Abstract
An H-2k MHC locus is critical for murine cytomegalovirus (MCMV) resistance in MA/My mice and virus control is abolished if H-2k is replaced with H-2b MHC genes from MCMV-susceptible C57L mice. Yet, H-2k resistance varies with genetic background; thus, modifiers of virus resistance must exist. To identify non-MHC resistance loci, spleen and liver MCMV levels and genome-wide genotypes were assessed in (C57L × MA/My) and (MA/My × C57L) F2 offspring (representing 550 meioses). Significantly, a non-Mendelian frequency of MHC genotypes was observed for offspring of the latter cross. Quantitative trait loci (QTL) and their interaction potential in MCMV resistance were assessed in R/qtl; QTL on chromosomes 17, 6, and 19 affected MCMV levels in infected animals. A chromosome 6 QTL was linked with the NK gene complex and acted in an additive fashion with an H-2k MHC QTL to mitigate spleen MCMV levels. We provide biological confirmation that this chromosome 6 QTL provided MCMV control independent of H-2k via NK cells. Importantly, both chromosome 6 and 19 QTLs contribute to virus control independent of H-2k. Altogether, MHC and non-MHC MCMV-resistance QTL contribute in early resistance to MCMV infection in this genetic system.
Keywords: Mouse, QTL, MHC, Cytomegalovirus
Introduction
Greater than half of the world population is likely infected with human cytomegalovirus. While asymptomatic in healthy adults, it can cause birth defects in infants as well as severe morbidity and mortality in immunocompromised persons (Landolfo et al. 2003), particularly in individuals with natural killer (NK) cell deficiencies (Orange 2002). MCMV has been used as an experimental model for the human pathogen and for the purpose of identifying host defense genes, especially those that are vital in the innate immune system (Beutler et al. 2005; Scalzo et al. 2007).
Cmv1r, a major MCMV-resistance locus in C57BL/6 mice, was initially genetically mapped to the NK gene complex (NKC) on distal chromosome 6 (Scalzo et al. 1992). The Ly49h gene has since been shown to confer Cmv1 resistance (Brown et al. 2001a; Daniels et al. 2001; Lee et al. 2001a). The Ly49H activation receptor expressed at the surface of NK cells can bind MCMV m157, an MHC class-I-related protein displayed by infected cells, to target virus specific NK cell-mediated immunity (Arase et al. 2002; Smith et al. 2002). Yet, Ly49H has no human ortholog and dominant virus resistance afforded by it is rare in wild mice (Scalzo et al. 2005). In earlier studies, Chalmer et al. found that MHC and non-MHC loci affected host survival after MCMV infection (Chalmer et al. 1977); however, substantial H-2k protection from lethal infection was never attributed to T cells. A possible role for NK cells was instead suggested (Grundy et al. 1981), and in accord with that hypothesis, Scalzo and Shellam showed that genetic factors also affect viral replication in infected spleen tissue and that NK cells are essential to MCMV resistance in MA/My (H-2k) mice (Scalzo et al. 1990, 1995). Nonetheless, major virus resistance loci were not previously mapped for the MA/My strain.
Using a genetic approach to analyze MCMV resistance and -susceptibility traits in (MA/My × BALB/c) F2 offspring, Vidal and coworkers observed that NKC and MHC loci could interact in an epistatic way in resistance to MCMV infection (Desrosiers et al. 2005). In separate work, we analyzed similar trait differences in backcross and F2 offspring of MA/My and MCMV-susceptible C57L mice (Dighe et al. 2005). It was reasoned that non-NKC, non-MHC genes could be uncovered because MA/My and C57L NKC-Ly49 haplotypes were previously shown to be highly related (Brown et al. 2001b). H-2k-linked resistance to infection observed in our initial analysis has since been validated using reciprocal H-2 congenic strains of mice and more precisely mapped to the MHC class I D locus (Dighe et al. 2005; Xie et al. 2007, 2009). Nonetheless, still-unknown loci must also contribute some resistance effects since the observed genetic variance in MCMV replication in F2 offspring was not fully accounted for by an MHC locus (Dighe et al. 2005). In agreement with that possibility, H-2k resistance varies in different genetic backgrounds and NK cells still became activated to produce IFN-γ in MCMV infected mice with H-2b MHC loci (Xie et al. 2007). Genetic and NK cell functional data therefore implicate additional non-MHC loci which could have a significant impact on virus immunity.
