Abstract
Kinase inhibitors are the largest class of new cancer drugs. However, it is already apparent that most tumours can escape from the inhibition of any single kinase. If it is necessary to inhibit multiple kinases, how do we choose which ones? In this Opinion article, we discuss some of the strategies that are currently being used to identify new therapeutic combinations of kinase targets.
More than 10,000 patent applications for kinase inhibitors have been filed since 2001 in the United States alone1. This massive investment has been fuelled by the realization that kinases are intimately involved in cancer cell growth, proliferation and survival. Indeed, kinases and their direct regulators are among the most frequently mutated oncogenes and tumour suppressors2–4. Well known examples include the oncogenic kinases PIK3CA (the p110α subunit of PI3K), epidermal growth factor receptor (EGFR) and BRAF; the Ras family of oncogenes, which activate both PI3Ks and Raf; and the PTEN tumour suppressor, which inhibits PI3K signalling.
Despite the excitement surrounding these targets, clinical progress has been uneven. Kinase inhibitors have revolutionized the treatment of a select group of diseases, such as chronic myeloid leukaemia (CML) and gastrointestinal stromal tumours (GIST), which are driven by a single oncogenic kinase; for these conditions, kinase inhibitors have achieved multi-year increases in survival5–7. Smaller but significant responses have been observed for some cancers that are highly dependent on angiogenesis, and therefore sensitive to inhibitors of vascular endothelial growth factor (VEGF) signalling, such as renal cell carcinoma8–11.
Kinase inhibitors have been least effective in treating the types of cancer that have the highest mortality rates, such as lung, breast, colorectal, pancreatic and prostate cancer. Clinical trials show that the most effective kinase inhibitors prolong survival by only a few months for these cancers12–17. Results have been improved by identifying markers for patients that are more likely to respond to kinase inhibitor therapy — such as EGFR mutations in lung cancer18, ERBB2 overexpression in breast cancer19, and wild-type KRAS in lung and colorectal cancer20,21 — but even among these subgroups, relapse is inevitable for patients with disseminated disease.
Why has clinical progress been so challenging? One reason is that most tumours can escape from the inhibition of any single kinase (FIG. 1). This first became clear when resistance mutations in BCR–ABL were discovered in patients with CML who were resistant to imatinib22; similar mutations have now been detected in other kinases following treatment with kinase inhibitors23–26. Alternatively, tumours can acquire drug resistance through mechanisms that do not involve mutation of the target (FIG. 1a). These mechanisms include the activation of surrogate kinases that substitute for the drug target27 and the inactivation of phosphatases to amplify the residual kinase activity that persists during drug treatment28. It is also clear that many tumours possess intrinsic resistance to kinase inhibitors at the time of initial therapy (FIG. 1b). This can result from the activation of multiple, redundant kinase signalling pathways29 or the presence of activating mutations in downstream pathway components, such as KRAS or PTEN, which enable the tumour to bypass the drug target20,21,30.
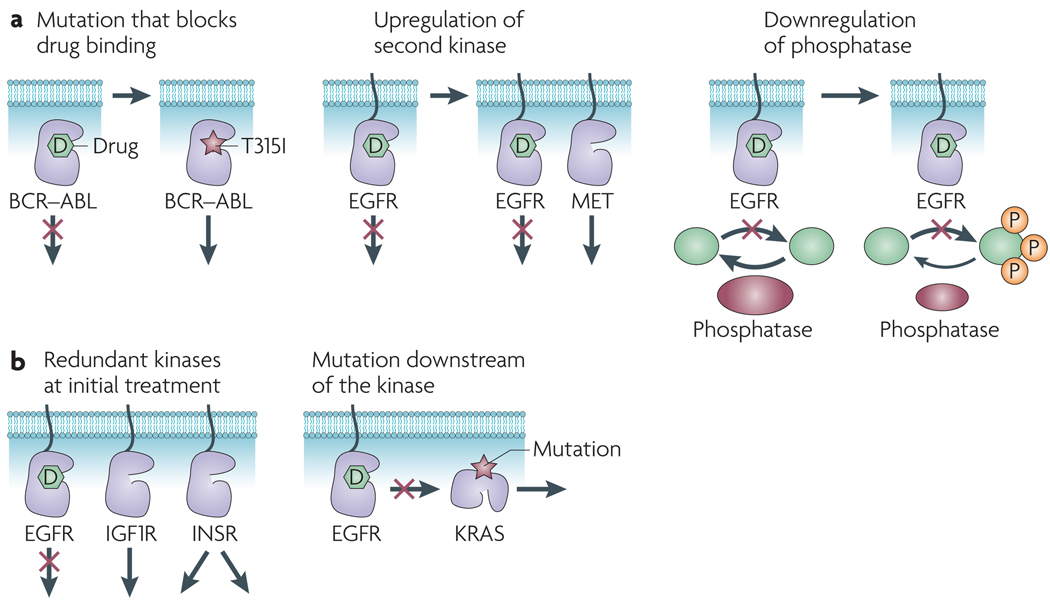
Figure 1. Resistance to kinase inhibitors.
a | Mechanisms of acquired resistance. treatment with kinase inhibitors can select for mutations that block drug binding (left panel). this was first demonstrated for the T315I mutation in BCR–ABL in chronic myeloid leukaemia22. treatment with a kinase inhibitor can induce upregulation of a second kinase that substitutes for the drug target (centre panel). The receptor tyrosine kinase MET (also known as hepatocyte growth factor receptor) has been shown to be overexpressed in lung cancer cells that acquire resistance to epidermal growth factor receptor (EGFR) inhibitors27. Tumour cells can respond to treatment with a kinase inhibitor by down-regulating the phosphatase that normally dephosphorylates the substrates of that kinase (right panel). This has the effect of decreasing the cellular potency of the kinase inhibitor. this mechanism has been observed in acquired resistance to EGFR inhibitors in breast cancer cells28. b | Mechanisms of intrinsic resistance. Many tumours express multiple oncogenic kinases that signal redundantly to promote cell survival (left panel). For example, some gliomas show constitutive activation of multiple receptor tyrosine kinases29. Mutational activation of a downstream pathway component can reduce the effectiveness of a kinase inhibitor (right panel). KRAS mutations are associated with resistance to EGFR inhibitors in lung and colorectal cancer20–21. IGF1R, insulin-like growth factor 1 receptor; INSR, insulin receptor; P, phosphorylation.
Overcoming these resistance mechanisms will require targeting tumour cells at multiple levels, through either single drugs that bind to multiple proteins31 or cocktails of highly selective inhibitors32. The challenge for the cancer research community is to learn how to predict the best combinations of targets and then prioritize those combinations for clinical testing. This is a daunting task, because the number of possible target combinations is almost limitless, but clinical trials are slow and expensive.
