Abstract
Gene inactivation studies of mammalian histone and DNA-modifying proteins have demonstrated a role for many such proteins in embryonic development. Post-implantation embryonic lethality implies a role for epigenetic factors in differentiation and in development of specific lineages or tissues. However a handful of chromatin-modifying enzymes have been found to be required in pre- or peri-implantation embryos. This is significant as implantation is the time when inner cell mass cells of the blastocyst exit pluripotency and begin to commit to form the various lineages that will eventually form the adult animal. These observations indicate a critical role for chromatin-modifying proteins in the earliest lineage decisions of mammalian development, and/or in the formation of the first embryonic cell types. Recent work has shown that the two major class I histone deacetylase-containing co-repressor complexes, the NuRD and Sin3 complexes, are both required at peri-implantation stages of mouse development, demonstrating the importance of histone deacetylation in cell fate decisions. Over the past 10 years both genetic and biochemical studies have revealed surprisingly divergent roles for these two co-repressors in mammalian cells. In this review we will summarise the evidence that the two major class I histone deacetylase complexes in mammalian cells, the NuRD and Sin3 complexes, play important roles in distinct aspects of embryonic development.
Keywords: Development, Histone deacetylase, Epigenetics, Chromatin, Stem cells
1. The NuRD complex
A decade ago the first purifications were reported of an abundant histone deacetylase complex that was distinct from the best-characterised deacetylase complex at the time, namely the Sin3 complex. The analogous Mi-2, NRD, NuRD and NuRD complexes were purified from Xenopus egg, HeLa cell and SW13 cell nuclear extracts, respectively (Tong et al., 1998; Wade et al., 1998; Xue et al., 1998; Zhang et al., 1998). The complex has become known as the NuRD (nucleosome remodelling and histone deacetylation) complex and, as the name suggests, couples the two main chromatin-modifying activities, chromatin remodelling and histone modification.
Independent purifications of the NuRD complex did not produce identical compositions, as there were subtle variations in each of the purifications. Nonetheless, the defining components of the complex are an Mi-2 chromatin remodelling subunit, an Mbd3 subunit and Mta subunit (Fig. 1A). Mta subunits (e.g. Mta1, Mta2 or Mta3) appear to be mutually exclusive within NuRD, possibly contributing to functional diversity of NuRD complexes (Bowen et al., 2004; Fujita et al., 2004). Mbd3 can be replaced by related protein Mbd2, forming the Mecp1 complex (Feng and Zhang, 2001; Le Guezennec et al., 2006a). The original purifications of NuRD did not identify Mbd2, indicating that Mecp1 accounts for only a small subfraction of the total NuRD in mammalian cells (Tong et al., 1998; Wade et al., 1998; Xue et al., 1998; Zhang et al., 1998). As Mbd2 has been shown to be dispensable for normal mammalian development (Hendrich et al., 2001), Mbd2 and Mecp1 will not be considered further in this review. NuRD also contains a core histone deacetylase complex comprised of Hdac1, Hdac2, Rbbp7 (formerly known as RbAp46) and Rbbp4 (formerly RbAp48). These core proteins are also found in the Sin3 complex. Additionally the presence of the Gatad2a and Gatad2b proteins (formerly kown as p66α and p66β) is often reported for NuRD purifications. While NuRD has been purified from mammalian, amphibian and insect cells, subunits of the complex have also been described in other organisms including plants and worms implying that NuRD is broadly conserved among plants and animals.
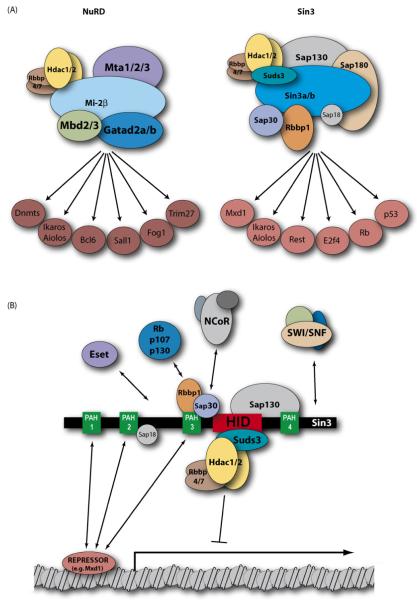
Fig. 1.
(A) Schematic of the NuRD and Sin3 co-repressor multiprotein complexes in mammalian cells. The individual components of the complexes are indicated above, with arrows indicating associations with transcriptional repressors. Each complex contains both Hdac1 and Hdac2 proteins and both Rbbp4 and Rbbp7 proteins. NuRD may contain both Gatad2a and Gatad2b, but Mta1, Mta2 and Mta3 are mutually exclusive within NuRD, as are Mbd2 or Mbd3. Similarly Sin3 complexes contain either Sin3a or Sin3b. Ikaros and Aiolos have been reported to associate with both complexes. “Dnmts” refers to reports that some NuRD components can interact with Dnmt1, Dnmt3a and Dnmt3b. (B) The co-repressor Sin3 serves as a scaffold, physically coupling class I histone deacetylases (via the Hdac-interaction domain, red) to sequence-specific transcriptional repressors (via PAH domains, green). In this way Sin3 can be recruited to diverse promoter sequences in many different cell types. The specificity and functionality of the complex are further increased by incorporating adaptor proteins that can recruit additional repressors and chromatin-modifying complexes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
2. The Mi-2 nucleosome remodelling proteins
Mi-2 is a key component of the NuRD complex as it contains both the ATPase, chromatin remodelling activity and physically associates with histone deacetylases. Originally identified as an autoantigen in dermatomyositis (Seelig et al., 1995), it is the largest subunit of the NuRD complex. Mammalian genomes are capable of encoding two Mi-2 proteins: Mi-2α (encoded by the Chd3 gene) and Mi-2β (encoded by the Chd4 gene) (Seelig et al., 1996). The latter is the form predominantly associated with the mammalian NuRD complex (Feng and Zhang, 2001; Zhang et al., 1998), although Mi-2α has been shown to be a member of the NuRD complex in a variety of human cell lines (Le Guezennec et al., 2006a; Tong et al., 1998; Xue et al., 1998). Whether functional or cell type-specific differences exist between Mi-2α- and Mi-2β-containing NuRD complexes remains to be determined. Mi-2 proteins belong to the CHD (chromo-helicase–ATP–DNA binding) family, which is conserved from yeast to humans. Structurally, Mi-2α and Mi-2β both contain two PHD (plant homeo domain)-zinc finger domains, two chromodomains and a SWI2/SNF2-type ATPase/helicase domain, the latter being responsible for the nucleosome remodelling activity (Wang and Zhang, 2001). The chromodomains of Drosophila melanogaster dMi-2 have been reported to have DNA-binding ability (Bouazoune et al., 2002), but the role of the chromodomains in the mammalian Mi-2 proteins remains to be determined. Similarly, the function(s) of the PHD fingers has not yet been demonstrated.