In the current work, we analyzed MCMV control traits in (C57L × MA/My)F2 and (MA/My × C57L)F2 offspring so that non-MHC linked effects, including a potential chromosome X-linked Cmv2 locus (Rodriguez et al. 2004, 2009) could be precisely mapped. Beyond clear-cut MHC genetic control, two MCMV-resistance loci were mapped on chromosomes 6 and 19. Our data provide biological confirmation for NKC genetic control in a system not previously known for its polymorphism. The data further support that the NKC can contribute additively to enhance MCMV resistance in the context of H-2k as well as provide H-2k-independent virus control in this system. Because recent genetic studies have implicated a potential role for NK cell receptors and particular MHC class I proteins with protection in human virus infections (Alter et al. 2007; Heeney et al. 2006; Khakoo et al. 2004; Martin et al. 2002, 2007), this genetic system may prove to be a highly relevant model to examine NK-mediated virus immunity under MHC regulation.
Materials and methods
Mouse care and handling
BALB/c, MA/My, and C57L breeder pairs were purchased from The Jackson Laboratory and housed in the MR-5 specific-pathogen-free vivarium at the University of Virginia, which is fully accredited by the American Association for Accreditation of Laboratory Animal Care. C57L.M-NKCmamy congenic mice were generated and have been maintained at UVA (Xie et al. 2007, 2009). F1 and F2 mice were bred and housed in the same vivarium. The direction of a given cross has been specified by the cross nomenclature in the text. C57L ♀ × MA/My ♂ outcross issued (C57L × MA/My)F1 offspring (LMF1). Likewise, a MA/My ♀ × C57L ♂ outcross issued (MA/My × C57L)F1 offspring (MLF1). All animal studies were approved by and conducted in accordance with Animal Care and Use Committee oversight.
Virus infection and assays
Salivary gland stock virus (SGV) was prepared after serial passage in BALB/c mice as described previously (Rodriguez et al. 2004). Average virus titer was determined in three to five independent titering experiments on NIH3T3 monolayers. Experimental mice (8–12 weeks of age) were infected intraperitoneally with 1×105 PFU MCMV, a dose which we have determined to be the LD50 for our virus stock in BALB/c mice (data not shown). To study the role of NK cells during MCMV infection, mice were i.p. injected with 200μg of mAb PK136 and depletion confirmed via flow cytometric analysis as described (Dighe et al. 2005). Mice were euthanized 3.5 days post-infection (84–90 h), and spleen and liver genomic DNA samples were purified using a kit (Gentra Systems, Minneapolis, MN). Virus levels were quantified using quantitative real-time PCR (QPCR) as previously described (Dighe et al. 2005; Wheat et al. 2003). All sample measurements were performed in triplicate. MCMV levels (average of triplicate measurements) were reported as log10 (number of MCMV genome copies per number of β-actin genomic copies). Minitab (v15; Minitab Inc., State College, PA) was used to generate box and whisker plots of virus levels.
Genotyping and quantitative trait mapping
A panel of 64 published (Rodriguez et al. 2004) and previously unpublished (Table 1) fluorescent-labeled simple sequence length polymorphism (SSLP) markers that distinguish C57L and MA/My alleles was used to determine genome-wide genotypes for genomic DNA samples described above. SSLP amplification products were analyzed using the 3130xl Genetic Analyzer using Data Collection (v3.0) and GeneMapper Software (v4.0; Applied Biosystems, Foster City, CA).