Targeting one kinase with multiple drugs
If a tumour depends on the activity of a single kinase, then using multiple drugs to target that kinase can be effective. This was first demonstrated in CML, in which early clinical trials showed that more than 90% of patients with chronic phase disease responded to the BCR–ABL inhibitor imatinib5 (TABLE 1), but that a subset of those patients relapsed while on the drug. Disease progression was associated with the emergence of leukaemic cells bearing mutations in BCR–ABL that block imatinib binding22, suggesting that drugs targeting these BCR–ABL mutants would be effective. Two second-generation BCR–ABL inhibitors were developed (dasatinib and nilotinib) that retain activity against most of the more than 50 clinically observed BCR–ABL resistance mutations, and these drugs are highly effective against imatinib-resistant disease33,34. However, a common BCR–ABL mutation (T315I) prevents the binding of all three drugs, and this has emerged as the default allele for many patients on long-term inhibitor therapy22,35. To address this problem, third-generation drugs have been developed that potently inhibit BCR–ABL T315I. These agents are effective in preclinical models of drug-resistant CML36–39, and four such compounds are currently in clinical trials. Some patients have now survived more than 10 years since starting treatment by undergoing sequential therapy with three generations of BCR–ABL inhibitors40, proving that it is possible to extend the therapeutic response in CML by repeatedly targeting the same kinase.
Table 1.
US FDA-approved kinase inhibitors
| Drug | Key targets for therapeutic activity | US FDA-approved indication |
|---|---|---|
| Imatinib | BCR–ABL, PDGFR and KIT | CML and GIST |
| Dasatinib | BCR–ABL | CML |
| Nilotinib | BCR–ABL | CML |
| Gefitinib | EGFR | Lung cancer |
| Erlotinib | EGFR | Lung and pancreatic cancers |
| Lapatinib | EGFR and ERBB2 | Breast cancer |
| Sunitinib | VEGFR2, PDGFR and KIT | Kidney cancer and GIST |
| Sorafenib | VEGFR2 and PDGFR | Kidney and liver cancers |
| Pazopanib | VEGFR2, PDGFR and KIT | Kidney cancer |
| Everolimus | mTOR | Kidney cancer |
| Antibody | ||
| Trastuzumab | ERBB2 | Breast cancer |
| Cetuximab | EGFR | Colorectal, and head and neck cancers |
| Panitumumab | EGFR | Colorectal cancer |
| Bevacizumab | VEGF | Colorectal, lung and breast cancers |
CML, chronic myeloid leukaemia; EGFR, epidermal growth factor receptor; FDA, Food and Drug Administration; GIST, gastrointestinal stromal tumour; PDGFR, platelet-derived growth factor receptor; VEGFR2, vascular endothelial growth factor receptor 2.
A similar approach has been used to target the ERBB2 receptor tyrosine kinase in breast cancer. Trastuzumab is a monoclonal antibody that binds to the extracellular domain of ERBB2, thereby both inhibiting ERBB2 signalling and recruiting immune cells to the tumour19; however, patients with metastatic cancer who are treated with trastuzumab invariably relapse. The mechanism of trastuzumab resistance is not understood, but it is clear that resistant tumours remain dependent on ERBB2 signalling. This is because patients with breast cancer who have progressed on trastuzumab therapy nonetheless respond to lapatinib14, a small molecule inhibitor of the tyrosine kinase domain of ERBB2. Therefore, it is possible to induce a second response in these patients by targeting ERBB2 with a drug that binds to a different site on the protein. Unlike CML, however, the clinical response to lapatinib in metastatic breast cancer is brief, and disease progression typically occurs within a few months14.
These examples show that in certain cases sequential targeting of a single kinase with multiple drugs can prolong the therapeutic response. It is unclear how broadly this model applies, because it is unclear how many tumours are truly dependent on a single oncogene (a state referred to as ‘oncogene addiction’ (REF. 41)). The strongest evidence in favour of this hypothesis is the discovery of resistance mutations after kinase inhibitor treatment in CML, GIST, lung cancer and a myeloproliferative disorder known as hypereosinophilic syndrome22–26. Such mutations are definitive proof that the mutated kinase was required for the survival of that tumour. The oncogene addiction model is also supported by many preclinical studies showing that tumour cell lines containing an activating mutation or amplification of a kinase can be more sensitive to inhibitors of that kinase in vitro42–44.
Conversely, the detection of an oncogenic kinase mutation does not guarantee sensitivity to the corresponding kinase inhibitor. For example, mutations in PIK3CA or PTEN are poor predictors of the sensitivity of tumour cell lines to PI3K inhibitors31,45. Mutations in KRAS do not, in general, sensitize tumour cells to inhibitors of Raf or Mek43,46. Indeed, the response of most tumours to inhibition of an oncogene is much less dramatic than the response in CML, in which even transient inhibition of BCR–ABL irreversibly commits cells to apoptosis44 (FIG. 2). In this respect, it is worth noting that the term oncogene addiction gained widespread use because it describes a paradox: inhibiting an oncogene would be predicted to reverse the gain of function caused by the oncogene, not kill all the tumour cells. It is only recently that this term has been conflated with the idea that oncogenes should be expected to be required for tumour survival. For this reason, there is clearly a need to identify additional vulnerabilities in tumours beyond the genes that are directly mutated.
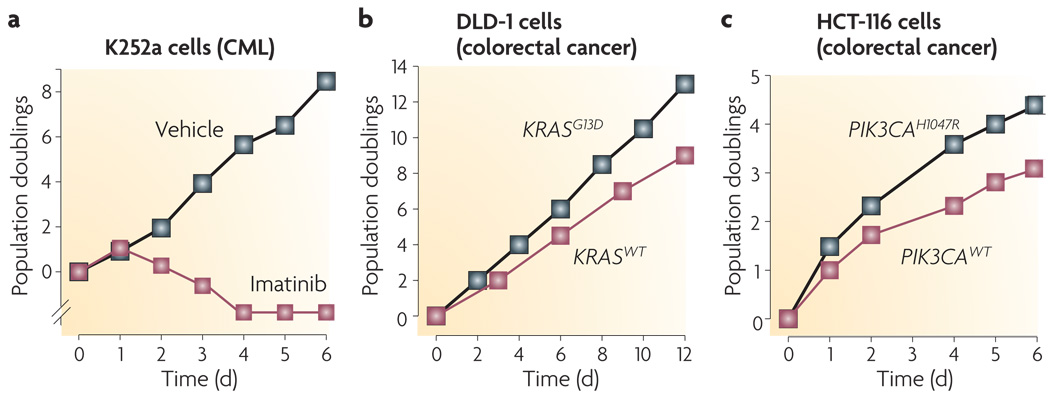
Figure 2. Degrees of oncogene addiction.