3. Transcriptional regulation and the role of Mi-2β and the NuRD complex
Since its purification in 1998, numerous different functions for NuRD have been postulated, although its main function is as a transcriptional repressor complex (Denslow and Wade, 2007). As histone deacetylation is generally correlated with transcriptional repression, the presence of two Hdacs in the complex implicates it in transcriptional repression. The remodelling activity of NuRD may be required to allow access of the deacetylases to the histone tails. Indeed, histone deacetylation of nucleosomal substrates by Mi-2 complexes is stimulated by ATP hydrolysis. Importantly, the NuRD complex represents one of the most abundant forms of a deacetylase complex in amphibian eggs and cultured mammalian cells (Wade et al., 1998; Zhang et al., 1998). This implicates it as a possible general co-repressor complex.
Evidence for a repressive function of NuRD also comes from physical interaction studies. In murine lymphocytes, Mi-2β was uncovered as interacting with Ikaros and Aiolos, which are zinc finger DNA-binding factors and potent repressors required for lymphoid cell development (Kim et al., 1999). Ikaros is able to bind DNA in a sequence-specific manner and is also associated with pericentric heterochromatin in cycling T-cells, indicating that it might be involved in a more general silencing mechanism. Indeed, mammalian NuRD has been shown to interact with a variety of other transcriptional repressors (Fig. 1A) implicating it as a widely used repressor in a variety of cell types.
Mi-2β has also been implicated in transcriptional activation. Biochemical analysis revealed that the amino-terminus of Mi-2β had transcriptional activating ability in reporter assays by interacting with Brg-1 (Shimono et al., 2003). Williams et al. (2004) showed that Mi-2β is required for several steps during T cell development, playing a direct role in promoting CD4 gene expression. This experiment defines a role for Mi-2β in gene activation in vivo. Whether this represents a NuRD-independent function of Mi-2β, or possibly implicates deacetylase function in transcriptional activation (e.g. Metivier et al., 2003; reviewed in Smith, 2008) remains to be demonstrated.
4. Developmental roles of the NuRD complex
Development requires the stable repression and activation of genes in different cell types. Transcriptional repression by the NuRD complex has been connected with developmental roles in numerous model systems (Table 1). In D. melanogaster, dMi-2 appears to function with Hb and Polycomb group proteins in HOX gene repression during embryo patterning (Kehle et al., 1998). dMi-2 is essential for embryogenesis as mutants arrest as first or second instar larvae, and is also required for germ cell development (Kehle et al., 1998). In Caenorhabditis elegans, mutations in the Mi-2 homologue let-418 revealed it to be required for proper vulval development (Solari and Ahringer, 2000; von Zelewsky et al., 2000). Genetic approaches have shown that let-418 is an essential gene and is required for larval viability. Additionally, let-418 is required for maintenance of germline-soma distinctions in C. elegans as let-418 deficient animals show inappropriate expression of germline-specific genes in somatic cells (Unhavaithaya et al., 2002). The Pickle (aka Gymnos) gene of Arabidopsis thaliana has been identified as a Mi-2 homologue (Eshed et al., 1999; Ogas et al., 1999). It has a similar role to let-418, as Pickle mutants express embryonic characteristics in root meristem cells (Ogas et al., 1997, 1999). Hence, Pickle is required to repress embryonic development to allow the transition to post-embryonic development. These studies highlight the importance of Mi-2 and NuRD in the development of various model organisms. Similarly, the involvement of the NuRD complex in mammalian development is now beginning to be understood.
Table 1.
