Obesity has become a major worldwide challenge to public health, due to the Western ‘obesogenic’ environment interacting with a strong genetic contribution1. Recent extensive genome-wide association studies (GWAS) have identified numerous single nucleotide polymorphisms (SNPs) associated with obesity, but these loci together account for only a small fraction of the known heritable component1. Thus, the “common disease, common variant” paradigm is increasingly under challenge2. We report a highly-penetrant form of obesity, initially observed in 31 subjects who were heterozygous for deletions of at least 593kb at 16p11.2 and whose ascertainment included cognitive deficits. Nineteen similar deletions were identified from GWAS data in 16053 individuals from 8 European cohorts. Such deletions were absent from healthy non-obese controls and accounted for 0.7% of our morbid obesity cases (body mass index, BMI ≥ 40 kg.m−2 or BMI standard deviation score ≥ 4; p = 6.4×10−8, OR = 43.0), demonstrating the potential importance in common disease of rare variants with strong effects. This highlights a promising strategy for identifying missing heritability in obesity and other complex traits: Cohorts with extreme phenotypes are likely to be enriched for rare variants, thereby improving power for their discovery. Subsequent analysis of the loci so identified may well reveal additional rare variants that further contribute to the missing heritability, as recently reported for SIM13. Thus, the most productive approach may be to combine the “power of the extreme”4 in small, well-phenotyped cohorts, with targeted follow-up in GWAS and population cohorts.
The extent to which copy number variants (CNVs) might contribute to the missing heritability of common disorders is currently much under debate2. Since the majority of common simple CNVs are well-tagged by SNPs, it has recently been suggested that common CNVs are unlikely to contribute substantially to the missing heritability5. However, rare variants or recurring CNVs that have arisen on multiple independent occasions are unlikely to be captured by SNP tagging, and their identification will require alternative approaches.
We have previously hypothesised that cohorts with extreme phenotypes that include obesity may be enriched for rare but very potent risk variants4,6. Here we have investigated 312 subjects, from three centres in the UK and France, presenting with congenital malformations and/or developmental delay in addition to obesity as previously defined6,7 (see Methods). Known syndromes (e.g. Prader-Willi, fragile X etc.) were excluded. A combination of array comparative genomic hybridisation (aCGH), genotyping arrays, quantitative PCR (qPCR) and multiplex ligation-dependent probe amplification (MLPA) was used to identify and confirm the presence of a heterozygous deletion on 16p11.2 in 9 individuals (2.9%). Such deletions, estimated to be a total of 740kb in size (one copy of a segmental duplication plus 593kb of unique sequences, Figure 1a), have previously been associated to varying extents with autism, schizophrenia and developmental delay8-11; however, the observed frequency of deletions in our cohort is appreciably higher than the reported frequencies in the cohorts from the previous studies (<1%), which did not include obesity as an inclusion criterion.
Figure 1. Identification and validation of deletions at 16p11.2.
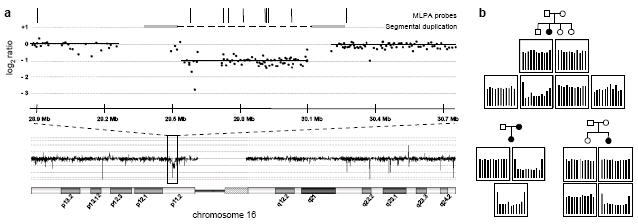
(a) aCGH data showing the location of the 16p11.2 deletion. The data show the log2 intensity ratio for a deletion carrier compared to an undeleted control sample. Grey bars connected by a broken line denote the segmental duplication flanking the deletion region. Vertical bars indicate the positions of the probe pairs used for MLPA validation. Note that CGH and genotyping array probes targeted against segmental duplications may not accurately report copy number due to the increased number of homologous sequences in the diploid state. Genome coordinates are according to the hg18 build of the reference genome. (b) MLPA validation of 16p11.2 deletions. Representative MLPA results are shown, illustrating one instance of maternal transmission and two instances of de novo deletions. Genotyping data excluded the possibility of non-paternity. Full results for MLPA validation and inheritance analysis are shown in Supplementary Figure S1. Each panel shows the relative magnitude of the normalised, integrated signal at each probe location, in order of chromosomal position of the MLPA probe pairs as indicated in (a). Each panel corresponds to its respective position on the associated pedigree, as shown.
A parallel, independent survey of aCGH and SNP-CGH data from 8 cytogenetic centres in France, Switzerland and Estonia, of 3,947 patients with developmental delay and/or malformations, but this time without selection for obesity, revealed 22 unrelated cases with similar deletions (0.6%). This is a frequency consistent with the previous studies8-11, but is significantly lower than for the above cohort which included only obese subjects (p=2.2×10−4, Fisher’s exact test).