Three examples of oncogene addiction drawn from the recent literature. a | Treatment of K252a chronic myeloid leukaemia (CML) cells with the BCR–ABL inhibitor imatinib results in complete cell death by day 4 (REF. 31). b | Disruption of the KRASG13D oncogene in DLD-1 colorectal cancer cells slows the rate of cell proliferation72. c | Disruption of the PIK3CAH1047R oncogene in HCT-116 colorectal cancer cells slows the rate of cell proliferation94. WT, wild type.
Targeting nodes in a signalling network
Three sets of targets collectively account for a large proportion of current efforts in kinase inhibitor drug discovery. These are the receptor tyrosine kinases (for example, EGFR, ERBB2, platelet-derived growth factor receptor (PDGFR) and VEGF receptor 2 (VEGFR2)), the kinases in the MAPK pathway (for example, BRAF, MEK1 and MEK2) and the kinases in the PI3K pathway (for example, PIK3CA, Akt and mTOR). These three groups of targets are mechanistically linked because most receptor tyrosine kinases activate the MAPK and PI3K pathways as their primary signalling function (FIG. 3).
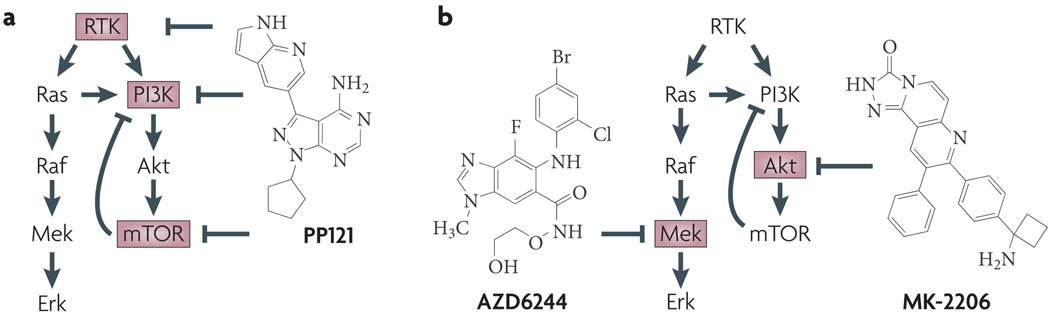
Figure 3. Strategies for multi-targeted kinase inhibition.
a | The single agent PP121 was shown to target both tyrosine kinases (such as vascular endothelial growth factor receptor 2, BCR–ABL and RET) and PI3K family members such as PIK3CA and mTOR (inhibitor targets are shown in red boxes). Note that the combined inhibition of mTOR and PI3K by PP121 disables a negative feedback loop in which mTOR inhibits PI3K. b | the combination of the MEK inhibitor AZD6244 and the Akt inhibitor MK-2206 results in the inhibition of both the MAPK and PI3K pathways. This combination is being evaluated in clinical trials. RTK, receptor tyrosine kinase.
There is a compelling biological rationale for targeting each of these groups in combination. For example, clinical resistance to tyrosine kinase inhibitors is often associated with reactivation of PI3K signalling28. Therefore, the effectiveness of tyrosine kinase inhibitors might be increased by combination with an inhibitor of the PI3K pathway. This combination has been shown to be effective in animal models and is undergoing extensive clinical testing: at least 21 clinical trials are currently evaluating the combination of a tyrosine kinase inhibitor and an mTOR inhibitor in several types of cancer. There has been a particular emphasis on the use of PI3K inhibitors to sensitize tumours to inhibitors of EGFR or ERBB2 (such as erlotinib47, lapatinib48 and trastuzumab49). This is because the anti-tumour activity of EGFR and/or ERBB2 inhibitors has been correlated with their ability to inhibit the phosphorylation of ERBB3, a kinase-inactive receptor that primarily functions to activate the PI3K pathway28.
Other target combinations are suggested by the connectivity of the signalling network. For example, mTOR activates a well-characterized negative feedback loop that inhibits the activity of PI3K (FIG. 3a). mTOR inhibitors such as rapamycin block this negative feedback loop, resulting in hyper-activation of PI3K that may counteract the anti-proliferative effect of mTOR inhibition. For this reason, it has been proposed that the dual inhibition of PI3K and mTOR may be more effective than inhibiting either target alone. Preclinical experiments support this idea45, and drugs such as PP121 that target multiple steps in this pathway have been designed31 (FIG. 3a). Several dual PI3K and mTOR inhibitors are currently being evaluated in clinical trials (for example, NVP-BEZ235, BGT226 and XL765) alongside agents that selectively target either PI3Ks (for example, XL147 and GDC-0941) or mTOR (for example, OSI-027, AZD8055 and rapamycin analogues). As there are practical challenges associated with developing both multi-targeted single agents and multi-drug cocktails50, it will be interesting to see which approach emerges as the most successful from these clinical trials.
Combination therapy can be used in other cases to target an otherwise undruggable protein. For example, KRAS is one of the most commonly mutated oncogenes, but efforts to find Ras inhibitors have been unsuccessful. It was long believed that the MAPK pathway was the primary Ras effector in most tumours51, but Raf and Mek inhibitors have inconsistent activity against tumour cells with Ras mutations43,46. Ras also directly binds to and activates PI3K52, and the disruption of this interaction prevents KRAS-driven tumorigenesis in the mouse53. For this reason, it may be necessary to inhibit both the MAPK and PI3K pathways to block the growth of tumours with Ras mutations. This conclusion is supported by data showing that resistance to Mek inhibitors in some KRAS-mutant cells is caused by mutations in PIK3CA or PTEN, and that this resistance is reversed by PI3K inhibition54. Moreover, the combination of PI3K and Mek inhibitors is active in a mouse model of KRAS-driven lung cancer55. The rationale for this combination is so compelling that Merck and AstraZeneca recently announced a joint Phase I clinical trial that will test the combination of an Akt inhibitor (MK-2206) and a Mek inhibitor (AZD6244) against solid tumours (FIG. 3b).
Limitations of rational drug combinations
The challenge associated with developing these types of rationally designed drug cocktails is that preclinical experiments do not predict their efficacy in humans. This is true even when the individual agents have already shown clinical anticancer activity. For example, preclinical experiments supported the combination of gefitinib and trastuzumab in breast cancer56,57, erlotinib and bevacizumab in renal cell carcinoma58, and cetuximab and bevacizumab in colorectal cancer59, but all of these failed in clinical trials58,60,61. In the case of cetuximab and bevacizumab, the drug combination reduced survival compared with the single agents60.