Table 1.
Microsatellite markers used for genotyping F2 offspring
| Locus | cMa | Mbpa |
|---|---|---|
| D1Mit21 | 32.8 | 66.9 |
| D2Mit56.1 | 41.0 | 70.5 |
| D2Mit338.1 | 73.9 | 130.6 |
| D2Uva155 | 155.2 | |
| D3Mit21 | 19.2 | 37.0 |
| D5Mit314 | 59.0 | 110.1 |
| D6Mit328 | 49.3 | 112.7 |
| D7Mit17.1 | 11.0 | 30.7 |
| D7Mit352.1 | 47.0 | 97.7 |
| SCOR8.15.1 | 15.1 | |
| SCOR8.65.171 | 65.2 | |
| SCOR8.85.14 | 85.1 | |
| SCOR8.95.15 | 95.1 | |
| SCOR8.124.21 | 124.2 | |
| D9Mit355 | 53.0 | 98.7 |
| SCOR10.49.00 | 49.0 | |
| D11Mit231 | 17.0 | 35.5 |
| D13Mit275 | 16.0 | 37.4 |
| SCOR14.39.2 | 39.2 | |
| SCOR14.54.1 | 54.1 | |
| SCOR14.100.2 | 100.2 | |
| D15Mit262 | 51.1 | 87.1 |
| D16Mit139 | 43.1 | 65.6 |
| D18Mit67 | 4.0 | 12.1 |
| D18Mit51 | 37.0 | 61.2 |
Mouse Genome Informatics (MGI) and Sequence Map locations based on build 37.1
R/qtl (version 1.07–12) was used to perform single-QTL genome scans by Haley–Knott regression using a step size of 1.0 cM (Broman et al. 2003). Virus levels were log10 transformed for the R/qtl analysis and regression was based on 64 markers spread throughout the genome. Since our genotype data was nearly complete (99.89%), Haley–Knott provides a very close approximation to expectation–maximization likelihood (Haley and Knott 1992). LOD scores were calculated by dividing the likelihood ratio statistic by 4.61 (twice the natural logarithm of 10). Significance thresholds were calculated via 10,000 permutation replicates using Haley–Knott regression at 1.0 cM step size (Churchill and Doerge 1994). Because of the presence of a large effect QTL, the scan was repeated using D17Mit16 as a covariate in a single-QTL scan (conditioning). Conditional LOD scores were obtained as log 10 likelihood ratios, with the specified locus (D17Mit16) included in both the null and alternative hypotheses (Broman and Speed 2003). We searched for loci that interact with the large effect locus by including D17Mit16 as an interactive covariate. Search of the model space was performed using forward selection followed by backward elimination, and the best model was chosen according to a penalized LOD score extension of the permutation-based model selection criterion proposed in Broman and Speed (2003). Models were fitted using Haley–Knott regression. To compare strength of evidence for epistatic and additive models on chromosomes 6 and 17, a test of epistasis was performed in which the epistasis LOD score was calculated as the difference of the best full two-QTL interaction model and the best two-QTL additive model on chromosomes 6 and 17, and compared to the empirical permutation distribution.
In total, the spleens of 275 animals were studied including a new cohort of 144 F2 offspring (includes LMF2 and MLF2 animals) and a cohort of 131 (C57L × MA/My) F2 offspring genotyped before (Dighe et al. 2005). The liver analysis includes 144 F2 offspring with 86 LMF2 mice (Dighe et al. 2005) for a total of 230 mice. As results were combined among studies, raw data was pooled and reanalyzed in toto. Genotyping was complete: from the new cohort of 144 F2 offspring, six animals were missing a single marker genotype and from the previously studied cohort of 131 F2 offspring, ten animals lacked a single marker genotype and one mouse with three missing.