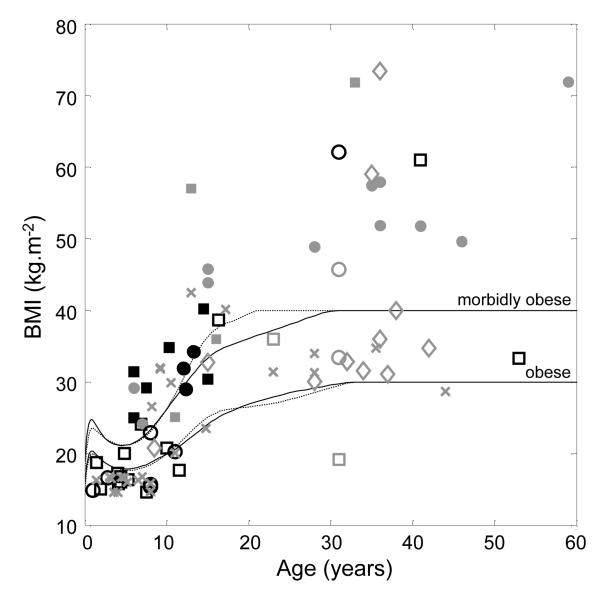
Analysis of the available clinical data for these 22 new carriers indicated that, in addition to the ascertained cognitive deficits or behavioural abnormalities (including hyperphagia, specifically identified in at least 9 cases; see Supplementary Table S1), a 16p11.2 deletion gave rise to a strongly-expressed obesity phenotype in adults, with a more variable phenotype in childhood. All 4 teenagers and adults carrying a deletion were obese, while child carriers were also frequently either obese (4/15) or overweight (2/15), a tendency that has previously been noted11; the very young (under 2 years) were of normal weight. This age-dependent penetrance was observed for all instances of deletions where phenotypic data were available, whether from this study or from previously published reports10-15, and regardless of ascertainment (Figure 2; see Supplementary Tables S2 and S3).
Figure 2. Dependence of BMI on age in subjects having a deletion at 16p11.2.
Data are for all individuals carrying a deletion for whom phenotypic data are available. Similar data from this study only are shown in Supplementary Figures S2 and S3. Lines denote the age- and gender-corrected thresholds (solid/broken – male/female) for obesity and morbid obesity. Symbols are: Square/circle – male/female; black/grey – ascertained/not ascertained for developmental delay; filled/open – ascertained/not ascertained for obesity; diamond – first-degree relative of proband; cross – previously published data10-15. The 31 year old male with BMI ~20 kg.m−2 was diabetic based on fasting blood glucose >7 mmol/L.
Taken together, the data from these parallel studies suggest a possible direct association of deletions at 16p11.2 with obesity, distinct from their cognitive phenotype. Also identified in these cohorts were instances of the reciprocal duplication, which has also been implicated in neurodevelopmental disorders, but with a variable phenotype and lower penetrance9,10,12. The frequency of the duplication in the two cohorts (12/4183, 0.3%) was consistent with previous reports for patients with cognitive deficits (0.3–0.7%)10,12. Carriers of the duplication were neither obese nor had reported hyperphagia.
To further investigate the association of 16p11.2 deletions with obesity, and to estimate the extent to which it is observed independently of ascertainment for neurodevelopmental symptoms, we carried out algorithmic and statistical analyses of genome-wide SNP genotyping data (see Table 1) from Swiss (CoLaus16), Finnish (NFBC6617) and Estonian (EGPUT18) general population cohorts (11,856 subjects in total), from child obesity and adult morbid obesity case-control cohorts6,19,20 (1,224 and 1,548 subjects respectively), from an extreme early-onset obesity cohort (SCOOP, 931 subjects) and from 141 patients undergoing bariatric weight-loss surgery (see online Methods); in total, we identified 17 instances of deletions (and 4 duplications) with no significant gender bias (Table 1). In addition, we identified 2 further unrelated carriers of a deletion from amongst 353 members of 149 families with sibling pairs discordant for obesity (SOS Sib Pair Study21). Where DNA was available for further analysis (15/19 samples), the presence of a deletion was validated using MLPA (Figure 1b) or qPCR; the remaining deletions were validated by applying a second independent algorithm to the data. With the exception of a single individual who is apparently diabetic (fasting glucose > 7 mmol/L), all adult carriers of such deletions were obese, the majority being morbidly obese; similarly, each of the 7 child/adolescent carriers had a BMI in the top 0.1% of the population range for their age and gender. None of the individuals ascertained on the basis of their obesity had any reported developmental delay or cognitive deficit; four subjects were reported as having hyperphagia.
Table 1. Frequency of detected 16p11.2 deletions in multiple cohorts.