In some cases, these discrepancies may be due to misinterpretation of the preclinical data, rather than a failure of the preclinical model itself. For example, careful studies have shown that the addition of gefitinib to trastuzumab therapy in xenograft models of breast cancer results in only modest additional efficacy62, and that this additional benefit requires gefitinib concentrations that may be toxic in humans63. In other cases, subtle changes in the dosing regimen can have a large effect on the activity of the combination. For example, preclinical studies of the cyclin-dependent kinase inhibitor flavopiridol and the topoisomerase inhibitor irinotecan showed that this combination can effectively induce apoptosis in colon cancer cells when administered in a specific sequence (irinotecan followed by flavopiridol, resulting in apoptosis in 43% of the cells)64. The reverse sequence of drugs (15% apoptosis) and concurrent therapy (30% apoptosis) were both less effective. This finding was rationalized by a model in which pretreatment with flavopiridol arrested cells in the G1 phase of the cell cycle and thereby reduced the number of cells progressing through S phase and therefore irinotecan sensitivity64. A subsequent clinical trial validated the safety and preliminary efficacy of this sequential dosing regimen65.
Preclinical studies of drug combinations are probably biased towards validating the targets that are already believed to be important, and this bias limits their ability to prioritize new drug combinations for clinical testing. For example, all kinase inhibitors have some toxicity to cells, and for this reason two kinase inhibitors can usually be shown to be more toxic than either compound alone. For these comparisons it is often unclear what should be used as the normal cell to measure therapeutic index66, and in many cases the survival of the mouse in a xenograft experiment is the only evidence of differential toxicity. This can be addressed to some degree by correlating lethality with genotype across many tumour cell lines42 or by using pairs of isogenic cell lines that differ at a single locus66, but this becomes challenging when comparing drug combinations.
For a small group of kinase targets with an undisputed role in cancer — such as the oncogenic receptor tyrosine kinases and the core components of the PI3K and MAPK pathways — numerous clinical trials of drug combinations are planned or underway. It is uncertain, however, that these kinases are the best cancer drug targets67, and the route to clinical testing for combinations of drugs that target other kinases is less straight-forward. One major obstacle is that it is difficult to conduct clinical trials combining two investigational drugs, and even more difficult if the two drugs originate from different pharmaceutical companies68,69. Companies are reluctant to conduct joint clinical trials of early-stage compounds because of fears about loss of intellectual property and the possibility of an unforeseen side effect from the combination68,69. This creates a Catch-22 scenario: many kinase inhibitors are likely to be effective only as part of a combination therapy, but it will be difficult to test those combinations until after the drugs are approved as single agents. Indeed, the joint venture mentioned above between AstraZeneca and Merck to test Akt and Mek inhibitors in combination was reported in national media, such as The Wall Street Journal, partly because such early stage collaborations are so rare70. In the field of AIDS research, this problem was addressed in 1993 by the formation of the Inter-Company Collaboration for AIDS Drug Development that coordinated the testing of drug cocktails by 15 pharmaceutical companies71. However, there is not yet a comparable mechanism for companies to collaborate to test new combinations of investigational drugs in oncology, where there is arguably the greatest need and opportunity.
Using RNAi to discover new targets
The development of RNA interference (RNAi) has made it possible to directly screen for the genes required for tumour proliferation in mammalian cells. These screens have two advantages. First, they can identify new drug targets, as any gene that selectively blocks tumour growth when knocked down by RNAi is a candidate. Second, these screens provide an unbiased test of models of tumour signalling, because they directly examine which genes are most important to the tumour. This perspective is valuable, because most combination therapies are based on simple models of tumour signalling; however, there is little evidence that such models capture the most crucial interactions in the tumour cell, which could be highly indirect and inaccessible to simple reasoning.
Three recent papers illustrate the power of large-scale RNAi screens to address this problem by looking for genes that are selectively required for the growth of tumour cells expressing an activated KRAS mutant72–74. Luo et al.72 screened ~75,000 short hairpin RNAs (shRNAs) and found 83 shRNAs targeting 77 genes that preferentially impaired the growth of KRASG13D cells compared with control cells in which the KRASG13D allele had been disrupted by homologous recombination72. Analysis of these hits revealed an unexpected enrichment of a network of genes involved in mitosis. A small molecule inhibitor of the mitotic kinase polo-like kinase 1 (PLK1) had increased cytotoxicity to KRAS-mutant cells in vitro and in vivo72.
Scholl et al.73 screened a smaller set of shRNAs (5,024 targeting 1,011 genes) against a broader panel of cells that included 8 tumour cell lines (4 KRASG13D mutant and 4 KRAS wild-type) and 2 control cell lines73. The top hit was STK33, a serine threonine kinase in the calmodulin kinase family with no previous connection to Ras signalling or cancer. shRNAs targeting STK33 induced KRAS mutation-dependent toxicity in a broad panel of tumour cell lines, through a mechanism that may involve modulation of S6K1 kinase activity73.
Barbie et al.74 screened a panel of shRNAs targeting kinases, phosphatases and oncogenes against a panel of 19 tumour cell lines and then extracted from these data the genes selectively required for the survival of KRAS-mutant cells74. The top hit from this screen was TBK1, a protein kinase that activates nuclear factor-κB (NF-κB) signalling by phosphorylating the NF-κB inhibitory protein IκBα. A companion paper showed that genetic inhibition of NF-κB signalling was sufficient to block tumour development in a mouse model of KRAS-driven lung adenocarcinoma75.
A common finding from all three papers was that, although many genes were required for the survival of KRAS-mutant cells, few of those genes could have been predicted in advance on the basis of known biochemical interactions or models of Ras signalling. Among the three kinases (PLK1, STK33 and TBK1) that were the focus of follow-up experiments, only TBK1 had been previously linked to Ras (through a pathway involving the exocyst complex, RALB and RALGDS), and this protein could hardly be described as a well-known Ras effector. This is even more remarkable when we consider that Ras and its downstream targets are among the most intensely studied proteins in biology.
Similar results were described in a series of papers that attempted to define the ‘essential kinome’ that is required for cell proliferation and survival67,76,77. This was done by carrying out kinome-wide shRNA screens on a large panel of tumour cell lines, primary cells and pairs of isogenic cells that differed in the expression of a single gene. The primary conclusion from these papers was that there was little overlap in the kinases that are required for cell proliferation across many different cell lines. Indeed, there was no correlation between the number of PubMed citations for a kinase and the likelihood that the kinase was important for tumour cell proliferation. In the words of the authors77: “Although the regulation of cell proliferation and survival are heavily studied areas, we did not see a bias in these screens toward the identification of previously known and well studied kinases, suggesting that our knowledge of the molecular events in these areas is still meager.” (D. A. Grueneberg et al, 2008).
Given the unpredictable sensitivities of tumour cells to shRNAs targeting a single kinase, it may be possible to identify new pairs of targets by screening shRNAs in the presence of a drug. An early experiment in this area looked for shRNAs that synergistically killed cancer cells in the presence of A-443654, a small molecule inhibitor of Akt78. This was motivated by the surprisingly weak anti-tumour activity of A-443654 as a single agent in preclinical models79. Two kinases were identified in this screen: casein kinase 1, γ3 (CSNK1G3) and inositol polyphosphate multikinase (IPMK). Neither of these kinases had previously been linked to Akt signalling or cancer. However, knock down of both genes potentiated the inhibition of phosphorylation of Akt and ribosomal protein S6, suggesting that these kinases may have a cryptic role in regulating signalling through the PI3K pathway.