Role of the NuRD and Sin3 complex subunits and their respective homologs in development of different model organisms, inferred from genetic (e.g. mutation or deletion) experiments
| Gene | Organism | Developmental role | References |
|---|---|---|---|
| Hdac1/2 | |||
| Hda-1 | Caenorhabditis elegans | Embryonic viability, vulval development | Shi and Mello (1998), Solari and Ahringer (2000) |
| Rpd3 | Drosophila melanogaster | Embryonic segregation, groucho-mediated transcriptional repression |
Chen et al. (1999), Mannervik and Levine (1999) |
| Hdac1 | Mus musculus | Embryonic viability (die at ~9.5 dpc), control of proliferation |
Lagger et al. (2002), Montgomery et al. (2007) |
| Hdac2 | M. musculus | Body size? | Montgomery et al. (2007), Zimmermann et al. (2007) |
| Rbbp4/7 | |||
| Lin-54 | C. elegans | Embryonic viability, vulval development | Lu and Horvitz (1998), Shi and Mello (1998), Solari and Ahringer (2000) |
| Rba-1 | C. elegans | Embryonic viability, vulval development | Shi and Mello (1998), Solari and Ahringer (2000) |
| Mi-2 | |||
| Pickle | Arabidopsis thaliana | Repression of embryonic and meristem genes | Eshed et al. (1999), Ogas et al. (1999) |
| let-418 | C. elegans | Larval viability, vulval development, maintenance of germline-soma distinctions |
Solari and Ahringer (2000), Unhavaithaya et al. (2002), von Zelewsky et al. (2000) |
| dMi-2 | D. melanogaster | Hox repression, larval and germ cell viability | Kehle et al. (1998) |
| Chd4 | M. musculus | Haematopoiesis, epidermal development, peri-implantation development |
Kashiwagi et al. (2007), Williams et al. (2004), Yoshida et al. (2008), Costello and Hendrich (unpublished) |
| Mta1/2 | |||
| Egl-27 | C. elegans | Embryonic patterning, Hox regulation, Wnt signalling, vulval development |
Ch’ng and Kenyon (1999), Herman et al. (1999), Solari and Ahringer (2000), Solari et al. (1999) |
| Egr-1 | C. elegans | Embryonic patterning, Hox regulation, vulval development |
Ch’ng and Kenyon (1999), Solari and Ahringer (2000), Solari et al. (1999) |
| Mta2 | M. musculus | Embryonic development and viability | Lu et al. (2008) |
| Mbd3 | |||
| dMbd2/3 | D. melanogaster | Stability of peri-centric heterochromatin, suppressor effect on peri-centric position effect variegation |
Marhold et al. (2004) |
| Mbd2/3 | C. elegans | Morphological defects (post-embryonic) |
Gutierrez and Sommer (2004) |
| Mbd2 | M. musculus | None detected | Hendrich et al. (2001) |
| Mbd3 | M. musculus | Lineage commitment of pluripotent cells, silencing of pre-implanation genes |
Kaji et al. (2006), Kaji et al. (2007) |
| Gatad2a/b | |||
| Simj (p66) | D. melanogaster | Embryonic viability, Wnt signalling | Kon et al. (2005) |
| Gatad2a (p66) | M. musculus | Pleiotrophic effects post-gastrulation, silencing of a few TE genes | Marino and Nusse (2007) |
| Sin3 | |||
| Sin3a | M. musculus | Peri-implantation development, cell proliferation | Cowley et al. (2005), Dannenberg et al. (2005) |
| Sin3a | D. melanogaster | Embryonic viability, Wnt signalling, cell cycle progression |
Pennetta and Pauli (1998), Pile et al. (2002), Tsai et al. (1999) |
| Sin3b | M. musculus | Embryonic viability, terminal differentiation | David et al. (2008) |
| Suds3 (Sds3) | M. musculus | Centromere function, cell viability | David et al. (2003) |
| Arid4a (Rbbp1) | M. musculus | Genomic imprinting? | Wu et al. (2006) |
| Arid4b (Sap180) | M. musculus | Peri-implantation development |
Wu et al. (2006) |
Alternate names for genes are given in parentheses. Modified and updated from Solari and Ahringer (2000).
5. Role of Mbd3 in early embryonic development
The first indication that NuRD may play an important role in early cell fate decisions was provided by the observation that murine embryos lacking Mbd3 die early in embryonic development (Hendrich et al., 2001). Maternally derived Mbd3 is absent by the blastocyst stage, yet at 4.5 dpc Mbd3 null embryos are morphologically normal and appear to have normal segregation of trophoblast, primitive endoderm and primitive ectoderm lineages, indicating that NuRD is not simply required for cell viability (Kaji et al., 2007). However by 5.5 dpc embryos lacking Mbd3 fail to develop recognisable extraembryonic ectoderm or embryonic ectoderm structures, and the few Oct4 expressing inner cell mass cells fail to expand in number and remain proximally located. The failure of Oct4 positive epiblast cells of Mbd3 null 5.5 dpc embryos to expand does not appear to be due to increased apoptosis or a reduction in the proliferative rate of Oct4 positive cells. Rather ICM cells continue to express both Oct4 and a marker of primitive endoderm, Gata4, as is seen in 4.5 dpc ICMs. Organised visceral endoderm also fails to form by 5.5 dpc, despite the presence of primitive endoderm at 4.5 dpc. While no extraembryonic ectoderm is detectable in Mbd3-mutant embryos, the ectoplacental cone continues to grow during post-implantation development, demonstrating that some extraembryonic lineages do not require Mbd3 function. Hence, Mbd3 is required for the development of many embryonic and extraembryonic tissues during early post-implantation development.
Ex vivo culture of Mbd3−/− ICMs revealed that Mbd3 is required for the expansion of the Oct4 expressing cell population, and no embryonic stem cell lines could be made from Mbd3−/− blastocysts (Kaji et al., 2007). Nevertheless, Mbd3null ES cells could be made by gene targeting, demonstrating that while Mbd3 function is required for ES cell derivation, it is dispensable for ES cell self-renewal (Kaji et al., 2006). The absence of Mbd3 results in a decrease in the abundance of some other subunits of the NuRD complex, including Mta1 and Mta2, implying that Mbd3 is required for the stability of other NuRD subunits. Accordingly, little, if any intact NuRD complex can be detected in Mbd3 null ES cells by immunoprecipitaiton (Kaji et al., 2006).
