellular substrates, including pro-apoptotic molecules such as Smac, so facilitating the execution of the apoptotic program by providing a feed-forward amplification loop.
- 122.Haddad JJE, Olver RE, Land SC. Antioxidant/pro-oxidant equilibrium regulates HIF-1α and NF-κB redox sensitivity. Evidence for inhibition by glutathione oxidation in alveolar epithelial cells. J Biol Chem. 2000;275(28):21130–21139. doi: 10.1074/jbc.M000737200. [DOI] [PubMed] [Google Scholar]
- 123.Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IκB kinase activity through IKKβ subunit phosphorylation. Science. 1999;284(5412):309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 124.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: The control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 125.Celec P. Nuclear factor κB - molecular biomedicine: The next generation. Biomed Pharmacother. 2004;58(6–7):365–371. doi: 10.1016/j.biopha.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 126.Epinat J-C, Gilmore TD. Diverse agents act at multiple levels to inhibit the Rel/NF-κB signal transduction pathway. Oncogene. 1999;18(49):6896–6909. doi: 10.1038/sj.onc.1203218. [DOI] [PubMed] [Google Scholar]
- 127.Phillips JB, Williams AJ, Adams J, Elliott PJ, Tortella FC. Proteasome inhibitor PS519 reduces infarction and attenuates leukocyte infiltration in a rat model of focal cerebral ischemia. Stroke. 2000;31(7):1686–1693. doi: 10.1161/01.str.31.7.1686. [DOI] [PubMed] [Google Scholar]
- 128.Salminen A, Liu PK, Hsu CY. Alteration of transcription factor binding activities in the ischemic rat brain. Biochem Biophys Res Commun. 1994;212(3):939–944. doi: 10.1006/bbrc.1995.2060. [DOI] [PubMed] [Google Scholar]
- 129.Williams AJ, Hale SL, Moffett JR, Dave JR, Elliott PJ, Adams J, Tortella FC. Delayed treatment with MLN519 reduces infarction and associated neurologic deficit caused by focal ischemic brain injury in rats via antiinflammatory mechanisms involving nuclear factor-κB activation, gliosis, and leukocyte infiltration. J Cereb Blood Flow Metab. 2003;23(1):75–87. doi: 10.1097/01.WCB.0000039285.37737.C2. [DOI] [PubMed] [Google Scholar]
- 130.Kisselev AF, Goldberg AL. Proteasome inhibitors: From research tools to drug candidates. Chem Biol. 2001;8(8):739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 131. Lee DH, Goldberg AL. Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 1998;8(10):397–403. doi: 10.1016/s0962-8924(98)01346-4.. • This paper describes the actions of these inhibitors, how they can be used to investigate cellular responses, the functions of the proteasome demonstrated by such studies and their potential applications in the future.
- 132.Groll M, Koguchi Y, Huber R, Kohno J. Crystal structure of the 20 S proteasome:TMC-95A complex: A non-covalent proteasome inhibitor. J Mol Biol. 2001;311(3):543–548. doi: 10.1006/jmbi.2001.4869. [DOI] [PubMed] [Google Scholar]
- 133.Kessler BM, Tortorella D, Altun M, Kisselev AF, Fiebiger E, Hekking BG, Ploegh HL, Overkleeft HS. Extended peptide-based inhibitors efficiently target the proteasome and reveal overlapping specificities of the catalytic β-subunits. Chem Biol. 2001;8(9):913–929. doi: 10.1016/s1074-5521(01)00069-2. [DOI] [PubMed] [Google Scholar]
- 134.Meng L, Kwok BH, Sin N, Crews CM. Eponemycin exerts its antitumor effect through the inhibition of proteasome function. Cancer Res. 1999;59(12):2798–2801. [PubMed] [Google Scholar]
- 135.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA. 1999;96(18):10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kroll M, Arenzana-Seisdedos F, Bachelerie F, Thomas D, Friguet B, Conconi M. The secondary fungal metabolite gliotoxin targets proteolytic activities of the proteasome. Chem Biol. 1999;6(10):689–698. doi: 10.1016/s1074-5521(00)80016-2. [DOI] [PubMed] [Google Scholar]
- 137.Nam S, Smith DM, Dou QP. Ester bond-containing tea polyphenols potently inhibit proteasome activity in vitro and in vivo. J Biol Chem. 2001;276(16):13322–13330. doi: 10.1074/jbc.M004209200. [DOI] [PubMed] [Google Scholar]
- 138.Nam S, Smith DM, Dou QP. Tannic acid potently inhibits tumor cell proteasome activity, increases p27 and Bax expression, and induces G1 arrest and apoptosis. Cancer Epidemiol Biomarkers Prev. 2001;10(10):1083–1088. [PubMed] [Google Scholar]
- 139.Andre P, Groettrup M, Klenerman P, de Giuli R, Booth BL, Jr, Cerundolo V, Bonneville M, Jotereau F, Zinkernagel RM, Lotteau V. An inhibitor of HIV-1 protease modulates proteasome activity, antigen presentation, and T cell responses. Proc Natl Acad Sci USA. 1998;95(22):13120–13124. doi: 10.1073/pnas.95.22.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kiyomiya K, Matsuo S, Kurebe M. Proteasome is a carrier to translocate doxorubicin from cytoplasm into nucleus. Life Sci. 1998;62(20):1853–1860. doi: 10.1016/s0024-3205(98)00151-9. [DOI] [PubMed] [Google Scholar]