Barriers to translating RNAi into drugs
RNAi screens can help challenge our assumptions about the genes that are most important in cancer. However, there are considerable obstacles to translating any hit from one of these screens into a new drug. Most RNAi screens measure only cell proliferation in vitro, which ignores most of the capabilities of a tumour. Therefore, it will be necessary to validate the large number of genes that emerge from these screens in more complex and time-consuming models. Once these hits are validated, they become subject to the same caveats that accompany potential drug targets identified in any other way.
In this respect, it is important to emphasize that there is not a direct correlation between RNAi knockdown of a gene and the identification of a potential drug target. Most drugs cannot be replicated by an shRNA because, for example, the drug interferes with multiple targets or inhibits a single domain of a multidomain protein only. Likewise, most shRNAs cannot be replicated by a drug, because most proteins are undruggable. Indeed, there are many examples in which an shRNA (or gene knockout) and a drug targeting the same protein give different phenotypes, and the reasons for these differences have been extensively documented (for a review of this topic see REF. 80). As a result, RNAi screens may be more likely to expose the gaps in our knowledge of cancer biology than to directly point the way to new therapeutic approaches.
Using drugs to discover kinase targets
Historically, most drugs were discovered because they possessed activity in cells or animals, and their targets and mechanism of action were elucidated only later. This is sometimes called ‘phenotype-based’ drug discovery because the phenotype was discovered before the target. By contrast, almost all modern drug discovery is ‘target-based’, meaning that the target is selected first, on the basis of a hypothesis about its role in disease.
Nonetheless, there are instances in which phenotype-based drug discovery has contributed to the development of kinase inhibitors for cancer, albeit unintentionally. One example is sorafenib, which was originally designed as an inhibitor of Raf based on the logic that Raf inhibition might be effective for Ras-mutant tumours81. Sorafenib has yet to show clinical benefit for tumours that contain frequent Ras mutations, such as lung and pancreatic cancer, and has also failed in clinical trials for the treatment of melanoma82, a disease that has a high rate of BRAF mutations43,83. However, in early clinical trials of sorafenib (which were designed to establish safety and therefore contained a diverse patient population) responses were observed in two unexpected tumour types84: renal cell (kidney) and hepatocellular (liver) cancer. One patient with kidney cancer in an early Phase I trial achieved stable disease for 2 years84, leading to the broader testing and approval of sorafenib for kidney cancer (and more recently liver cancer). The efficacy of sorafenib in kidney cancer is now attributed to the inhibition of VEGFR2 in endothelial cells, which blocks angiogenesis, rather than the inhibition of Raf in the tumour. Preclinical studies have shown that the inhibition of an additional target, PDGFR in pericytes, may be important85. Therefore, sorafenib probably blocks tumour growth through the inhibition of two kinases, expressed in different tissues, neither of which was the intended target of the drug.
Imatinib provides a second example of serendipitious target discovery. After its initial approval for the treatment of CML, imatinib was tested in five patients with hypereosinophilic syndrome, a disease of unknown molecular origin, based on the reasoning that treatments that are effective in CML are sometimes also effective in patients with hypereosinophilia86 (even though the mechanism of action of those other treatments, such as hydroxyurea and interferon-α, is unrelated to the mechanism of imatinib). Remarkably, four of the five patients treated with imatinib showed a complete haematological response (normalized eosinophil counts), such that they were able to discontinue other therapies. Analysis of DNA from the leukocytes of these patients led to the discovery of a chromosomal rearrangement that generated a fusion between PDGFRA and FIP1L1, producing a constitutively active PDGFR kinase24. As PDGFR is one of a small number of kinases inhibited by imatinib, this suggested that PDGFR activation was probably the cause of the disease. This was confirmed by the discovery of a T674I resistance mutation in PDGFR in a patient who had relapsed from imatinib therapy24.
As these examples show, the advantage of using drugs to identify cancer targets is that they can reveal in an unbiased way the proteins most essential to the tumour. The major limitation of this approach is that it is difficult to identify the targets of a molecule that has an unknown mechanism of action87. If the target is unknown, then it is difficult to increase the potency of the compound by medicinal chemistry. It can also be challenging to determine whether the efficacy and toxicity of the drug are linked (because they reside in the same target) or separable (because they reside in different targets). For these reasons, it is often impossible to improve compounds that are identified in a screen but have an unknown mechanism of action.
Targeted polypharmacology
In the case of sorafenib and imatinib, it was straightforward to identify the relevant targets of those drugs, because the targets were almost certain to be kinases. As these two drugs have a relatively small number of high-affinity targets in the human kinome (fewer than 20), the possibilities could be rapidly tested. Could this approach be generalized, so that kinase inhibitors could be used to search in an unbiased way for new combinations of therapeutic targets?
A unique feature of kinase inhibitors is that they have the potential for greater target promiscuity than almost any other type of drug. This is because the kinase superfamily (including the structurally related protein, lipid and small molecule kinases) is the largest family of druggable genes that binds to a common substrate (ATP). Kinases differ in this respect from other large gene families, such as G protein-coupled receptors, which interact in their druggable site with a wide range of structurally diverse ligands, including both peptides and small molecules. This fact has been emphasized50 by noting that the kinase inhibitor sunitinib inhibits at least 79 kinases at low micromolar concentrations, whereas all the other approved drugs combined target only 320 proteins. Therefore, individual kinase inhibitors have an enormous potential for unpredicted target combinations and so new biological activities.
Despite this potential for promiscuity, it is increasingly feasible to enumerate the targets of kinase inhibitors in a systematic way. This is because most kinases can be heterologously expressed, either as a soluble kinase domain or on the surface of phage, and assayed for drug binding in a purified format. Although there are exceptions, the activity of most kinase inhibitors in cells correlates with biochemical parameters that can be measured in vitro, such as the dissociation constant (KD) of the drug and the Michaelis–Menten constant for ATP binding (KM,ATP) of the kinase88. As kinases have become increasingly important drug targets, the measurement of these biochemical parameters has been industrialized, and there are now many vendors that offer to screen compounds against panels of kinases that approach or exceed half of the kinome (FIG. 4a). As the cost of assaying compounds against these panels has decreased, it has transformed the types of experiments that are feasible (FIG. 4b). For example, a widely cited paper from 2000 reported the specificity of 24 commonly used kinase inhibitors against 28 kinases (approximately 700 kinase–drug pairs)89. In 2007, the same group published a follow-up paper that analysed the specificity of 65 common kinase inhibitors against 70 kinases (approximately 4,500 kinase–drug pairs)90. In 2008, scientists from GlaxoSmithKline reported the testing of a panel of 577 diverse kinase inhibitors against 203 kinases (more than 117,000 unique kinase–drug pairs)91; in this case, the aim was not to evaluate any specific compound but to characterize the selectivity properties of kinase inhibitors as a drug class.