For each cohort, 16p11.2 deletions were identified and validated using the indicated technologies. Where full phenotypic data was available, members of cohorts were categorised according to the appropriate obesity criteria (see Supplementary Information)
| Cohort | Deletions/Total | Technology | ||||
|---|---|---|---|---|---|---|
| Lean/ Normal |
Overweight | Obese | Morbidly Obese |
Total | ||
| Ascertained for cognitive deficits/malformations and obesity | ||||||
| Lille/Strasbourga | 8/279 | qPCR, aCGH | ||||
| Londona | 1/33 | aCGH, MLPA | ||||
| Ascertained for cognitive deficits/malformations | ||||||
| French-Swiss cytogenetic clinical diagnostic groupa | 21/3870 | aCGH, QMPSF, qPCR, FISH | ||||
| Estonian cases of cognitive deficita | 1/77 | Illumina CNV370-Duo, qPCR | ||||
| Ascertained for obesity | ||||||
| Swedish families with discordant siblingsb,d | 0/140 | 0/54 | 0/115 | 2/44 | 2/353 | Illumina 610K-Quad, MLPA |
| French adult case-controlb | 0/669 | 0/174 | - | 4/705 | 4/1548 | Illumina CNV370-Duo, MLPA |
| French child case-controlc | 0/530 | 0/51 | 1/260 | 3/383 | 4/1224 | Illumina CNV370-Duo, MLPA |
| British extreme early-onset obesity (SCOOP)c | 3/931 | 3/931 | Affymetrix 6.0, MLPA | |||
| French bariatric weight-loss surgeryb | - | - | 0/15 | 2/126 | 2/141 | Illumina 1M-duo, MLPA |
| Population cohorts (origin) | ||||||
| NFBC66 (Finnish)b | 1/3148 | 0/1622 | 1/434 | 1/42 | 3/5246 | Illumina CNV370-Duo |
| CoLaus (Swiss)b | 0/2675 | 0/2049 | 0/830 | 0/58 | 0/5612 | Affymetrix 500K |
| EGPUT (Estonian)b | 0/412 | 0/358 | 1/213 | 0/15 | 1/998 | Illumina CNV370-Duo, qPCR |
| Total without ascertainment for cognitive deficits/malformationsd |
1/7434 | 0/4254 | 3/1742 | 13/2260 | ||
Not categorised, complete phenotypic data not available.
BMI thresholds for overweight, obese, morbidly obese were ≥25 kg.m−2, ≥30 kg.m−2, ≥40 kg.m−2 respectively.
BMI thresholds for overweight, obese, morbidly obese were the age- and gender-corrected 90th precentile, 97th precentile, +4 standard deviations above the mean, respectively.
Discordant siblings not included in totals due to relatedness.
To enable sufficient statistical power to give robust conclusions, we combined data from the population and obesity cohorts in an overall case-control association analysis (the samples from sib pair families were excluded to avoid complications due to their relatedness). Compared to lean/normal weight subjects (see Table 1 and Methods), 16p11.2 deletions were associated with obesity (p = 5.7×10−7, Fisher’s exact test; odds ratio = 29.8, 95% confidence limits = 4.0, 225) and morbid obesity (p = 6.4×10−8; OR = 43.0 [5.6, 329]) at or near genome-wide levels of significance. Expanding the control group to include all non-obese individuals increased the significance to p = 4.1×10−9 (obese) and p = 6.1×10−10 (morbidly obese).
Previous reports have indicated that these deletions are frequently not inherited from either parent but arises de novo, possibly by non-allelic homologous recombination between the >99% sequence identical segmental duplications flanking the deleted region11,14. Therefore, where possible we investigated the parents of carriers of deletions, identifying 11 cases of maternal transmission and 4 of paternal transmission. The available data showed that all first-degree relatives carrying a deletion were also obese (Supplementary Table S1). In 10 instances the deletion was apparently de novo (see Figure 1b). Extrapolation to our full dataset indicates that ~0.4% of all morbidly obese cases are due to an inherited 16p11.2 deletion. The frequency of de novo events is consistent with the previous report where ascertainment was for developmental delay and/or congenital anomalies11; by contrast, deletions are reported to be almost exclusively de novo in autistic subjects8-10.
Although they may be heterogeneous in nature, these deletions are highly likely to be the causal variants, representing the second most frequent genetic cause of obesity after point mutations in MC4R22,23. Their repeated de novo occurrence is likely to result in lack of linkage disequilibrium with any other flanking variant – no consistent haplotype has been identified by analysis of the available surrounding genotypes. To assess the effect of a deletion on the expression of nearby genes (e.g. the obesity GWAS-associated SH2B1 locus 800kb away24), we analysed available transcript data for subcutaneous adipose tissue samples from the discordant sibling cohort. Comparisons of the 2 subjects carrying a deletion with their corresponding non-obese siblings, and with other obese and non-obese subjects (Supplementary Figure S4 and Supplementary Tables S4 and S5), showed that many though not all transcripts from within the deletion had markedly reduced abundance (0.4-0.7 fold). In contrast, no clear evidence was found for consistent cis effects of the deletion on the abundance of mRNAs encoded by genes flanking the deletion. In addition, global analysis of this dataset has not identified any trans expression quantitative trait loci either within or nearby the deletion.
