Abstract
Although dimorphic sexes have evolved repeatedly in multicellular eukaryotes, their origins are unknown. The mating locus (MT) of the sexually dimorphic multicellular green alga, Volvox carteri, specifies the production of eggs and sperm and has undergone a remarkable expansion and divergence relative to MT from Chlamydomonas reinhardtii, a closely related unicellular species that has equal-sized gametes. Transcriptome analysis revealed a rewired gametic expression program for Volvox MT genes relative to Chlamydomonas, and identified multiple gender-specific and sex-regulated transcripts. The retinoblastoma tumor suppressor homolog MAT3 is a Volvox MT gene that displays sexually regulated alternative splicing and evidence of gender-specific selection, both indicative of cooption into the sexual cycle. Thus, sex-determining loci impact the evolution of both sex-related and non-sex-related genes.
Sexually dimorphic gametes have evolved in every major group of eukaryotes, and are thought to be selected when parents can differentially allocate resources to progeny (1). However, the origins of oogamy (large eggs and small sperm) and the contribution of sex determining loci to such evolution are largely unknown (2, 3)(See Glossary of Terms (4) for further explanation of terminology).
The Volvocine algae are a group of chlorophytes comprising unicellular species such as Chlamydomonas reinhardtii (hereafter Chlamydomonas) and a range of multicellular species of varying complexity such as Volvox carteri (hereafter Volvox). Volvox has a vegetative reproductive form containing 16 large germ cells (gonidia) and ~2000 terminally differentiated somatic cells (4,5)(Fig. S1).
Chlamydomonas and other Volvocine algae also undergo a sexual cycle where a large, haploid mating locus (MT) controls sexual differentiation, mating compatibility, and zygote development (6). MT in Chlamydomonas is a 200–300 kb multigenic chromosomal region (Fig. 1A) within which gene order is rearranged between the two sexes (MT+ and MT−) and meiotic recombination is suppressed, thus leading to its inheritance as a single Mendelian trait. Within each MT allele are gender-limited genes (allele present in only one of the two sexes) required for the sexual cycle as well as shared genes (alleles present in both sexes), most of which have no known function in sex or mating (7). The rearrangements that suppress recombination serve to maintain linkage of gender-limited genes, but they also reduce genetic exchange between shared genes leading to their meiotic isolation. Thus, Chlamydomonas MT bears similarity to sex chromosomes and to expanded mating type regions of some fungi and bryophytes (8–10)
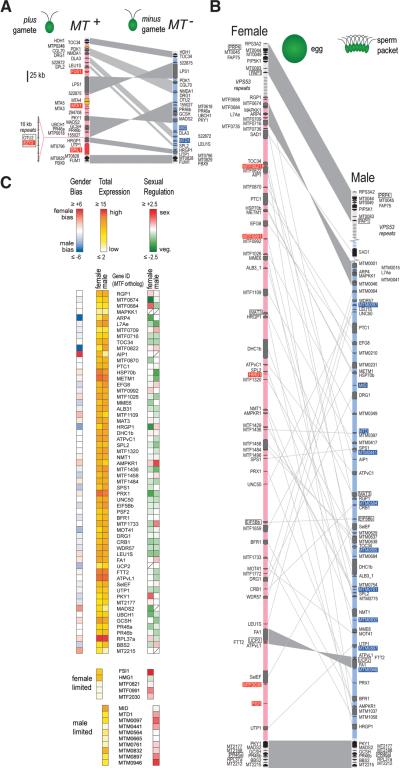
Figure 1.
Expansion of Volvox MT and sex-regulated gene expression. (A) Schematic of Chlamydomonas mating locus with rearranged domains in light blue or pink. MT+ limited genes are shaded red if unique or orange if they have an autosomal copy. MT− limited genes are shaded blue. Flanking and shared genes are shaded black and gray respectively. Synteny is indicated by gray shading. (B) Schematic of Volvox MT scaled as in (A). Boxed genes were used for mapping. The broken segment represents a transposon repeat region containing copies of VPS53. (C) Expression heat maps of Volvox MT genes. Left panel, female/male expression ratio; middle panel, total expression; right panel, sexual induction (Sex) or repression (Veg). Diagonal hatch, insufficient data.
While Chlamydomonas is isogamous (producing equal-sized gametes), Volvox and several other Volvocine genera have evolved oogamy that is under the control of female and male MT loci (11)(Fig. S1). Moreover, the Volvox sexual cycle is characterized by a suite of other traits not found in Chlamydomonas—such as a diffusible sex-inducer protein rather than nitrogen deprivation (−N) as a trigger for gametogenesis (Table S1). A detailed characterization of MT in Volvox would be expected to shed light on the transition from isogamy to oogamy and on other properties of the sexual cycle that evolved in this multicellular species (Table S1).