In the absence of Mbd3, the differentiation potential of ES cells is compromised. In a number of different contexts Mbd3 null ES cells were shown to be able to initiate differentiation but could not commit to specific cell fates (Kaji et al., 2006). Correspondingly, Mbd3 null ES cells were capable of LIF-independent self-renewal even after prolonged periods in culture conditions without LIF. Unexpectedly, Mbd3 null ES cells can differentiate when treated with retinoic acid, indicating that ES cells lacking Mbd3 can differentiate in response to certain signals, and that Mbd3 is not an absolute requirement for differentiation (Kaji et al., 2006). Overall, these studies indicate that Mbd3/NuRD is required to create an epigenetic state that allows cells to commit to specific lineages in response to particular developmental cues, but is not absolutely required for differentiation.
NuRD appears to be required neither for maintenance of global histone acetylation levels (Kaji et al., 2006) nor as a global regulator of gene silencing, as only about 200 genes were found to be misregulated ≥2× in Mbd3−/− ES cells (Kaji et al., 2007; K. Kaji and B. Hendrich, unpublished). Analysis of a handful of genes mis-expressed in Mbd3 null ES cells identified at least 11 genes that are misregulated in pre-implantation Mbd3 null embryos. A subset of these genes were found to be inappropriately expressed in ICM cells of null embryos, while others were not appropriately silenced in the transition of ICM cells of 3.5 dpc null embryos to the early epiblast cells of 4.5 dpc embryos (Kaji et al., 2007). This indicates a role for NuRD-mediated transcriptional repression in the transition of pluripotent cells from a pre-implantation, potentially differentiation-resistant state, to that of the post-implantation embryo where cells are competent for lineage commitment. This role in silencing transcription of gene expression in pluripotent cells in vivo prior to developmental transitions is strikingly similar to the functions ascribed to the Arabidopsis pickle and C. elegans let-418 genes, which were both shown to facilitate silencing of germline transcripts upon the initiation of embryogenesis.
6. Role of Gatad2a in murine development
Gatad2a and 2b were not identified in the initial mammalian NuRD purifications, however it now appears that these zinc finger proteins are bona fide NuRD components (Brackertz et al., 2002; Le Guezennec et al., 2006a). The single Drosophila homologue of these proteins was independently identified in a genetic screen for modifiers of Wnt signalling, and homozygous mutant flies displayed metamorphosis defects consistent with misregulation of ecdysone mediated transcription (Kon et al., 2005). In mice, Gatad2a null embryos are capable of implantation but die at around 10.5 dpc (Marino and Nusse, 2007). Homozygous embryos have a range of defects detectable from 7.5 to 10.5 dpc including delayed development, smaller size, abnormal tissue development, failure to turn and increased amounts of apparent necrotic tissues. Gatad2a null ES cells were found to be viable and capable of normal differentiation in embryoid bodies (Marino and Nusse, 2007). Thus while Gatad2a is not required for early lineage decisions, a degree of redundancy during these early developmental decisions with Gatad2b remains a possibility.
7. Mi-2β and somatic stem cells
Recently evidence has emerged for an important role of Mi-2β in somatic stem cells. By inducing deletion of a conditional Chd4 allele in the haematopoietic system, Yoshida et al. (2008) provided evidence that Mi-2β plays important roles in both homeostasis and lineage choice of haematopoitic stem cells in vivo . Further, this group has provided extensive evidence for Mi-2β function in lymphopoiesis (Williams et al., 2004; Yoshida et al., 2008). By crossing these same conditional Chd4 mice with a keratin 14-Cre transgenic line, the Georgopoulos lab also found that Mi-2β functions during transition stages of normal skin development. Mi-2β appears to be required in the conversion of ectodermal progenitor cells to become basal cells, and the subsequent reprogramming of basal cells to follicle progenitors and later, matrix stem cells (Kashiwagi et al., 2007). This requirement for a NuRD component during transition stages of development observed in the epidermis is reminiscent of the role of Mbd3 in developmental transitions during development of pluripotent cells in vivo. However some evidence exists that some of the functions attributed to Mi-2β during haematopoiesis does not involve the NuRD complex (Williams et al., 2004; Yoshida et al., 2008). Whether a constitutive Chd4 deletion would reveal similar functions during peri-implantation development as did the Mbd3-knockout mice, or possibly a more severe phenotype, consistent with Mi-2β functioning both within and independently of NuRD, remains to be seen.
8. The mammalian Sin3 complex
The other class I histone deacetylase-containing complex present in mammalian cells, the Sin3 complex, was first described in yeast as a general repressor of transcription via histone deacetylation (Vidal and Gaber, 1991; Vidal et al., 1991). The mammalian homologues of the yeast Sin3 protein, Sin3a and Sin3b, serve as co-repressor scaffold proteins that physically bridge connections between histone deacetylases and sequence-specific transcription factors (Fig. 1B), thereby bringing histone deacetylase activity within proximity of target genes (for further review see Silverstein and Ekwall, 2005). Repressive Sin3/Hdac complexes have also been described in worms (Choy et al., 2007), flies (Tsai et al., 1999), amphibians (Vermaak et al., 1999), and plants (Hill et al., 2008), suggesting this mechanism of co-repression is widespread throughout metazoans.
The highly conserved domains shared by Sin3a and Sin3b are the Hdac interaction domain (HID), required for Hdac1 and Hdac2 recruitment via the Suds3 protein (Laherty et al., 1997) (Fig. 1B; Table 2), and four paired amphipathic helix (PAH) domains that directly bind diverse transcriptional repressors (Brubaker et al., 2000; Le Guezennec et al., 2006b; Sahu et al., 2008), Although Sin3a and Sin3b complexes share many core components, transcription factor binding partners, and gene targets in mammalian somatic cells both paralogs are independently required for mouse embryonic development (Cowley et al., 2005; Dannenberg et al., 2005; David et al., 2008), indicating that the functions of Sin3a and Sin3b cannot be completely redundant. However genetic evidence for some redundancy of the proteins was provided by the observation that deletion of Suds3 results in cell death due to defects in centromere function (David et al., 2003), whereas neither Sin3a nor Sin3b is essential for cell viability (see Table 1).