Thus, while we cannot completely exclude that a 16p11.2 deletion affects the expression of nearby genes (for instance, its impact may be different in other tissues), the above expression analysis strongly indicates that the observed phenotypes are likely to be due to haploinsufficiency of one or more of the ~30 genes within the deleted region. Indeed, rather than being due to a single haploinsufficiency, the phenotype may well result from the deletion of multiple genes that impact on pathways central to the development of obesity (see Supplementary Table S5). Functional network analysis of the deleted genes has led to the suggestion of a similar multi-gene effect for the cognitive phenotype8. The extent to which there is overlap between the genes involved in the obesity and cognitive phenotypes remains to be elucidated.
There is a strong correlation between developmental and cognitive disabilities and the prevalence of obesity: Patients with autism or who have learning disabilities have a greatly increased risk of obesity25; and the severely obese exhibit significant cognitive impairment26. Possible explanations include a direct causal relationship between obesity and developmental delay; the involvement of the same or related regulatory pathways; or different outcomes of the same set of behavioural disorders with complex pleiotropic effects and variable ages of onset and expressivities. The higher frequency of 16p11.2 deletions in the cohort ascertained for both phenotypes (2.9%), compared to cohorts ascertained for either phenotype alone (0.4%, 0.6% respectively), confirms their impact on both obesity and developmental delay, adding to the evidence that these two phenotypes may be fundamentally interrelated.
Methods Summary
Obesity
Definitions for overweight, obesity and morbid obesity were based on previous studies6,7: for adults, BMI ≥ 25, 30 and 40 kg.m−2 respectively; for children, BMI respectively above the 90th, 97th percentiles and ≥ 4 standard deviations above the mean, calculated according to their age and gender from a French reference population27,28.
Statistics
All reported statistical tests used Fisher’s exact test29, carried out on contingency tables constructed for the number of subjects carrying/lacking a 16p11.2 deletion versus the obesity status/ascertainment of the individual. Since no homozygous deletions were observed, it was unnecessary to make a prior distinction between recessive, additive and dominant models of disease risk. Odds ratios and 95% confidence limits were calculated as described30.
CNV discovery
Subjects ascertained for cognitive deficit/malformations with or without obesity were selected from those clinically referred for genetic testing; 16p11.2 deletions were identified in these individuals by standard clinical diagnostic procedures. Algorithmic analyses of GWAS data were variously carried out using the cnvHap algorithm; a moving window average intensity procedure; a Gaussian Mixture Model; QuantiSNP; PennCNV; BeadStudio GT module; and Birdseed. Where experimental validation was not possible, at least two independent algorithms were used for each dataset.
Online Methods
Obesity phenotype
We have used previously-defined criteria for to define overweight, obesity and morbid (class III) obesity6,7; in adults, the thresholds were BMI ≥ 25, 30 and 40 kg.m−2 respectively. In children and adolescents, we used age- and sex-specific percentiles of BMI, calculated from a French reference population27,28, that approximately correspond to these thresholds: overweight and obesity were defined by thresholds at the 90th and 97th percentiles respectively. Childhood morbid obesity was defined as BMI ≥ 4 SDs above the age- and sex-specific mean, which corresponds to a BMI of 40 kg.m−2 between the ages of 20 and 30 years for both men and women; this threshold was used in the recruitment of the SCOOP severe early-onset obesity cohorts7. The age- and sex-specific thresholds use to define obesity and morbid obesity are shown in Figure 1 and Supplementary Figures 1-2. No carriers of a 16p11.2 deletion were reported to be taking atypical antipsychotics (known to be associated with weight gain).
Patient and population cohorts
Patients referred for cognitive delay and obesity
A group of 33 patients was selected from those referred for genetic testing at the North West Thames Regional Genetics Service, based at Northwick Park Hospital in Harrow, UK, with approval from the Harrow Research Ethics Committee. Inclusion was based on 3 criteria: mental retardation; dysmorphology; and weight >97th percentile for age and gender. Abnormal karyotype, Fragile X and Prader Willi Syndrome were previously excluded.
A second group of 279 French children were selected from those referred to 2 centres (Laboratoire de Diagnostic Génétique, Nouvel Hôpital Civil, Strasbourg; Centre de Génétique Chromosomique, Hôpital Saint-Vincent de Paul, GHICL, Lille). Inclusion was based on obesity plus at least one Prader Willi-like syndromic feature (neonatal hypotonia and difficulty to thrive, mental retardation, developmental delay, behavioural problems, skin picking, facial dysmorphism, hypogenitalism or hypogonadism). Chromosomal abnormalities and Prader Willi Syndrome were excluded by karyotyping and DNA methylation analysis.