The MT+ allele of Chlamydomonas was previously sequenced and resides on chromosome 6 (Figs. 1A, S2)(12). To enable a comparison of mating loci evolution between two related species with markedly different sexual cycles, we sequenced Chlamydomonas MT− and both alleles of Volvox MT (Fig. 1, Table S2)(4). Volvox MT was previously assigned to Linkage Group I (LG I)(5) but the locus had not been further characterized. We mapped Volvox MT to the genome sequence and assembled most of LG I (Table S3)(4). Extensive synteny with Chlamydomonas chromosome 6 indicates that MT has remained on the same chromosome in both lineages for ~200 million years since their estimated divergence, despite numerous intra-chromosomal rearrangements between the two (Fig. S2)(13).
While the haploid Volvox genome is ~17% larger than that of Chlamydomonas (138 Mb versus 118 Mb) and the two have very similar predicted proteomes (12, 14), Volvox MT is ~500% larger than Chlamydomonas MT and contains over 70 protein coding genes in each allele (Fig. 1B, Tables S4,S5). Compared to autosomes Volvox MT is unusually repeat-rich (>3X the genomic average), has lower gene density, and has genes with more intronic sequence (Table S6)—all properties that suggest an unusual evolutionary history and distinguish it from Chlamydomonas MT.
Only two gender-limited genes from Chlamydomonas MT− have recognizable homologs in Volvox—MID and MTD1—that are both in male MT (Figs.1,S3,S4). MID is a conserved RWP-RK family transcription factor whose expression in other Volvocine algae is induced by −N (15–17) as is also the case for MTD1 (18, 19). Surprisingly, both MTD1 and MID are expressed constitutively in Volvox (Fig. S5) indicating that their transcription is uncoupled from sexual differentiation. This result suggests that additional MT genes might play a role in gametogenesis.
We used differential deep transcriptome sequencing (Tables S7,S8)(4) to identify MT genes in Volvox, a method that helped to mitigate problems associated with automated gene prediction in atypical genomic regions such as MT. We identified transcripts for five new female-limited and eight new male-limited genes that do not have detectable homologs in Chlamydomonas, and found that most of these gender-limited genes are sex-regulated (expression induced or repressed during sexual differentiation)(Fig. 1C,Table S9)(4). HMG1 encodes a female-limited HMG domain protein (Figs. S6,S7,Table S10) that belongs to a family of DNA binding proteins whose members regulate mammalian and fungal sex determination (20, 21). However, HMG proteins had not been previously implicated in the sexual cycles of green algae or plants. A second novel female-limited gene, FSI1, is strongly induced during gametogenesis and encodes a small predicted transmembrane protein with no identifiable homologs (Figs. 1C,S7).
Besides identification of new gender-limited genes, our transcriptome data provided empirical support for 51 of 52 single-copy shared genes in Volvox MT that previously had limited EST support for the female allele (33 of 52), and no EST support for the male allele. Moreover, some of these shared genes showed patterns of expression suggesting cooption into the Volvox sexual cycle. These patterns include gender-biased expression (male:female expression ratio ≠ 1) and sex-regulated expression (Figs. 1C,S7)(4). This set of genes encodes putative signaling, extracellular matrix, and chromatin-associated proteins with known or potential roles in gametogenesis and fertilization, and are candidates for further investigation (Fig. S7).
In diploid species heterogametic sex chromosomes evolve rapidly (22) and lose genes that are not related to sex (23). Genes within large haploid mating loci are predicted to accumulate mutations more rapidly than genes in autosomal regions due to suppressed recombination, but they are continuously exposed to selection (24). Suppressed recombination also appears to have played a role in diversification of mating-locus linked genes in haploid fungi and bryophytes (8–10). Our data allowed us to compare the evolutionary history of Volvox MT genes from this oogamous species to each other, and to genes from MT of its isogamous relative Chlamydomonas.