Table 2.
Sin3 complex components and their proposed functions
| Component | Function |
|---|---|
| Sin3a/b | Scaffold |
| Suds3 (mSds3) | Mediates Hdac1/2 interaction with Sin3 |
| Rbbp4/7 (RbAp48/46) | Mediates Hdac1/2 interaction with Sin3 and acetylated histones |
| Hdac1/2 | Histone deacetylase activity |
| Sap30 | Mediates interaction with NCoR, p33ING1b, and Rb family proteins |
| Sap130 | Contributes to repression |
| Arid4a (Rbbp1) | Mediates interaction with Rb family proteins |
| Arid4b (Sap180) | Contributes to repression |
| Sap18 | Mediates interaction with hedgehog signalling targets |
Alternate names for proteins are given in parentheses.
9. Sin3 binds repressors in diverse developmental pathways
Mammalian Sin3a and Sin3b were initially identified through their interactions with Mxd1 (aka Mad)-Max and Mxd1-Mxi1 heterodimeric complexes, which bind to and repress Myc target genes, thereby blocking Myc-induced proliferation and promoting cellular differentiation (Ayer et al., 1995; Laherty et al., 1997; Schreiber-Agus and DePinho, 1998). Subsequent purifications of Sin3 complexes from numerous mammalian cell types have revealed that they are recruited by a wide range of repressors, many of which, as with Myc/Mxd1 network proteins, control the critical processes of cell cycle progression, arrest, and exit throughout development. Notable examples of Sin3 interactors that promote cellular differentiation via cell cycle exit include Rb (Lai et al., 2001) and E2F family members (Rayman et al., 2002) that control the G1/G0 restriction point in many cell types (Halaban, 2005) and Hbp1 which blocks proliferation by repressing targets of the Wnt/β-catenin pathway (Sampson et al., 2001; Swanson et al., 2004). In a contrasting role, Sin3a and Hdac1/2 have been shown to interact with Dach1 to repress CDK inhibitor targets of Six6, thereby driving proliferation during organogenesis (Li et al., 2002). Furthermore, the tumour suppressor p53 requires Sin3/Hdac complexes to block cell cycle progression and induce apoptosis in response to genotoxic stress (Lagger et al., 2003; Murphy et al., 1999).
Sin3/Hdac complexes have also been shown to interact with a variety of developmentally important factors that are not directly involved in cell cycle control. For example, Sin3/Hdac recruitment is required for repression by Rest/Nrsf (Grimes et al., 2000; Huang et al., 1999; Nomura et al., 2005), which silences neuronal gene expression programmes in non-neuronal tissues (Chong et al., 1995; Schoenherr and Anderson, 1995) and fetal cardiac genes in adult cardiac myocytes (Bingham et al., 2007). It has also been proposed that the unliganded nuclear hormone receptors thyroid hormone receptor (TR) and retinoic acid receptor (RAR) can repress their targets by recruiting co-repressor supercomplexes containing Sin3, Hdacs, and the closely related co-repressors NCoR and Smrt (Alland et al., 1997; Heinzel et al., 1997; Nagy et al., 1997). Further evidence for this mechanism of repression is provided by genetic and physical interaction studies in Drosophila showing that ecdysone receptor (EcR)-mediated repression requires recruitment of both dSin3 and the NCoR/Smrt orthologue Smrter (Tsai et al., 1999). As a final example, Ikaros and Aiolos, which are crucial for proper proliferation and differentiation throughout B- and T-lymphocyte development, recruit Sin3a/b, Mi-2β, and Hdac1/2, all of which are required for complete repression by Ikaros (Koipally and Georgopoulos, 2002; Koipally et al., 1999).
In addition to possessing Hdac activity, Sin3 complexes have been found to associate with other chromatin-modifying factors in mammalian cells, such as Swi/Snf complex subunits (Sif et al., 2001), and the histone methyltransferase Eset (Yang et al., 2003). Furthermore, Sin3a complexes have been shown to possess potent histone deacetylase-independent transcriptional repression activity (Laherty et al., 1997; Shi et al., 2006; Vermeulen et al., 2006). Taken together, all of these interactions in diverse contexts link Sin3-mediated repression to many critical roles in mammalian development.
10. Sin3a is required for peri-implantation development, MEF proliferation, and T-cell differentiation
The essential role for Sin3a in mouse development is evident from the absence of homozygous null pups born from Sin3a+/− intercrosses (Cowley et al., 2005; Dannenberg et al., 2005). Analysis of early embryos from Sin3a+/− matings revealed that all genotypes are present at E3.5, but null embryos cannot be detected at E6.5. Instead, null embryos appear to die shortly after implantation leaving behind empty decidua, indicating that they survive until at least embryonic day 4.5–5 and are capable of implantation. At this time in development, cell cycle dynamics in the epiblast are relatively stable compared to the profound changes in regulation observed after E6.5 (White and Dalton, 2005; White et al., 2005). Also at this time, however, signals from the pluripotent ICM begin to stimulate proliferation of the trophectoderm (Kunath et al., 2004). Therefore, it is possible that in the developing embryo a lack of repression by Sin3a is lethal due to a trophoblast defect rather than an epiblast defect. Nevertheless, E3.5 null blastocysts appear morphologically normal but show severely retarded proliferation when cultured ex vivo, indicating a requirement for Sin3a in the ICM as well as the trophectoderm (Cowley et al., 2005).