Patients referred for cognitive delay
Patients with cognitive deficits are routinely referred to clinical genetics for etiological work-ups including aCGH. We surveyed 7 cytogenetic centres in France and Switzerland, identifying 3870 patients ascertained for developmental delay and/or malformations. Also included in the study were a further 77 patients, ascertained on similar criteria, who were referred to the Department of Genetics, University of Tartu. These analyses were performed for clinical diagnostic purposes, all available phenotypic data (weight, height) being those provided anonymously by the clinician ordering the analysis. Consequently, research-based informed consent was not required by the institutional review board that approved the study.
CoLaus
This prospective population cohort was described previously16: 6188 white individuals aged 35–75 years were randomly selected from the general population in Lausanne, Switzerland. These individuals underwent a detailed phenotypic assessment, and were genotyped using the Affymetrix Mapping 500K array; 5612 samples passed genotyping quality control. This study was approved by the institutional review boards of the University of Lausanne, and written consent was obtained from all participants. Because recruitment of this cohort required the ability to give informed consent, it is possible that the (statistically non-significant) lack of 16p11.2 deletions/duplications is due to an ascertainment bias. However, any such bias, if it exists, is very small and affects the identification of only 1-2 subjects carrying a deletion.
NFBC1966
The Northern Finland Birth Cohort 1966 is a prospective birth cohort of almost all individuals born in 1966 in the two northernmost provinces of Finland. Expectant mothers were enrolled and clinical data collection took place prenatally, at birth, and at ages six months, one year, 14 years and 31 years. Biochemical and DNA samples were collected with informed consent at age 31 years. Genotyping using the Illumina Infinium 370cnvDuo array and phenotypic characteristics of the cohort were as previously described17. Phenotypic and genotyping data was available for 5246 subjects after quality control.
EGPUT
The Estonian Genome Project is a biobank coordinated by the University of Tartu (EGPUT)18. The project is conducted according to Estonian Gene Research Act and all participants have given written informed consent. The cohort includes more than 39000 individuals older than 18 years of age and reflects closely the age distribution in the Estonian population (33% male, 67% female; 83% Estonians, 14% Russians, 3% other). Subjects are recruited by general practitioners (GP) and hospital physicians and are then randomly selected. Computer Assisted Personal interview (CAPI) was filled during 1-2 hours at the doctor’s office. The data included personal data (place of birth, place(s) of living, nationality etc.), family history (four generations), educational and occupational history, lifestyle and anthropometric data. 1090 randomly-selected subjects were genotyped using the Illumina 370cnvDuo array, 998 passing the required criteria (nationality, genotyping call rate, phenotype availability).
Case-Control familial obesity
The adult-obesity case-control groups and the child-obesity case control groups were as previously published6, and were genotyped using the Illumina Human CNV370-duo array. 643 children with familial obesity (BMI ≥ 97th percentile corrected for gender and age, at least one obese first degree relative, age < 18 yr), 581 non-obese children (BMI ≤ 90th percentile), 705 morbidly obese adults with familial obesity (BMI ≥ 40 kg/m2, at least one obese first degree relative with BMI ≥ 35 kg/m2, age ≥ 18 yr) and 197 lean adults (BMI ≤ 25 kg/m2) passed quality control; this cohort included a further 646 control subjects from the DESIR prospective cohort19 (age at exam ≥ 45 yr, normal fasting glucose according to 1997 ADA criteria, BMI < 27 kg/m2) genotyped using the Illumina Hap300 array20. All participants or their legal guardians gave written informed consent, and all local ethics committees approved the study protocol.
Severe early-onset obesity cohort
The Genetics of Obesity Study (GOOS) cohort consists of over 3000 patients ascertained for severe obesity, defined as a BMI ≥ 4 SDs above the age- and sex-specific mean, and onset of obesity before 10 years of age. In this study, we selected a discovery set of 1000 UK Caucasian patients from this cohort in whom developmental delay had been excluded by routine clinical examination by experienced Physicians (this cohort is referred to as SCOOP). Mutations in LEPR, POMC and MC4R were excluded by direct nucleotide sequencing and a karyotype was performed. DNA samples were analysed using Affymetrix Genome-Wide Human SNP Array 6.0 by Aros, Inc (Århus, Denmark), of which 931 passed quality control.
Bariatric surgery cohort
Patients undergoing elective bariatric weight-loss surgery for were recruited for the ABOS study at Lille Regional University Hospital. Genotyping was carried out using the Illumina Human 1M-duo array, and data from 141 adults passed quality control. All participants gave written informed consent, and the study protocol was approved by the local ethics committee.