- 141.Piccinini M, Tazartes O, Mezzatesta C, Ricotti E, Bedino S, Grosso F, Dianzani U, Tovo PA, Mostert M, Musso A, Rinaudo MT. Proteasomes are a target of the anti-tumour drug vinblastine. Biochem J. 2001;356(3):835–841. doi: 10.1042/0264-6021:3560835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Berti R, Williams AJ, Velarde LC, Moffett JR, Elliott PJ, Adams J, Yao C, Dave JR, Tortella FC. Effect of the proteasome inhibitor MLN519 on the expression of inflammatory molecules following middle cerebral artery occlusion and reperfusion in the rat. Neurotox Res. 2003;5(7):505–514. doi: 10.1007/BF03033160. [DOI] [PubMed] [Google Scholar]
- 143.Williams AJ, Berti R, Dave JR, Elliot PJ, Adams J, Tortella FC. Delayed treatment of ischemia/reperfusion brain injury: extended therapeutic window with the proteosome inhibitor MLN519. Stroke. 2004;35(5):1186–1191. doi: 10.1161/01.STR.0000125721.10606.dc. [DOI] [PubMed] [Google Scholar]
- 144.Buchan AM, Li H, Blackburn B. Neuroprotection achieved with a novel proteasome inhibitor which blocks NF-κB activation. Neuroreport. 2000;11(2):427–430. doi: 10.1097/00001756-200002070-00041. [DOI] [PubMed] [Google Scholar]
- 145.Zhang L, Zhang ZG, Zhang RL, Lu M, Adams J, Elliott PJ, Chopp M. Postischemic (6-hour) treatment with recombinant human tissue plasminogen activator and proteasome inhibitor PS-519 reduces infarction in a rat model of embolic focal cerebral ischemia. Stroke. 2001;32(12):2926–2931. doi: 10.1161/hs1201.100207. [DOI] [PubMed] [Google Scholar]
- 146. Di Napoli M, Papa F. MLN-519. Millennium/PAION. Curr Opin Invest Drugs. 2003;4(3):333–341.. • This paper reports that MLN-519 is currently undergoing phase I clinical trials in acute stroke and myocardial infarction, and is poised to enter phase II trials.
- 147.Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2002;11(5):515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 148.Halliwell B. Hypothesis: Proteasomal dysfunction: A primary event in neurogeneration that leads to nitrative and oxidative stress and subsequent cell death. Ann NY Acad Sci. 2002;962:182–194. doi: 10.1111/j.1749-6632.2002.tb04067.x. [DOI] [PubMed] [Google Scholar]
- 149.Jenner P. Oxidative stress in Parkinson's disease. Ann Neurol. 2003;53 Suppl 3:S26–S36. doi: 10.1002/ana.10483. [DOI] [PubMed] [Google Scholar]
- 150.Mayer RJ. From neurodegeneration to neurohomeostasis: The role of ubiquitin. Drug News Perspect. 2003;16(2):103–108. doi: 10.1358/dnp.2003.16.2.829327. [DOI] [PubMed] [Google Scholar]
- 151.Elliott PJ, Zollner TM, Boehncke WH. Proteasome inhibition: A new anti-inflammatory strategy. J Mol Med. 2003;81(4):235–245. doi: 10.1007/s00109-003-0422-2. [DOI] [PubMed] [Google Scholar]
- 152.Keller JN, Markesbery WR. Proteasome inhibition results in increased poly-ADP-ribosylation: Implications for neuron death. J Neurosci Res. 2000;61(4):436–442. doi: 10.1002/1097-4547(20000815)61:4<436::AID-JNR10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 153.Kikuchi S, Shinpo K, Takeuchi M, Tsuji S, Yabe I, Niino M, Tashiro K. Effect of geranylgeranylaceton on cellular damage induced by proteasome inhibition in cultured spinal neurons. J Neurosci Res. 2002;69(3):373–381. doi: 10.1002/jnr.10298. [DOI] [PubMed] [Google Scholar]
- 154. Wojcik C. Regulation of apoptosis by the ubiquitin and proteasome pathway. J Cell Mol Med. 2002;6(1):25–48. doi: 10.1111/j.1582-4934.2002.tb00309.x.. • This paper is a well written and referenced review of the role of UPS in apoptosis.
- 155.Zund G, Uezono S, Stahl GL, Dzus AL, McGowan FX, Hickey PR, Colgan SP. Hypoxia enhances induction of endothelial ICAM-1: Role for metabolic acidosis and proteasomes. Am J Physiol. 1997;273(5 Pt 1):C1571–C1580. doi: 10.1152/ajpcell.1997.273.5.C1571. [DOI] [PubMed] [Google Scholar]
- 156.Adams J. The proteasome: A suitable antineoplastic target. Nat Rev Cancer. 2004;4(5):349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 157.Shah IM, Lees KR, Pien CP, Elliott PJ. Early clinical experience with the novel proteasome inhibitor PS-519. Br J Clin Pharmacol. 2002;54(3):269–276. doi: 10.1046/j.1365-2125.2002.01638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Millennium Pharmaceuticals Inc. Research and Development day. Company Presentation. 2004 November 16; [Google Scholar]
- 159.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 160. Lindsten K, Menendez-Benito V, Masucci MG, Dantuma NP. A transgenic mouse model of the ubiquitin/proteasome system. Nat Biotechnol. 2003;21(8):897–902. doi: 10.1038/nbt851.. •• This paper describes an innovative transgenic animal model that provides a tool for monitoring the status of the UPS in physiological or pathological conditions.