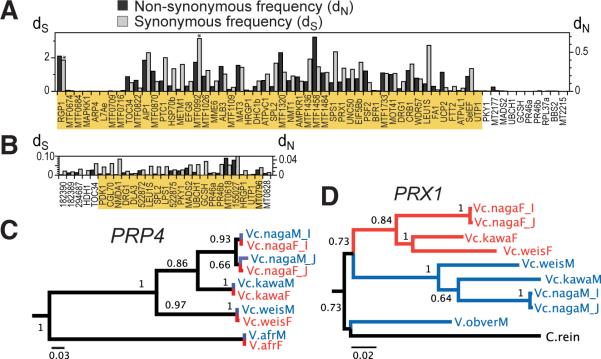
Divergence was measured from synonymous (dS) and non-synonymous (dN) substitutions (Figs. 2A,2B), and from total nucleotide distances for shared genes (Tables S11,S12)(4). Unexpectedly, divergence for Volvox MT allelic pairs is up to two orders of magnitude larger than for allelic pairs in Chlamydomonas MT, suggesting that Volvox MT alleles may have been subject to more intense and/or more prolonged recombinational suppression than Chlamydomonas MT alleles have been. In contrast, two internal syntenic blocks within Volvox MT are relatively similar (Figs. 1B,2A) suggesting that they were acquired more recently in an ongoing stratification process as first described for the human X chromosome(25). Volvox MT genes also showed reduced codon usage bias relative to autosomal genes (Fig. S8), most likely due to suppressed recombination(26).
Figure 2.
Divergence of MT genes. (A,B) dN and dS for shared Volvox (A) or Chlamydomonas (B) genes within MT (orange shading) or flanking MT. Asterisks mark saturated dS values. (C,D) Maximum likelihood phylogenies for PRP4 (C) and MT gene PRX1 (D). Red/blue signify female/male strains and clades.
Three MT genes and a flanking gene, PRP4, were sequenced from a set of related Volvox species to determine the extent of MT gene isolation (4). Phylogenies revealed the expected pattern for PRP4 that grouped by species and geographical location (Fig. 2C). In contrast, the MT genes grouped by gender (Figs. 2D,S9). These data demonstrate that the shared genes in Volvox MT have essentially become gender specific and have remained genetically isolated during speciation. Thus, the MT locus in Volvox has become a repository of genetic diversity that is linked to the sexual cycle.
In Chlamydomonas the retinoblastoma (RB) tumor suppressor pathway controls cell division in response to cell size (27), and the RB homolog encoded by MAT3 is adjacent to MT (28). Volvox MAT3, on the other hand, is within MT (Fig. 1B) and we investigated its evolution and expression as a candidate regulator of sexually dimorphic cell divisions (Fig. S1). The Volvox male and female MAT3 proteins are exceptionally diverged from each other (Figs. S9,S10,S11). Moreover male and female Volvox MAT3 have different structures: the female allele contains an intron that is absent from males while the male allele contains an unusually large fourth intron compared to females (Fig. 3). Although MAT3 shows signs of having undergone purifying selection (dN/dS=0.23, Table S11), several short sequences in the male and female proteins are asymmetric in their conservation pattern, suggesting that the two alleles are under different selective constraints (Figs. S10,S11). We also found dozens of alternatively processed MAT3 mRNAs from both Volvox sexes, representing most types of alternative splicing (29)(Figs. 3,S12). In addition, sex-regulated pre-mRNA splicing of MAT3 was found for both genders, and might be controlled by the MT encoded splicing factor, SPL2, whose expression level is sex-regulated in males (Fig.1C, S13). Intriguingly, the predominant MAT3 isoform in sexual males retains the first two introns, leading to inclusion of an early termination codon (Figs. 3,S12). mat3 mutants in Chlamydomonas produce tiny gametes (28), and down-regulation of MAT3 in Volvox males through alternative splicing may be linked to the production of small-celled sperm.
Figure 3.
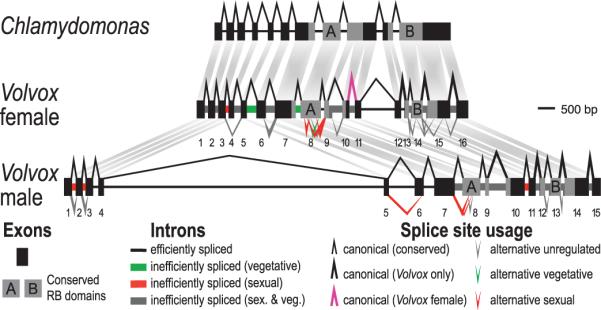
Gender-specific divergence and splicing of MAT3. Schematic of MAT3 from Chlamydomonas (top), Volvox female (middle) and Volvox male (bottom). Volvox exons are numbered.
The accelerated divergence of sex chromosomes is usually associated with gene loss and degeneration (23), though adaptive evolution of sex chromosomes is an emerging theme (30). Our data suggest that expansion, loss of recombination, and rapid divergence can be mutually reinforcing properties of sex determining regions that facilitate cooption into the sexual cycle and provide novel sources of developmental innovation (Fig. S14).
Summary.