The early lethal phenotype in Sin3a−/− embryos prompted functional studies of conditional Sin3a deletion in somatic cells (Cowley et al., 2005; Dannenberg et al., 2005). As suggested by the interaction data implicating Sin3 in the regulation of proliferation and cell cycle progression, loss of Sin3a in mouse embryonic fibroblasts (MEFs) results in profound growth arrest, apoptosis, and G2/M arrest. This type of arrest is also observed upon Sin3 depletion in Drosophila cells (Pile et al., 2002), and coincides with unscheduled DNA replication followed by a functional S-phase checkpoint. Interestingly, the observed apoptosis and arrest in Sin3−/− MEFs are independent of p53 despite pronounced upregulation of the primary p53 target p21Cip1, as these phenotypes were not alleviated by loss or functional inactivation of p53.
Consistent with the observed phenotypes, transcriptional profiling revealed significant de-repression of numerous genes controlling cell cycle progression (including especially the G2/M transition), DNA replication and repair, as well as known and predicted targets of E2F, Rb, p53, and Myc upon loss of Sin3a. Substantial overlap exists between the affected processes in MEFs and the results of a similar experiment in Drosophila (Pile et al., 2003). This de-repression appears to be the result of local changes in chromatin structure rather than a global change in histone acetylation, although the deletion of Sin3a did cause a general delocalization of the heterochromatic protein HP1α throughout the nucleus when compared to wild-type MEFs (Cowley et al., 2005; Dannenberg et al., 2005).
The observation that some older Sin3a+/− mice developed enlarged spleens with an unusually high CD4/CD8 positive T-cell ratio prompted an investigation into the role of mSin3a in T-cell development (Cowley et al., 2005). Deletion of Sin3a using a T-cell-specific cre led to decreased thymic cellularity and reduced number of circulating T-cells. This was found to be due, at least in part, to an apparent partial block at the beta-checkpoint leading to reduced proliferation consistent with a cell cycle defect. Together these genetic studies in mice indicate that Sin3A-mediated transcriptional repression is important for maintaining proliferative potential in a wide variety of cell types.
11. Sin3b is required at late stages of development
The two mammalian Sin3 paralogs are both widely expressed and highly homologous, and biochemical evidence for distinct Sin3a- and Sin3b-containing co-repressor complexes is scarce. Nevertheless, the fact that Sin3a−/− embryos are not rescued by the presence of Sin3b provides clear genetic evidence for distinct functions for these two proteins. Indeed, recently Sin3b was also demonstrated to be essential for embryogenesis in mice, although the consequences of Sin3b deletion are not manifested until after midgestation (David et al., 2008). Notably, Sin3b−/− embryos show defects in erythrocyte and granulocyte maturation similar to those seen in embryos deficient for known Sin3 interactors E2F4 and Mxd1 (Mad) (Foley et al., 1998; Humbert et al., 2000; Rempel et al., 2000), thereby providing evidence that Sin3b functions to promote E2F4- and Mxd1-dependent terminal differentiation of these lineages. Similarly, Sin3b−/− embryos were found to display defects in skeletal development reminiscent of those shown to be dependent upon Rb, Rbl1 (p107) and/or Rbl2 (p130) (Cobrinik et al., 1996), thus establishing a genetic link to the Rb family of proteins. Finally, a further link to E2F4 and the Rb family of repressors was provided by evidence that Sin3b mutant cells display defects in G0/G1 restriction point control (Dannenberg et al., 2000; David et al., 2008; Gaubatz et al., 2000; Sage et al., 2000).
12. Conclusions
The NuRD and Sin3 complexes are abundant in mammalian cells, and both have been linked to repression by a number of different sequence-specific transcription factors. Contrary to what might have initially been expected, neither appears to be functioning as a ‘global’ transcriptional repressor. Genetic analyses in mammals are now revealing that, like in flies, worms, and some plants, NuRD is particularly important for developmental transitions. Work with mice and cell lines mutant for NuRD components has indicated that this co-repressor complex plays an important role in maintaining stem cell homeostasis and lineage choice in both the haematopoietic system and in skin, and during maturation and lineage commitment of pluripotent cells in early embryos. In contrast Sin3a and Sin3b complexes appear to be used by the cell to control cell cycle progression and proliferation, terminal differentiation of some somatic cell types, and possibly centromere function. Hence NuRD and Sin3 provide a mechanism for the cell to apply one silencing mechanism, namely histone deacetylation by class 1 deacetylases, to two different ends. However it is worth keeping in mind that most studies of NuRD and Sin3 function have been directed towards studying deacetylation of chromatin, while the non-histone targets of this deacetylation, and the effects that may have upon transcription, remain largely ill-defined (for review see Smith, 2008). Defining the targets of NuRD- and Sin3-mediated chromatin modification and protein deacetylation, and the proteins that target these actions, will allow us to further elucidate the molecular pathways controlling lineage commitment and control of proliferation during mammalian development, and inevitably, in cancer.
Acknowledgements
PEM is the recipient of an EMBO Long Term Postdoctoral Fellowship, IC was funded by a Wellcome Trust 4-Year PhD Fellowship, and BH is a Wellcome Trust Senior Research Fellow in the Basic Biomedical Sciences.