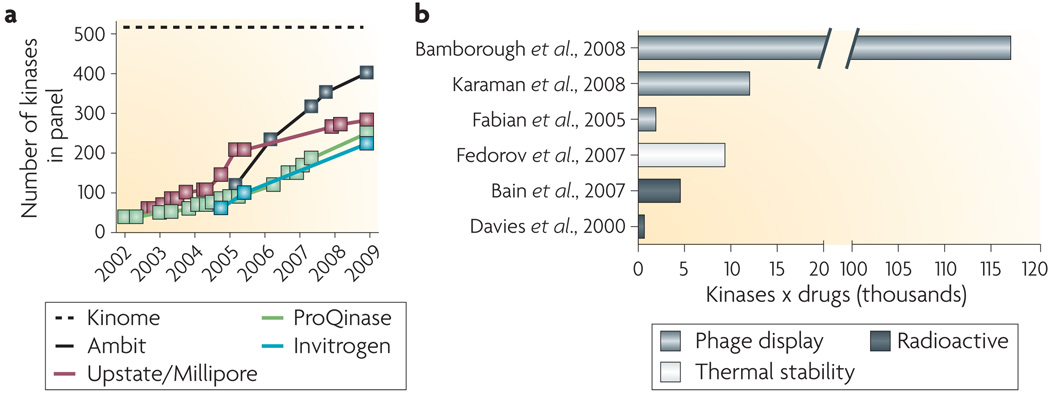
Figure 4. Selectivity profiling of kinase inhibitors.
a | The number of kinases available for screening from commercial vendors by year. the complete human kinome includes approximately 520 protein kinases95 and a smaller number of lipid and small molecule kinases. b | Landmark papers in kinase inhibitor selectivity profiling89–91,93,96,97, plotted against the number of selectivity measurements (kinases × drugs) that were reported. Representatives from three different approaches that measure inhibitor binding are shown.
Extrapolating from these trends, it is plausible that some drug discovery programmes in the near future will profile every kinase inhibitor that is synthesized against most of the kinome. This would occur before any biological testing, as a component of routine compound characterization. The availability of selectivity data on this scale would enable medicinal chemistry to focus on optimizing drug profiles against complex patterns of kinases that gave a desired phenotype, rather than attempting to maximize specificity for a single target. It is likely that drug discovery at some pharmaceutical companies already operates in this way to some degree, although it may not be explicitly acknowledged.
What would be the advantage of this approach? The primary advantage is that it allows for target serendipity — the discovery of target combinations that could not have been predicted, but that are optimal for killing tumour cells — while allowing medicinal chemists to optimize compounds based on biochemical measurements against purified proteins. This has the potential to address the limitations of both target-based drug discovery, which often fails because the target is wrong, and phenotype-based drug discovery, which often fails because the compounds cannot be optimized.
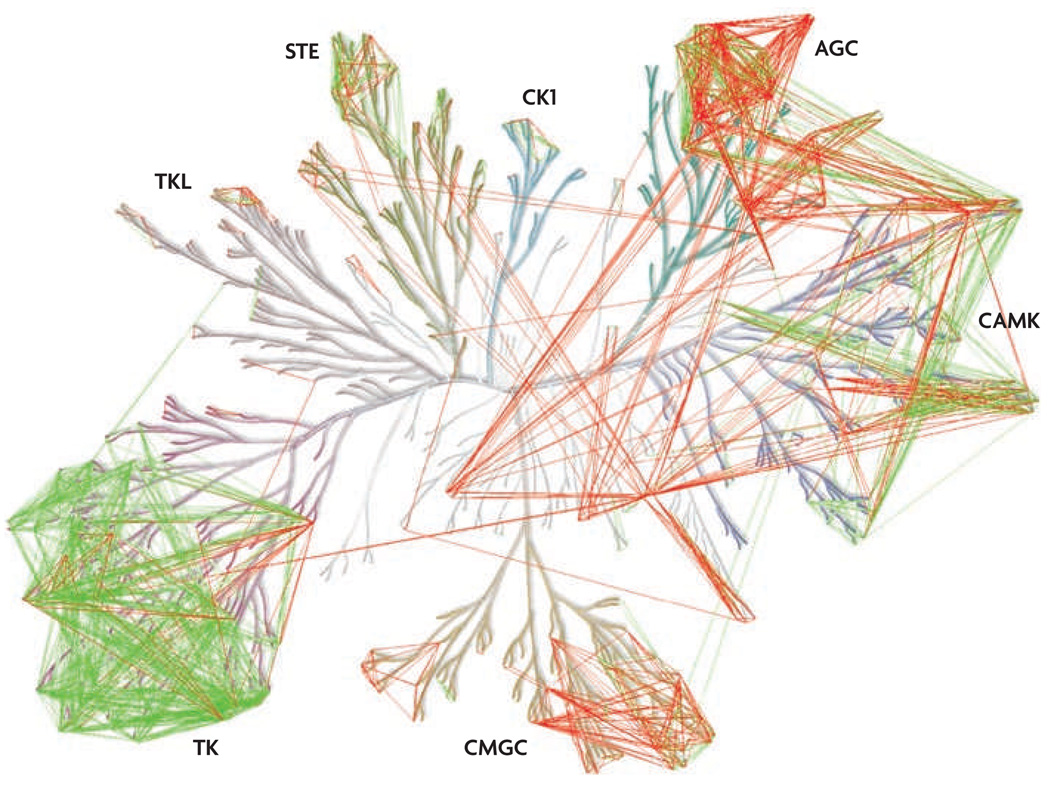
This type of ‘targeted polypharmacology’ would represent a considerable challenge to medicinal chemists, who would be asked to carry out chemical optimization against multi-dimensional target profiles. However, there is already evidence that this is possible for certain target combinations31,92, and kinase drug discovery seems to be the ideal setting to test this model. We analysed a large database of kinase inhibitor selectivity data93 to discover whether certain combinations of kinase targets are enriched among known kinase inhibitors; whether the preference for these target combinations could be rationalized on the basis of sequence analysis; and whether this could be used to estimate the combinatorial druggability of most of the kinome that has not yet been targeted by a small molecule (FIG. 5; see Supplementary information S1 (figure)). We have found, consistent with previous analyses93, that there are clearly clusters of kinases that tend to be inhibited by similar drugs, but that there are also many target combinations that should be accessible but remain undiscovered. We interpret this to mean that there is an important opportunity to discover multi-targeted kinase inhibitors with new biological activities.
Figure 5. Polypharmacology in the protein kinome.