The Volvox mating locus illuminates the evolutionary origin of dimorphic sexes
Supplementary Material
Acknowledgments
We thank David Kirk for DNA from Volvox mapping populations. We thank Vicki Lundblad and Sabeeha Merchant for advice on the manuscript. This work was supported by the Coypu Foundation and from grants NIH R01 GM078376 to JGU, NIH F32 GM086037 to BO, JSPS Grant S05750/L06701 to PF, Grant-in-Aid for Scientific Research (No. 20247032) from the Ministry of Education, Culture, Sports, Science and Technology, Japan to HN, NIH grant T32-HG002536 to SD, and DE-FC02-02ER63421 and AFOSR to MP. The U.S. Department of Energy Joint Genome Institute provided sequencing and analyses for algal mating loci under the Community Sequencing Program (No. 776835) supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Sequencing of the Volvox carteri genome was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. Sequences generated in this study have been deposited in Genbank under accession numbers GU814014, GU814015, GU784915, GU784916, GU735444-GU735478. Materials used in this study will be made available on request with the completion of a Materials Transfer Agreement from Salk Institute.
REFERENCES AND NOTES
- 1.Parker GA, Baker RR, Smith VG. J. Theor. Biol. 1972;36:529. doi: 10.1016/0022-5193(72)90007-0. [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth B. J. Theor. Biol. 1978;73:347. doi: 10.1016/0022-5193(78)90195-9. [DOI] [PubMed] [Google Scholar]
- 3.Williams TM, Carroll SB. Nat Rev Genet. 2009;10:797. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- 4.Information on Materials and Methods is available on Science Online.
- 5.Kirk DL. Volvox. In: Bard JBL, Barlow PW, Green PB, Kirk DL, editors. Developmental and Cell Biology Series. Cambridge University Press; Cambridge, U.K.: 1998. [Google Scholar]
- 6.Goodenough U, Lin H, Lee J-H. Semin. Cell Dev. Biol. 2007;18:350. doi: 10.1016/j.semcdb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Ferris PJ, Armbrust EV, Goodenough UW. Genetics. 2002;160:181. doi: 10.1093/genetics/160.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser JA, et al. Plos Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menkis A, Jacobson DJ, Gustafsson T, Johannesson H. PLoS Genet. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamato KT, et al. Proc Natl Acad Sci USA. 2007;104:6472. [Google Scholar]
- 11.Nozaki H. J Plant Res. 1996;109:353. [Google Scholar]
- 12.Merchant SS, et al. Science. 2007;318:245. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herron MD, Hackett JD, Aylward FO, Michod RE. Proc. Natl. Acad. Sci. USA. 2009;106:3254. doi: 10.1073/pnas.0811205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. http://genomeportal.jgi-psf.org/Volca1/Volca1.home.html.
- 15.Ferris PJ, Goodenough UW. Genetics. 1997;146:859. doi: 10.1093/genetics/146.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamaji T, et al. Genetics. 2008;178:283. doi: 10.1534/genetics.107.078618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nozaki H, Mori T, Misumi O, Matsunaga S, Kuroiwa T. Curr Biol. 2006;16:R1018. doi: 10.1016/j.cub.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Hamaji T, Ferris PJ, Nishii I, Nozaki H. J Phycol. 2009:1. doi: 10.1111/j.1529-8817.2009.00744.x. [DOI] [PubMed] [Google Scholar]
- 19.Lin H, Goodenough UW. Genetics. 2007;176:913. doi: 10.1534/genetics.106.066167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Idnurm A, Walton FJ, Floyd A, Heitman J. Nature. 2008;451:193. doi: 10.1038/nature06453. [DOI] [PubMed] [Google Scholar]
- 21.Waters PD, Wallis MC, Marshall Graves JA. Semin. Cell Dev. Biol. 2007;18:389. doi: 10.1016/j.semcdb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Hughes JF, et al. Nature. 2010;463:536. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlesworth B, Charlesworth D. Philos Trans R Soc Lond, B, Biol Sci. 2000;355:1563. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bull J. The American Naturalist. 1978;112:245. [Google Scholar]
- 25.Lahn BT, Page DC. Science. 1999;286:964. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 26.Kliman RM, Hey J. Mol. Biol. Evol. 1993;10:1239. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- 27.Fang S-C, de los Reyes C, Umen JG. PLoS Genet. 2006;2:e167. doi: 10.1371/journal.pgen.0020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umen JG, Goodenough UW. Genes Dev. 2001;15:1652. doi: 10.1101/gad.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matlin A, Clark F, Smith C. Nat Rev Mol Cell Biol. 2005;6:386. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 30.Bachtrog D. Curr. Opin. Genet. Dev. 2006;16:578. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.