Abbreviations
- Hdac
histone deacetylase
- ESCells
embryonic stem cells
- ICM
inner cell mass
- dpc
days post coitum
- MEF
mouse embryonic fibroblast
References
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, et al. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homolog of yeast repressor Sin3. Cell. 1995;80:767–76. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Bingham AJ, Ooi L, Kozera L, White E, Wood IC. The repressor element 1-silencing transcription factor regulates heart-specific gene expression using multiple chromatin-modifying complexes. Mol Cell Biol. 2007;27:4082–92. doi: 10.1128/MCB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21:2430–40. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta. 2004;1677:52–7. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Brackertz M, Boeke J, Zhang R, Renkawitz R. Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J Biol Chem. 2002;277:40958–66. doi: 10.1074/jbc.M207467200. [DOI] [PubMed] [Google Scholar]
- Brubaker K, Cowley SM, Huang K, Loo L, Yochum GS, Ayer DE, et al. Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell. 2000;103:655–65. doi: 10.1016/s0092-8674(00)00168-9. [DOI] [PubMed] [Google Scholar]
- Ch’ng Q, Kenyon C. egl-27 generates anteroposterior patterns of cell fusion in C. elegans by regulating Hox gene expression and Hox protein function. Development (Cambridge, Engl) 1999;126:3303–12. doi: 10.1242/dev.126.15.3303. [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Gene Dev. 1999;13:2218–30. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–57. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Choy SW, Wong YM, Ho SH, Chow KL. C. elegans SIN-3 and its associated HDAC corepressor complex act as mediators of male sensory ray development. Biochem Biophys Res Commun. 2007;358:802–7. doi: 10.1016/j.bbrc.2007.04.194. [DOI] [PubMed] [Google Scholar]
- Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, et al. Shared role of the pRB-related p130 and p107 proteins in limb development. Gene Dev. 1996;10:1633–44. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, et al. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Gene Dev. 2005;19:1581–95. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Gene Dev. 2000;14:3051–64. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci USA. 2008;105:4168–72. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Turner GM, Yao Y, Protopopov A, DePinho RA. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Gene Dev. 2003;17:2396–405. doi: 10.1101/gad.1109403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/s0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Gene Dev. 2001;15:827–32. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley KP, McArthur GA, Queva C, Hurlin PJ, Soriano P, Eisenman RN. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–85. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Gaubatz S, Lindeman GJ, Ishida S, Jakoi L, Nevins JR, Livingston DM, et al. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6:729–35. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, et al. The co-repressor mSin3A is a functional component of the REST-coREST repressor complex. J Biol Chem. 2000;275:9461–7. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- Gutierrez A, Sommer RJ. Evolution of dnmt-2 and mbd-2-like genes in the free-living nematodes Pristionchus pacificus, Caenorhabditis elegans and Caenorhabditis briggsae. Nucleic Acid Res. 2004;32:6388–96. doi: 10.1093/nar/gkh982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R. Rb/E2F: a two-edged sword in the melanocytic system. Cancer Metast Rev. 2005;24:339–56. doi: 10.1007/s10555-005-1582-z. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–8. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Gene Dev. 2001;15:710–23. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Ch’ng Q, Hettenbach SM, Ratliff TM, Kenyon C, Herman RK. EGL-27 is similar to a metastasis-associated factor and controls cell polarity and cell migration in C. elegans. Development (Cambridge, Engl) 1999;126:1055–64. doi: 10.1242/dev.126.5.1055. [DOI] [PubMed] [Google Scholar]
- Hill K, Wang H, Perry SE. A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 2008;53:172–85. doi: 10.1111/j.1365-313X.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–72. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Humbert PO, Rogers C, Ganiatsas S, Landsberg RL, Trimarchi JM, Dandapani S, et al. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol Cell. 2000;6:281–91. doi: 10.1016/s1097-2765(00)00029-0. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development (Cambridge, Engl) 2007;134:1123–32. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Kashiwagi M, Morgan BA, Georgopoulos K. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development (Cambridge, Engl) 2007;134:1571–82. doi: 10.1242/dev.001750. [DOI] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, et al. dMi-2, a hunchback-interacting protein that functions in Polycomb repression. Science (New York, NY) 1998;282:1897–900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–55. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Koipally J, Georgopoulos K. A molecular dissection of the repression circuitry of Ikaros. J Biol Chem. 2002;277:27697–705. doi: 10.1074/jbc.M201694200. [DOI] [PubMed] [Google Scholar]
- Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon C, Cadigan KM, da Silva SL, Nusse R. Developmental roles of the Mi-2/NURD-associated protein p66 in Drosophila. Genetics. 2005;169:2087–100. doi: 10.1534/genetics.104.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Strumpf D, Rossant J. Early trophoblast determination and stem cell maintenance in the mouse—a review. Placenta. 2004;25(Suppl. A):S32–38. doi: 10.1016/j.placenta.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Lagger G, Doetzlhofer A, Schuettengruber B, Haidweger E, Simboeck E, Tischler J, et al. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol. 2003;23:2669–79. doi: 10.1128/MCB.23.8.2669-2679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–81. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–56. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge MC, et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol. 2001;21:2918–32. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006a;26:843–51. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X, Vermeulen M, Stunnenberg HG. Molecular characterization of Sin3 PAH-domain interactor specificity and identification of PAH partners. Nucleic Acid Res. 2006b;34:3929–37. doi: 10.1093/nar/gkl537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science (New York, NY) 2002;297:1180–3. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–91. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Lu X, Kovalev GI, Chang H, Kallin E, Knudsen G, Xia L, et al. Inactivation of NuRD component Mta2 causes abnormal T cell activation and lupus-like autoimmune disease in mice. J Biol Chem. 2008;283:13825–33. doi: 10.1074/jbc.M801275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik M, Levine M. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc Natl Acad Sci USA. 1999;96:6797–801. doi: 10.1073/pnas.96.12.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold J, Kramer K, Kremmer E, Lyko F. The Drosophila MBD2/3 protein mediates interactions between the MI-2 chromatin complex and CpT/A-methylated DNA. Development (Cambridge, Engl) 2004;131:6033–9. doi: 10.1242/dev.01531. [DOI] [PubMed] [Google Scholar]