Pairs of kinases were associated based on kinase selectivity data for a broad range of inhibitor scaffolds93 and the sequence conservation between aligned sequences of an expanded ATP binding site. Pairs of kinases that were potently inhibited by a common inhibitor (common dissociation constant (KD)<10 nM) (green lines) were used to determine a sequence similarity cut-off to predict pairs of kinases that can be inhibited by a common inhibitor (red lines). A much higher sequence similarity in the binding pockets was observed for those pairs of kinases with a common potent inhibitor compared with pairs of kinases with a common but less potent inhibitor or no common inhibitor at all. The level of sequence conservation necessary for the predictions was determined based on the sequence conservation distribution of the experimentally determined pairs. The predicted pairs of kinases (red lines) represent potential target combinations that may be more easily accessible with new kinase inhibitors. The image is shown in more detail in supplementary information s1 (figure). AGC, kinases from the protein kinase A, protein kinase G and protein kinase C families; CAMK, calcium/calmodulin-dependent protein kinases; CK1, casein kinase 1; CMGC, kinases from the cyclin-dependent kinase, MAP kinase, glycogen synthase kinase and casein kinase II families; STE, homologues of yeast sterile 7, sterile 11 and sterile 20 kinases; TK, tyrosine kinases; TKL, tyrosine kinase-like kinases.
We have focused on approaches to identify combinations of kinase targets with increased anticancer activity, but understanding the basis for kinase inhibitor-mediated toxicity to normal cells is also valuable, as this information will improve efforts to increase therapeutic index. The broad kinome profiling of clinically approved and investigational kinase inhibitors is likely to help identify such problematic kinase targets. Removing these toxicity-associated kinases from new drug candidates may allow for more complete inhibition of cancer cell targets while avoiding systemic toxicity.
Conclusions
Many different approaches will be necessary to identify the best combinations of targeted therapies for cancer. However, it is important to begin to consider the challenges that may be faced in the near future, when drugs targeting every kinase linked to cancer have been tested in clinical trials, but survival rates for most types of cancer have only marginally improved. It will not be sufficient in this case to simply pursue the next set of oncogenes, because tumour sequencing projects have already shown that such oncogenes do not exist, at least among the genes that are mutated with high frequency2–4. Therefore the burden will be on the cancer research community to think of more creative ways to target important proteins such as kinases that have already been identified.
Supplementary Material
Acknowledgements
This work was partly supported by the US National Institutes of Health grants DK083531 (Z.A.K.) and EB001987 (K.M.S.).
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
National cancer institute Drug Dictionary: http://www.cancer.gov/drugdictionary/
bevacizumab | cetuximab | dasatinib | erlotinib | flavopiridol | imatinib | irinotecan | lapatinib | nilotinib | rapamycin | sorafenib | trastuzumab
UniProtKB: http://www.uniprot.org
BRAF | EGFR | PIK3CA | PLK1 | PTEN | STK33 | TBK1 | VEGFR2
FURTHER INFORMATION
PubMed database: http://www.ncbi.nlm.nih.gov/pubmed
SUPPLEMENTRAY INFORMATION
See online article: S1 (figure)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Zachary A. Knight, The Rockefeller University, New York, New York 10065, USA.
Henry Lin, Howard Hughes Medical Institute and Department of Cellular and Molecular Pharmacology, University of California, San Francisco, California 94158, USA..
Kevan M. Shokat, Howard Hughes Medical Institute and Department of Cellular and Molecular Pharmacology, University of California, San Francisco, California 94158, USA.
References
- 1.Akritopoulou-Zanze I, Hajduk PJ. Kinase-targeted libraries: the design and synthesis of novel, potent, and selective kinase inhibitors. Drug Discov. Today. 2009;14:291–297. doi: 10.1016/j.drudis.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Parsons DW, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 3.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 4.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 6.Heinrich MC, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 7.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 8.Motzer RJ, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 9.Escudier B, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N. Engl. J. Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 10.Yang JC, et al. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N. Engl. J. Med. 2003;349:427–434. doi: 10.1056/NEJMoa021491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N. Engl. J. Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 12.Shepherd FA, et al. Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 13.Sandler A, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N. Engl. J. Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 14.Geyer CE, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N. Engl. J. Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 15.Miller K, et al. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N. Engl. J. Med. 2007;357:2666–2676. doi: 10.1056/NEJMoa072113. [DOI] [PubMed] [Google Scholar]
- 16.Jonker DJ, et al. Cetuximab for the treatment of colorectal cancer. N. Engl. J. Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 17.Moore MJ, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 18.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 19.Hudis CA. Trastuzumab — mechanism of action and use in clinical practice. N. Engl. J. Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 20.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 21.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 23.Tamborini E, et al. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–299. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Cools J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N. Engl. J. Med. 2003;348:1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 25.Chen LL, et al. A missense mutation in KIT kinase domain 1 correlates with imatinib resistance in gastrointestinal stromal tumors. Cancer Res. 2004;64:5913–5919. doi: 10.1158/0008-5472.CAN-04-0085. [DOI] [PubMed] [Google Scholar]
- 26.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 28.Sergina NV, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 30.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 31.Apsel B, et al. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nature Chem. Biol. 2008;4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawyers CL. Cancer: mixing cocktails. Nature. 2007;449:993–996. doi: 10.1038/449993a. [DOI] [PubMed] [Google Scholar]
- 33.Talpaz M, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 34.Kantarjian H, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N. Engl. J. Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 35.Shah NP, et al. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J. Clin. Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Hare T, et al. SGX393 inhibits the CML mutant Bcr-AblT315I and preempts in vitro resistance when combined with nilotinib or dasatinib. Proc. Natl Acad. Sci. USA. 2008;105:5507–5512. doi: 10.1073/pnas.0800587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gontarewicz A, et al. Simultaneous targeting of Aurora kinases and BCR-ABL kinase by the small molecule inhibitor PHA-739358 is effective against imatinib-resistant BCR-ABL mutations including T315I. Blood. 2008;111:4355–4364. doi: 10.1182/blood-2007-09-113175. [DOI] [PubMed] [Google Scholar]
- 38.Carter TA, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl Acad. Sci. USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Hare T, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giles FJ, et al. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–502. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- 41.Weinstein IB, et al. Disorders in cell circuitry associated with multistage carcinogenesis: exploitable targets for cancer prevention and therapy. Clin. Cancer Res. 1997;3:2696–2702. [PubMed] [Google Scholar]