Swedish discordant sibling cohort
The SOS Sib Pair Study cohort was as previously published21. It includes 154 nuclear families, each with BMI discordant sibling pairs (BMI difference >10 kg.m−2), giving a total of 732 subjects. Genotyping data using the Illumina 610K-Quad array was available for 353 siblings from 149 families. Expression data from subcutaneous adipose tissue (sampled after overnight fasting) were available for 360 siblings from 151 families. Subjects received written and oral information before giving written informed consent. The Regional Ethics Committee in Gothenburg approved the studies.
Statistical Methods
In view of the low frequency of the 16p11.2 deletions, all reported statistical tests were carried out using Fisher’s exact test29. This was applied to comparisons of separately-ascertained cohorts or categories and was carried out on contingency tables constructed for the number of subjects carrying/lacking a 16p11.2 deletion (zero or one copies, as no homozygous deletions were observed) versus the obesity status/ascertainment of the individual. Since no homozygous deletions were observed, it was unnecessary to make a prior distinction between recessive, additive and dominant models of disease risk. For overall analysis of the obesity risk resulting from a deletion, cohorts were pooled according to their obesity status determined according to the criteria described above, and the described tests were then applied to the pooled data. Odds ratios and 95% confidence limits were calculated as described30.
CNV discovery and validation
Clinical identification of 16p11.2 deletions
All diagnostic procedures (aCGH, qPCR, QMPSF, FISH) were carried out according to the relevant guidelines of good clinical laboratory practice for the respective countries. All rearrangements in probands were confirmed by a second technique and karyotyping was performed in all cases to exclude a complex rearrangement.
cnvHap
CNVs were detected in the child/adult case-control, bariatric surgery, SOS sibpair and NFBC cohorts using the cnvHap algorithm (Coin et al., manuscript submitted); this method is based on an Hidden Markov Model which models transitions between copy number states at the haplotype level, improving sensitivity and accuracy by capturing LD information between CNVs and SNPs. The compiled JAR and associated parameter files can be downloaded from http://www.imperial.ac.uk/medicine/people/l.coin/. Sample data from the algorithm applied to the NFBC cohort is illustrated in Supplementary Figure S5a.
After clustering of genotyping data using the internal Illumina BeadStudio cluster files, values for logR ratio (LRR) and B allele frequency (BAF) were exported from each project and normalised: Effects of %GC content on LRR were removed by regressing on GC and GC2, while wave effects31 were removed by fitting a loess function. Normalised data for probes within 2.5Mb of the 16p11.2 deletion were analysed using cnvHap, and CNV calls intersecting the single-copy sequences within the deletion (chr16:29514353-30107356, build hg18) were extracted. 16p11.2 deletions were identified by a minimum 90% of probes within the deleted region being called as having reduced copy number.
All called 16p11.2 deletions were validated by direct analysis of LRR. Data for each probe were normalised by first subtracting the median value across all samples (so that the distribution of LRR for each probes was centred on zero), and then dividing by the variance across all samples (to correct for variation in the sensitivity of different probes to copy number variation). The normalised data were then smoothed by application of a 9-point moving average and visualised graphically (see Supplementary Figure S6); putative deletions were checked by subsequent manual confirmation of loss-of-heterozygosity across the entire region. Equally, all deletions called by this method were confirmed by cnvHap.
Gaussian Mixture Model
For the CoLaus cohort, raw genotyping data were normalized using the aroma.affymetrix framework32. Normalization steps included allelic cross-talk calibration33,34, intensity summarization using robust median average and correction for any PCR amplification bias. CN ratios for a given sample, at a given SNP or CN probe, were computed as the log2 ratio of the normalized intensity of this probe divided by the median across all the samples. CN ratios were subsequently smoothed by fitting a Loess function31. CNV calling was done using a new method based on a Gaussian mixture model (Valsesia et al., manuscript in preparation). This Gaussian mixture model fits four components (deletion, copy neutral, 1 and 2 additional copy) to CN ratios. The final copy number at each probe location is determined as the expected (dosage) copy number. The method has been validated by comparing test datasets with results from the CNAT35 and CBS36,37 algorithms and by replicating a subset of CoLaus subject on Illumina arrays. All calls at the 16p11.2 locus made by the highly stringent CBS algorithm were replicated by the Gaussian mixture model. Principal components analysis detected no significant batch effects. Sample data from the algorithm applied to the CoLaus cohort is illustrated in Supplementary Figure S5b.