Results
MHC and non-MHC genetic factors affect MCMV resistance in LMF2 and MLF2 offspring
MA/My and C57L mice (multiple breeder pens) were outcrossed in both directions. The offspring generated for analysis include 73 (C57L × MA/My)F2 (hereafter referred to as LMF2) and 71 (MA/My × C57L)F2 (hereafter referred to as MLF2). All LMF2 and MLF2 offspring were typed for several H-2 loci. As expected, a normal Mendelian frequency of MHC genotypes was observed in the LMF2 offspring (Table 2). Curiously, a 1:1:1 MHC genotype distribution was sometimes observed for MLF2 offspring such that H-2 heterozygotes were underrepresented. Comparison of the observed MLF2 distribution with an expected Mendelian genotype distribution of 1:2:1 revealed a statistically significant difference (Table 2). Although this occurrence was somewhat irregular, when H-2 haplotype skewing among MLF2 offspring was evident, it was seen in litters from different breeder pens produced in a given round of breeding and in multiple rounds of breeding.
Table 2.
MHC genotypes for F2 offspring through three rounds of breeding
| Crossa | H-2 genotypeb |
Number F2 offspring produced | ||||
|---|---|---|---|---|---|---|
| Round 1c | Round 2 | Round 3 | Totals | |||
| MA/My × | kk | 12 | 8 | 5 | 25 | p <0.05d |
| C57L | kb | 10 | 9 | 6 | 25 | |
| bb | 8 | 9 | 4 | 21 | ||
| C57L × | kk | 7 | N/A | 11 | 18 | NS |
| MA My | kb | 13 | 28 | 41 | ||
| bb | 3 | 11 | 14 | |||
The initial outcross to produce F1 offspring
H-2 genotype is based on typing with D17Mit16, D17Uva06, and D17Mit10 markers
A round is group of animals (from three separate breeders) with a similar date of birth
Chi-square test was used to determine the statistical significance by comparing the observed and expected (Mendelian 1:2:1) genotype distribution
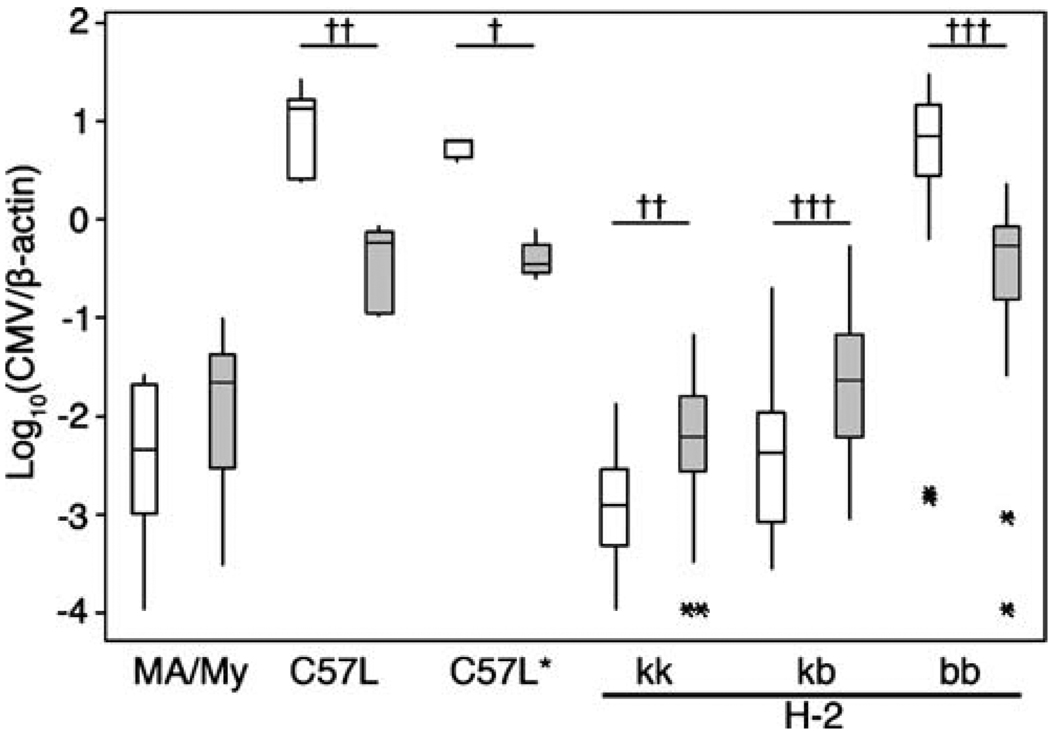
LMF2 and MLF2 offspring were infected with MCMV and quantitative real-time PCR (QPCR) was used to measure spleen and liver virus levels. Consistent with previous data (Dighe et al. 2005), spleen MCMV levels generally corresponded with H-2 genotypes in the F2 offspring and further highlighted the importance of a strong QTL on chromosome 17 (Fig. 1; Table 3). Also significant, H-2k homozygosity corresponded with additive protection by comparison with H-2 heterozygotes (spleen, p≤0.001; liver, p≤0.00005). Thus, an H-2k MHC locus provided substantial genetic protection in LMF2 and MLF2 offspring infected with MCMV.