- Marino S, Nusse R. Mutants in the mouse NuRD/Mi2 component P66alpha are embryonic lethal. PLoS ONE. 2007;2:e519. doi: 10.1371/journal.pone.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Gene Dev. 2007;21:1790–802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Gene Dev. 1999;13:2490–501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, et al. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–80. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nomura M, Uda-Tochio H, Murai K, Mori N, Nishimura Y. The neural repressor NRSF/REST binds the PAH1 domain of the Sin3 corepressor by using its distinct short hydrophobic helix. J Mol Biol. 2005;354:903–15. doi: 10.1016/j.jmb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science (New York, NY) 1997;277:91–4. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:13839–44. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennetta G, Pauli D. The Drosophila Sin3 gene encodes a widely distributed transcription factor essential for embryonic viability. Dev Genes Evol. 1998;208:531–6. doi: 10.1007/s004270050212. [DOI] [PubMed] [Google Scholar]
- Pile LA, Schlag EM, Wassarman DA. The SIN3/RPD3 deacetylase complex is essential for G2 phase cell cycle progression and regulation of SMRTER corepressor levels. Mol Cell Biol. 2002;22:4965–76. doi: 10.1128/MCB.22.14.4965-4976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pile LA, Spellman PT, Katzenberger RJ, Wassarman DA. The SIN3 deacetylase complex represses genes encoding mitochondrial proteins: implications for the regulation of energy metabolism. J Biol Chem. 2003;278:37840–8. doi: 10.1074/jbc.M305996200. [DOI] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, et al. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Gene Dev. 2002;16:933–47. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel RE, Saenz-Robles MT, Storms R, Morham S, Ishida S, Engel A, et al. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol Cell. 2000;6:293–306. doi: 10.1016/s1097-2765(00)00030-7. [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, et al. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Gene Dev. 2000;14:3037–50. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu SC, Swanson KA, Kang RS, Huang K, Brubaker K, Ratcliff K, et al. Conserved themes in target recognition by the PAH1 and PAH2 domains of the Sin3 transcriptional corepressor. J Mol Biol. 2008;375:1444–56. doi: 10.1016/j.jmb.2007.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson EM, Haque ZK, Ku MC, Tevosian SG, Albanese C, Pestell RG, et al. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–11. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science (New York, NY) 1995;267:1360–3. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, DePinho RA. Repression by the Mad(Mxi1)-Sin3 complex. Bioessays. 1998;20:808–18. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Seelig HP, Moosbrugger I, Ehrfeld H, Fink T, Renz M, Genth E. The major dermatomyositis-specific Mi-2 autoantigen is a presumed helicase involved in transcriptional activation. Arthritis Rheum. 1995;38:1389–99. doi: 10.1002/art.1780381006. [DOI] [PubMed] [Google Scholar]
- Seelig HP, Renz M, Targoff IN, Ge Q, Frank MB. Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheum. 1996;39:1769–71. doi: 10.1002/art.1780391029. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mello C. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Gene Dev. 1998;12:943–55. doi: 10.1101/gad.12.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y, Murakami H, Kawai K, Wade PA, Shimokata K, Takahashi M. Mi-2 beta associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J Biol Chem. 2003;278:51638–45. doi: 10.1074/jbc.M309198200. [DOI] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Gene Dev. 2001;15:603–18. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Smith CL. A shifting paradigm: histone deacetylases and transcriptional activation. Bioessays. 2008;30:15–24. doi: 10.1002/bies.20687. [DOI] [PubMed] [Google Scholar]
- Solari F, Ahringer J. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr Biol. 2000;10:223–6. doi: 10.1016/s0960-9822(00)00343-2. [DOI] [PubMed] [Google Scholar]
- Solari F, Bateman A, Ahringer J. The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development (Cambridge, Engl) 1999;126:2483–94. doi: 10.1242/dev.126.11.2483. [DOI] [PubMed] [Google Scholar]
- Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, et al. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nature Struct Mol Biol. 2004;11:738–46. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–21. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Yao TP, McKeown M, Evans RM. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell. 1999;4:175–86. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol Cell Biol. 1999;19:5847–60. doi: 10.1128/mcb.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Walter W, Le Guezennec X, Kim J, Edayathumangalam RS, Lasonder E, et al. A feed-forward repression mechanism anchors the Sin3/histone deacetylase and N-CoR/SMRT corepressors on chromatin. Mol Cell Biol. 2006;26:5226–36. doi: 10.1128/MCB.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–27. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Strich R, Esposito RE, Gaber RF. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–16. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Müller F. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development (Cambridge, Engl) 2000;127:5277–84. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–6. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wang HB, Zhang Y. Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acid Res. 2001;29:2517–21. doi: 10.1093/nar/29.12.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–8. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16:2018–27. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–33. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Wu M-Y, Tsai T-F, Beaudet AL. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Gene Dev. 2006;20:2859–70. doi: 10.1101/gad.1452206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yang L, Mei Q, Zielinska-Kwiatkowska A, Matsui Y, Blackburn ML, Benedetti D, et al. An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and transcription co-repressors mSin3A/B. Biochem J. 2003;369:651–7. doi: 10.1042/BJ20020854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, et al. The role of the chromatin remodeler Mi-2{beta} in hematopoietic stem cell self-renewal and multilineage differentiation. Gene Dev. 2008;22:1174–89. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig H-P, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activites. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Kiefer F, Prudenziati M, Spiller C, Hansen J, Floss T, et al. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67:9047–54. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]