PennCNV, QuantiSNP and Birdsuite
CNV discovery in the EGPUT cohort was carried out using QuantiSNP38, PennCNV39 and BeadStudio GT module (Illumina Inc.). All analyses were carried out using the recommended settings, except changing EMiters to 25 and L to 1,000,000 in QuantiSNP. For PennCNV, the Estonian population-specific B-allele frequency file was used. Data from the SCOOP cohort were analysed using Affymetrix Power Tools and Birdsuite software40
Multiplex ligation-dependent probe amplification (MLPA)
MLPA was carried out according to standard methods41 using reagents obtained from MRC-Holland (Amsterdam NL). The SALSA MLPA Kit P343-B1 Autism-1 probemix was used, which contained 9 probes within the deleted region on 16p11.2, plus one probe upstream and one downstream of this locus (see Figure 1a). MLPA products were separated using an AB3130 Genetic Analyser (Applied Biosystems) and outputs were analysed using GeneMarker software (Soft Genetics) and Microsoft Excel. Data normalisation was carried out by dividing the peak areas for each of the 11 test probes by the mean of 9 control probe peak areas. Normalised peak area data were then compared across the tested samples to determine which ones carried the 16p11.2 deletion.
Supplementary Material
Acknowledgements
AJW, AIFB, PF are supported by grants from Wellcome Trust and Medical Research Council. JSB is supported by a grant from the Swiss National Foundation (310000-112552). LJMC is supported by an RCUK Fellowship. SJ is funded by Swiss National Fund 320030_122674 and Synapsis Foundation, University of Lausanne. AV is funded by the Ludwig Institute for Cancer Research. SB is supported by the Swiss Institute of Bioinformatics. ISF and MEH are funded by the Wellcome Trust and MRC. We thank the DHOS (Direction de l’Hospitalisation et de l’Organisation des Soins) from the French Ministry of Health for their support in the development of several array CGH platforms in France. We thank “le Conseil Regional Nord Pas de Calais/FEDER” for their financial support. Part of the CoLaus computation was performed at the Vital-IT center for high performance computing of the Swiss Institute of Bioinformatics. The CoLaus authors thank Yolande Barreau, Mathieu Firmann, Vladimir Mayor, Anne-Lise Bastian, Binasa Ramic, Martine Moranville, Martine Baumer, Marcy Sagette, Jeanne Ecoffey, and Sylvie Mermoud for data collection. The CoLaus study was supported by grants from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne and by the Swiss National Foundation (33CSCO-122661). KM, AK, TE, MN, AM received support from targeted financing from Estonian Government SF0180142s08 and PP from SF0180026s09; and from the EU via the European Regional Development Fund. TE, MN and AM received support from FP7 grants (201413 ENGAGE, 212111 BBMRI, ECOGENE (#205419, EBC)). The genotyping of the Estonian Genome Project samples were performed in Estonian Biocentre Genotyping Core Facility. EGPUT authors thank Mr. Viljo Soo for technical help in genotyping. The Northwick Park authors acknowledge support from NIHR Biomedical Research Centre Scheme and the Hammersmith Hospital Charity Trustees. Genome Canada and Genome Quebec funded genotyping of DESIR subjects. Work on the SOS sib pair cohort was supported by grants from the Swedish Research Council (K2008-65X-20753-01-4, K2007-55X-11285-13, 529-2002-6671), Swedish Foundation for Strategic Research to Sahlgrenska Center for Cardiovascular and Metabolic Research, Swedish Diabetes Foundation, Åke Wiberg Foundation, Foundations of the National Board of Health and Welfare, Jeansson Foundations, Magn Bergvall Foundation, Tore Nilson Foundation, Royal Physiographic Society (Nilsson-Ehle Foundation), VINNOVA-VINNMER, Swedish federal government under the LUA/ALF agreement. The DESIR study has been supported by INSERM, CNAMTS, Lilly, Novartis Pharma and Sanofi-Aventis, by INSERM (Réseaux en Santé Publique, Interactions entre les determinants de la santé), by the Association Diabète Risque Vasculaire, the Fédération Française de Cardiologie, La Fondation de France, ALFEDIAM, ONIVINS, Ardix Medical, Bayer Diagnostics, Becton Dickinson, Cardionics, Merck Santé, Novo Nordisk, Pierre Fabre, Roche, Topcon. Northern Finland Birth Cohort 1986 (NFBC1986) was supported by the Academy of Finland (project grants 104781, 120315 and Center of Excellence in Complex Disease Genetics), University Hospital Oulu, Biocenter, University of Oulu, Finland, the European Commission (EURO-BLCS, Framework 5 award QLG1-CT-2000-01643), NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), NIH/NIMH (5R01MH63706:02), ENGAGE project and grant agreement HEALTH-F4-2007-201413, and the Medical Research Council, UK (studentship grant G0500539).. The NFBC authors thank Prof. Paula Rantakallio (launch of NFBC1966 and initial data collection), Ms Sarianna Vaara (data collection), Ms Tuula Ylitalo (administration), Mr Markku Koiranen (data management), Ms Outi Tornwall and Ms Minttu Jussila (DNA biobanking).