Fig. 1.
The role of MHC and non-MHC genes in MCMV resistance in F2 offspring of C57L and MA My mice. F2 mice were generated in both LMF2 and MLF2 outcross directions. Shown are box and whisker plots for spleen (white) and liver (gray) virus levels determined by QPCR 3.5 d.p.i. of MA/My (n=12), C57L (n=9), *C57L (n=5) and F2 (n=144) mice infected with 1 × 105 PFU SGV. Plotted asterisks for virus levels in several F2 mice designate outliers for a given genotype class. *C57L denotes a littermate control mouse (H-2b haplotype). H-2 genotypes for F2 mice are indicated at the bottom of the graph. Significance as determined by Wilcoxon test are indicated (†††p≤0.00001, ††p≤0.00005, †p≤0.01)
Table 3.
Haley–Knott regression analysis of F2 intercross cohort, spleen (n=275)
| Chr | Position (cM)a | Nearest marker | LOD | % Variance | pb |
|---|---|---|---|---|---|
| 6 | 114 | D6Wum25 | 2.8 | 4.6 | 0.11 |
| 17 | 1 | D17Mit16 | 33.1 | 42.5 | <0.001 |
| 19 | 6 | D19Mit68 | 2.57 | 4.2 | 0.18 |
Putative chromosome QTL locations determined by R/qtl are listed
p values are calculated based on the genome-wide scan and are estimated from 10,000 permutation replicates
Importantly, there were indications of H-2-independent control. First, a non-normal distribution of virus levels (p< 0.005, Anderson–Darling Test) was observed in the H-2k/b offspring. By examining the distribution of the number of animals versus virus level, we have observed a bimodal distribution (data not shown), indicating that additional MCMV-resistance loci beyond the MHC afforded some virus protection. Secondly, two of 35 (5.7%) H-2b homozygous F2 offspring had very low spleen virus levels, which we have observed previously (Dighe et al. 2005). In fact, genotype skewing (discussed below) might have reduced the frequency of H-2b F2 animals with low MCMV. A sex or cross-direction specific difference in virus control was not observed (data not shown). Thus, MHC and non-MHC loci likely affected MCMV resistance in the LMF2 and MLF2 offspring.