- 42.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc. Natl Acad. Sci. USA. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah NP, et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008;14:485–493. doi: 10.1016/j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Fan QW, et al. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nature Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan QW, et al. A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res. 2007;67:7960–7965. doi: 10.1158/0008-5472.CAN-07-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eichhorn PJ, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Junttila TT, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 50.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nature Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 51.Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004;14:639–647. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-Viciana P, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 53.Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110α is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 54.Wee S, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69:4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 55.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nature Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Normanno N, et al. Cooperative inhibitory effect of ZD1839 (Iressa) in combination with trastuzumab (Herceptin) on human breast cancer cell growth. Ann. Oncol. 2002;13:65–72. doi: 10.1093/annonc/mdf020. [DOI] [PubMed] [Google Scholar]
- 57.Moulder SL, et al. Epidermal growth factor receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in vivo. Cancer Res. 2001;61:8887–8895. [PubMed] [Google Scholar]
- 58.Bukowski RM, et al. Randomized phase II study of erlotinib combined with bevacizumab compared with bevacizumab alone in metastatic renal cell cancer. J. Clin. Oncol. 2007;25:4536–4541. doi: 10.1200/JCO.2007.11.5154. [DOI] [PubMed] [Google Scholar]
- 59.Tonra JR, et al. Synergistic antitumor effects of combined epidermal growth factor receptor and vascular endothelial growth factor receptor-2 targeted therapy. Clin. Cancer Res. 2006;12:2197–2207. doi: 10.1158/1078-0432.CCR-05-1682. [DOI] [PubMed] [Google Scholar]
- 60.Tol J, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N. Engl. J. Med. 2009;360:563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- 61.Arteaga CL, et al. A phase I–II study of combined blockade of the ErbB receptor network with trastuzumab and gefitinib in patients with HER2 (ErbB2)-overexpressing metastatic breast cancer. Clin. Cancer Res. 2008;14:6277–6283. doi: 10.1158/1078-0432.CCR-08-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warburton C, et al. Treatment of HER-2/neu overexpressing breast cancer xenograft models with trastuzumab (Herceptin) and gefitinib (ZD1839): drug combination effects on tumor growth, HER-2/neu and epidermal growth factor receptor expression, and viable hypoxic cell fraction. Clin. Cancer Res. 2004;10:2512–2524. doi: 10.1158/1078-0432.ccr-03-0244. [DOI] [PubMed] [Google Scholar]
- 63.Normanno N, Campiglio M, Perrone F, De Luca A, Menard S. Is the gefitinib plus trastuzumab combination feasible in breast cancer patients? Ann. Oncol. 2005;16:1709. doi: 10.1093/annonc/mdi325. [DOI] [PubMed] [Google Scholar]
- 64.Motwani M, et al. Augmentation of apoptosis and tumor regression by flavopiridol in the presence of CPT-11 in Hct116 colon cancer monolayers and xenografts. Clin. Cancer Res. 2001;7:4209–4219. [PubMed] [Google Scholar]
- 65.Shah MA, et al. A phase I clinical trial of the sequential combination of irinotecan followed by flavopiridol. Clin. Cancer Res. 2005;11:3836–3845. doi: 10.1158/1078-0432.CCR-04-2651. [DOI] [PubMed] [Google Scholar]
- 66.Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nature Biotechnol. 2001;19:940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- 67.Manning BD. Challenges and opportunities in defining the essential cancer kinome. Sci. Signal. 2009;2:15. doi: 10.1126/scisignal.263pe15. [DOI] [PubMed] [Google Scholar]
- 68.Goldman B. For investigational targeted drugs, combination trials pose challenges. J. Natl Cancer Inst. 2003;95:1744–1746. doi: 10.1093/jnci/95.23.1744. [DOI] [PubMed] [Google Scholar]
- 69.Chen H. Optimising strategies for clinical development of combinations of targeted agents. Eur. J. Cancer Suppl. 2007;5:46–52. [Google Scholar]
- 70.Winslow R. AstraZeneca, Merck to Test Cancer Drugs in ‘Cocktail’. The Wall Street Journal. 2009 [online], http://online.wsj.com/article/SB124380640803770139.html. [Google Scholar]
- 71.Barry DW, Distlerath LM. History and accomplishments of the inter-company collaboration for AIDS drug development. Drug Inf. J. 2000;34:741–752. [Google Scholar]
- 72.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 74.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meylan E, et al. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baldwin A, et al. Kinase requirements in human cells: II. Genetic interaction screens identify kinase requirements following HPV16 E7 expression in cancer cells. Proc. Natl Acad. Sci. USA. 2008;105:16478–16483. doi: 10.1073/pnas.0806195105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grueneberg DA, et al. Kinase requirements in human cells: I. Comparing kinase requirements across various cell types. Proc. Natl Acad. Sci. USA. 2008;105:16472–16477. doi: 10.1073/pnas.0808019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morgan-Lappe S, et al. RNAi-based screening of the human kinome identifies Akt-cooperating kinases: a new approach to designing efficacious multitargeted kinase inhibitors. Oncogene. 2006;25:1340–1348. doi: 10.1038/sj.onc.1209169. [DOI] [PubMed] [Google Scholar]
- 79.Luo Y, et al. Potent and selective inhibitors of Akt kinases slow the progress of tumors in vivo. Mol. Cancer Ther. 2005;4:977–986. doi: 10.1158/1535-7163.MCT-05-0005. [DOI] [PubMed] [Google Scholar]
- 80.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 81.Lyons JF, Wilhelm S, Hibner B, Bollag G. Discovery of a novel Raf kinase inhibitor. Endocr. Relat. Cancer. 2001;8:219–225. doi: 10.1677/erc.0.0080219. [DOI] [PubMed] [Google Scholar]
- 82.Hauschild A, et al. Results of a phase III, randomized, placebo-controlled study of sorafenib in combination with carboplatin and paclitaxel as second-line treatment in patients with unresectable stage III or stage IV melanoma. J. Clin. Oncol. 2009;27:2823–2830. doi: 10.1200/JCO.2007.15.7636. [DOI] [PubMed] [Google Scholar]
- 83.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 84.Strumberg D, et al. Results of phase I pharmacokinetic and pharmacodynamic studies of the Raf kinase inhibitor BAY 43-9006 in patients with solid tumors. Int. J. Clin. Pharmacol. Ther. 2002;40:580–581. doi: 10.5414/cpp40580. [DOI] [PubMed] [Google Scholar]
- 85.Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J. Clin. Invest. 2003;111:1287–1295. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gleich GJ, Leiferman KM, Pardanani A, Tefferi A, Butterfield JH. Treatment of hypereosinophilic syndrome with imatinib mesilate. Lancet. 2002;359:1577–1578. doi: 10.1016/S0140-6736(02)08505-7. [DOI] [PubMed] [Google Scholar]
- 87.Burdine L, Kodadek T. Target identification in chemical genetics: the (often) missing link. Chem. Biol. 2004;11:593–597. doi: 10.1016/j.chembiol.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 88.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 89.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bain J, et al. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bamborough P, Drewry D, Harper G, Smith GK, Schneider K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J. Med. Chem. 2008;51:7898–7914. doi: 10.1021/jm8011036. [DOI] [PubMed] [Google Scholar]
- 92.Hopkins AL, Mason JS, Overington JP. Can we rationally design promiscuous drugs? Curr. Opin. Struct. Biol. 2006;16:127–136. doi: 10.1016/j.sbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 93.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nature Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 94.Samuels Y, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 95.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 96.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nature Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 97.Fedorov O, et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl Acad. Sci. USA. 2007;104:20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.