References
- 1.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat. Rev. Genet. 2009;10:431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stutzmann F, et al. Loss-of-function mutations in SIM1 cause a specific form of Prader-Willi-like syndrome. Diabetologia. 2009;52:S104. [Google Scholar]
- 4.Froguel P, Blakemore AIF. The Power of the Extreme in Elucidating Obesity. New Eng. J. Med. 2008;359:891–893. doi: 10.1056/NEJMp0805396. [DOI] [PubMed] [Google Scholar]
- 5.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009;461 doi: 10.1038/nature08516. doi:10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyre D, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 7.Farooqi IS, O’Rahilly S. Recent advances in the genetics of severe childhood obesity. Arch. Dis. Child. 2000;83:31–34. doi: 10.1136/adc.83.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum. Molec. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 9.Marshall CR, et al. 16p11.2 Deletion Syndrome. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss LA, et al. Association between Microdeletion and Microduplication at 16p11.2 and Autism. New Eng. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 11.Bijlsma EK, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur. J. Med. Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 12.McCarthy SE, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 2009 doi: 10.1038/ng.474. doi:10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghebranious N, Giampietro PF, Wesbrook FP, Rezkalla SH. A novel microdeletion at 16p11.2 harbors candidate genes for aortic valve development, seizure disorder, and mild mental retardation. Am. J. Med. Genet. A. 2007;143A:1462–1471. doi: 10.1002/ajmg.a.31837. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez BA, et al. Phenotypic Spectrum Associated with De Novo and Inherited Deletions and Duplications at 16p11.2 in Individuals Ascertained for Diagnosis of Autism Spectrum Disorder. J. Med. Genet. 2009 doi: 10.1136/jmg.2009.069369. doi:10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- 15.Shimojima K, Inoue T, Fujii Y, Ohno K, Yamamoto T. A familial 593-kb microdeletion of 16p11.2 associated with mental retardation and hemivertebrae. Eur. J. Med. Genet. 2009 doi: 10.1016/j.ejmg.2009.09.007. doi:10.1016/j.ejmg.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Firmann M, et al. Prevalence of obesity and abdominal obesity in the Lausanne population. BMC Cardiovasc. Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [Google Scholar]
- 17.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2008;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelis M, et al. Genetic Structure of Europeans: A View from the North–East. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. doi:10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balkau B, Eschwege E, Tichet J, Marre M. Proposed criteria for the diagnosis of diabetes: evidence from a French epidemiological study (D.E.S.I.R.) Diabetes Metab. 1997;23:428–34. [PubMed] [Google Scholar]
- 20.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 21.Jernås M, et al. Regulation of carboxylesterase 1 (CES1) in human adipose tissue. Biochem. Biophys. Res. Comm. 2009;383:63–67. doi: 10.1016/j.bbrc.2009.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeo GS, et al. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nature Genet. 1998;20:111–112. doi: 10.1038/2404. [DOI] [PubMed] [Google Scholar]
- 23.Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nature Genet. 1998;20:113–4. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 24.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen AY, Kim SE, Houtrow AJ, Newacheck PW. Prevalence of Obesity Among Children With Chronic Conditions. Obesity. 2009 doi: 10.1038/oby.2009.185. doi:10.1038/oby.2009.185. [DOI] [PubMed] [Google Scholar]
- 26.Boeka AG, Lokken KL. Neuropsychological performance of a clinical sample of extremely obese individuals. Arch. Clin. Neuropsychol. 2008;23:467–474. doi: 10.1016/j.acn.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Poskitt EM. European Childhood Obesity Group. Defining childhood obesity: the relative body mass index (BMI) Acta. Paediatr. 1995;84:961–963. doi: 10.1111/j.1651-2227.1995.tb13806.x. [DOI] [PubMed] [Google Scholar]
- 28.Rolland-Cachera MF, et al. Body mass index variations: centiles from birth to 87 years. Eur. J. Clin. Nutr. 1991;45:13–21. [PubMed] [Google Scholar]
- 29.Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J. Roy. Stat. Soc. 1922;85:87–94. [Google Scholar]
- 30.Woolf B. On estimating the relation between blood group and disease. Ann. Hum. Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 31.Marioni JC, et al. Breaking the waves: improved detection of copy number variation from microarray-based comparative genomic hybridization. Genome Biol. 2007;8:R228. doi: 10.1186/gb-2007-8-10-r228. doi:10.1186/gb-2007-8-10-r228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bengtsson H, Simpson K, Bullard J, Hansen K. Department of Statistics, University of California; Berkeley: 2008. aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. (Technical Report 745). [Google Scholar]
- 33.Bengtsson H, Irizarry R, Carvalho B, Speed TP. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–767. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- 34.Bengtsson H, Ray A, Spellman P, Speed TP. A single-sample method for normalizing and combining full-resolution copy numbers from multiple platforms, labs and analysis methods. Bioinformatics. 2009;25:861–867. doi: 10.1093/bioinformatics/btp074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, et al. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum. Genomics. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olshen AB, Venkatraman ES. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 37.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 38.Collela S, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, et al. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korn JN, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.