Non-MHC-linked MCMV resistance genetically mapped to chromosomes 6 and 19
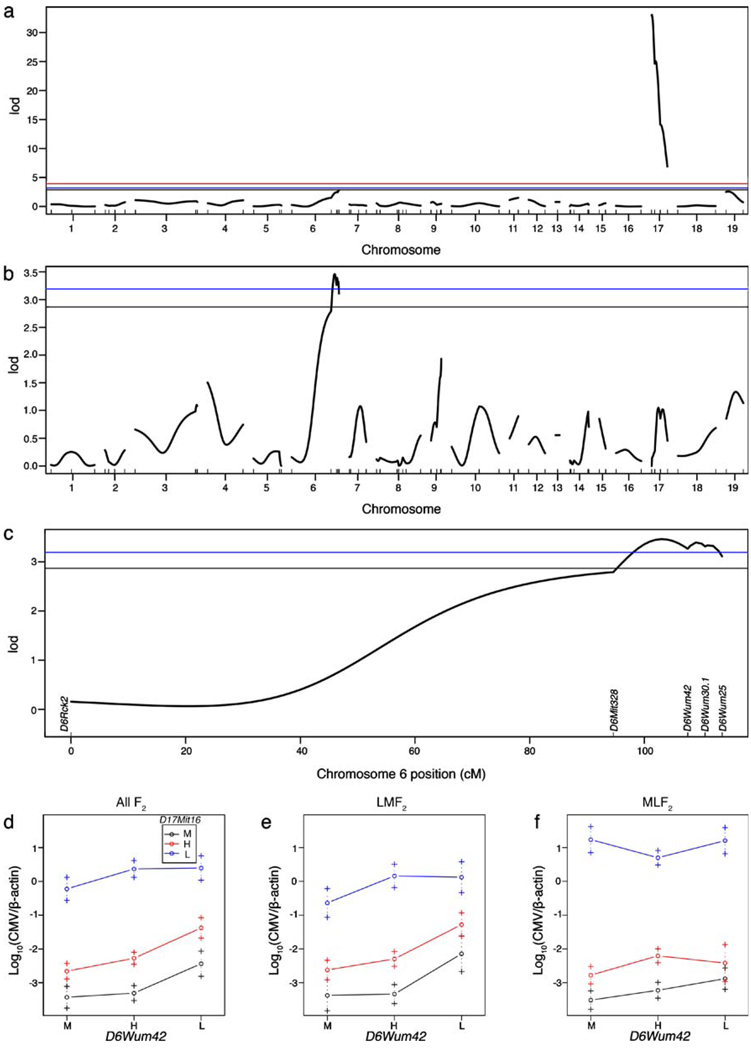
To map chromosome locations for non-MHC MCMV-resistance loci, F2 animals were genotyped with a panel of genome-wide SSLP markers for MA/My and C57L alleles (Table 1). For greater statistical power in the analysis, the current F2 cohort (n=144) was combined with another LMF2 cohort (n=131) genotyped previously for a total of 275 F2 mice for linkage analysis. The data were analyzed using Haley–Knott regression in R/qtl and the genome-wide logarithm of the odds (LOD) scores plotted (Fig. 2a; Broman et al. 2003). As seen by LOD score, two non-MHC loci on chromosomes 6 and 19 were suggestive (90% confidence).
Fig. 2.
MHC and NKC alleles in spleen MCMV resistance in F2 offspring of C57L and MA/My mice. a R/qtl single-QTL genome scan confirms previously reported linkage to D17Mit16 for spleen virus control (Dighe et al. 2005). b and c Chromosome 6 linkage is revealed via single-QTL scan by conditioning on D17Mit16. For a, lines represent 99% (highly significant), 95% (significant), and 90% (suggestive) confidence and b and c, lines for 95%, and 90% confidence are shown. d–f Protective effect of MA/My alleles visualized by effect plots showing the log10-transformed spleen virus levels for various combinations of MA/My (M, black), heterozygous (H, red), or C57L (L, blue), alleles at D17Mit16 with D6Wum42 for all F2 (d), LMF2 (e), and MLF2 (f)
A suggestive linkage (LOD<4.3; Lander and Kruglyak 1995) for the strongest non-MHC QTL was found on chromosome 6 near the distal NKC marker D6Wum25 (Table 3), with MA/My alleles affording increased resistance. Because of a very strong main effect QTL, the data set was further analyzed after conditioning on D17Mit16 as a covariate in a single-QTL scan so as to identify other potential QTL (Broman and Speed 2003). A strong genetic association with MCMV levels by conditional LOD score coincided with a proximal NKC marker D6Wum42, with the highest LOD score between it and D6Mit328 (Fig. 2b and c). As LOD scores can be inflated at positions between markers, the NKC is a prime candidate to affect virus control since its clustered genes for cell surface receptors contribute to NK cell effector functions (Brown and Scalzo 2008).
Further analysis using effect plots revealed a D6Wum42 effect, with protection afforded by MA/My alleles. The observed protection was greatest in homozygous D17Mit16mamy F2 offspring with even one D6Wum42mamy allele; a similar trend was observed in D17Mit16het F2 animals (Fig. 2d). Curiously, when MCMV levels were considered separately for LMF2 and MLF2 offspring, a D6Wum42 resistance effect was evident in LMF2 offspring (Fig. 2e), but was missing in homozygous D17Mit16c57l MLF2 offspring (Fig. 2f). In fact, virus levels in MLF2 offspring with a D17Mit16c57l D6Wum42mamy combined genotype were 15-fold higher than in LMF2 offspring with the same genotype. Nonetheless, even modest resistance in F2 offspring homozygous for D17Mit16c57l and D6Wum42mamy (Fig. 2d) suggested that an NKC-linked QTL might provide some H-2-independent virus resistance.
We next searched for potential interacting loci by including D17Mit16 as an interactive covariate in R/qtl analysis. A model was generated in order to examine how non-MHC loci contribute to virus control in the context of H-2k. Both interaction (epistatic) and additive effects were considered and models were fitted with Haley–Knott regression, as our genotyping was complete (see “Materials and methods” section; Broman and Speed 2003). Different from genetic epistasis between MHC and NKC loci in MCMV resistance observed before (Desrosiers et al. 2005), there was no evidence for such an interaction in the current cross. To formally assess a possibility of genetic epistasis, a likelihood ratio test comparing the best full two-QTL interaction model to the best additive model involving QTL on chromosomes 6 and 17 was performed. The LOD score for the full model, which takes into account both epistatic and additive effects was 37.1. The epistasis LOD score was only 0.64, which corresponds to a genome-wide p value of >0.99. An additive model for MHC and NKC loci on chromosomes 6 and 17 was highly significant (Table 4); the best model had a LOD score of 36.48 and accounted for 45.7% of the phenotypic variation. Thus, in this strain combination, MHC and NKC loci appear to contribute additively to virus resistance.
Table 4.
Additive model for chromosomes 6 and 17 in spleen virus control
| Map position (cM) | LOD | % Variance | p | |
|---|---|---|---|---|
| Chra | ||||
| 6 | 103 | 3.42 | 3.2 | <0.0005 |
| 17 | 1 | 34.17 | 41.9 | <0.00001 |
| Modelb | ||||
| Chr 6. & 17 | 36.48 | 45.7 | <0.00001 |
Individual contributions to the model from each QTL are calculated using ANOVA
Additive model generated as described in “Materials and methods” section
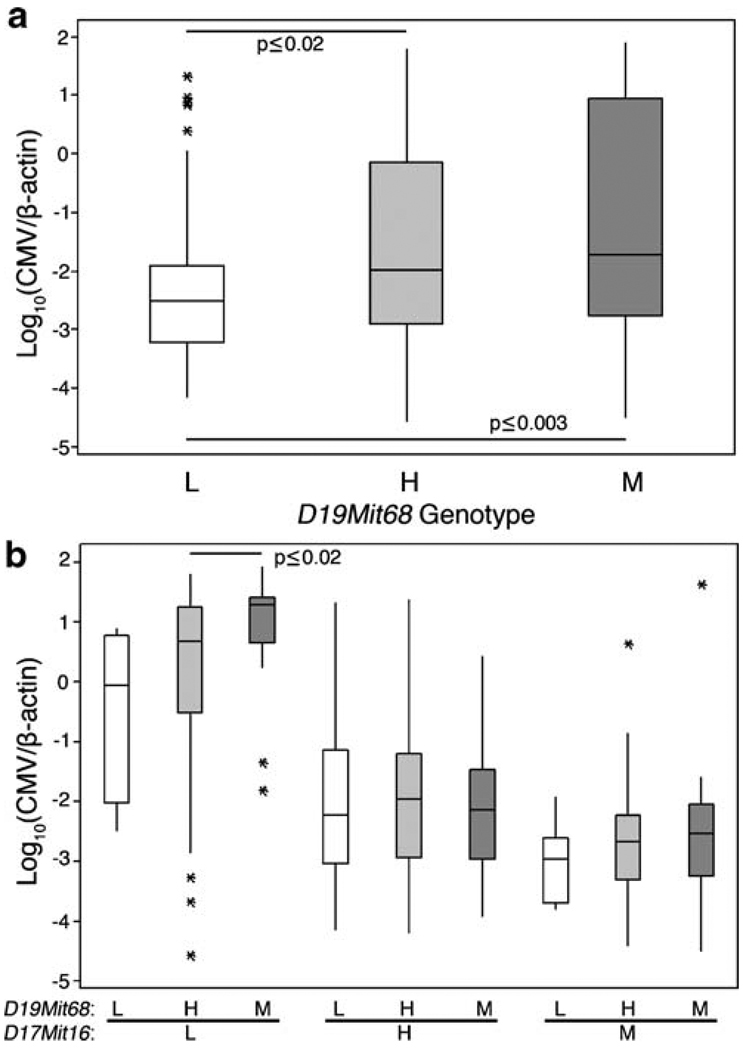
Another suggestive non-H-2 association was linked with D19Mit68 on chromosome 19 (Table 3). To further interrogate this association, LMF2 and MLF2 animals were stratified by D19Mit68 genotypes. We found that virus levels were significantly lower in D19Mit68c57l F2 spleens than in D19Mit68het or D19Mit68mamy F2 spleens (Fig. 3a). Curiously, further stratification of the data set by D17Mit16 revealed that D19Mit68c57l D17Mit16c57l F2 offspring were significantly underrepresented (Table 5), an outcome not unlike that of H-2 skewing seen with MLF2 offspring (Table 2). Despite this, a D19Mit68-linked QTL apparently extended MHC protection in F2 offspring homozygous for H-2b or H-2k since MCMV levels were generally or significantly lower when one or two D19Mit68c57l alleles were also available (Fig. 3b). Thus, a D19Mit68-linked QTL confers MCMV resistance and enhanced MHC protection.
Fig. 3.
A protective role for a chromosome 19 QTL in MCMV infected F2 mice. Spleen virus level data stratified in a by D19Mit68 genotype and in b by D19Mit68 and D17Mit16 genotypes shows a protective effect of C57L alleles and a generally protective effect in combination with MA/My alleles on chromosome 17. Significance as determined by Wilcoxon test is indicated. For comparison, a similar stratification was performed for the adjacent marker D19Mit88 and no protective effect was seen (data not shown)
Table 5.
D17Mit16 and D19Mit68 genotypes for F2 offspring (n=275)
| D17Mit16 genotype | |||||
|---|---|---|---|---|---|
| LL | ML | MM | |||
| LL | 4 | 33 | 19 | ||
| D19Mit68 genotype | ML | 45 | 78 | 42 | p <0.001* |
| MM | 19 | 18 | 17 | ||
p<0.001; Chi-square test was used as described in Table 2
The NKC contributes MHC-independent MCMV resistance
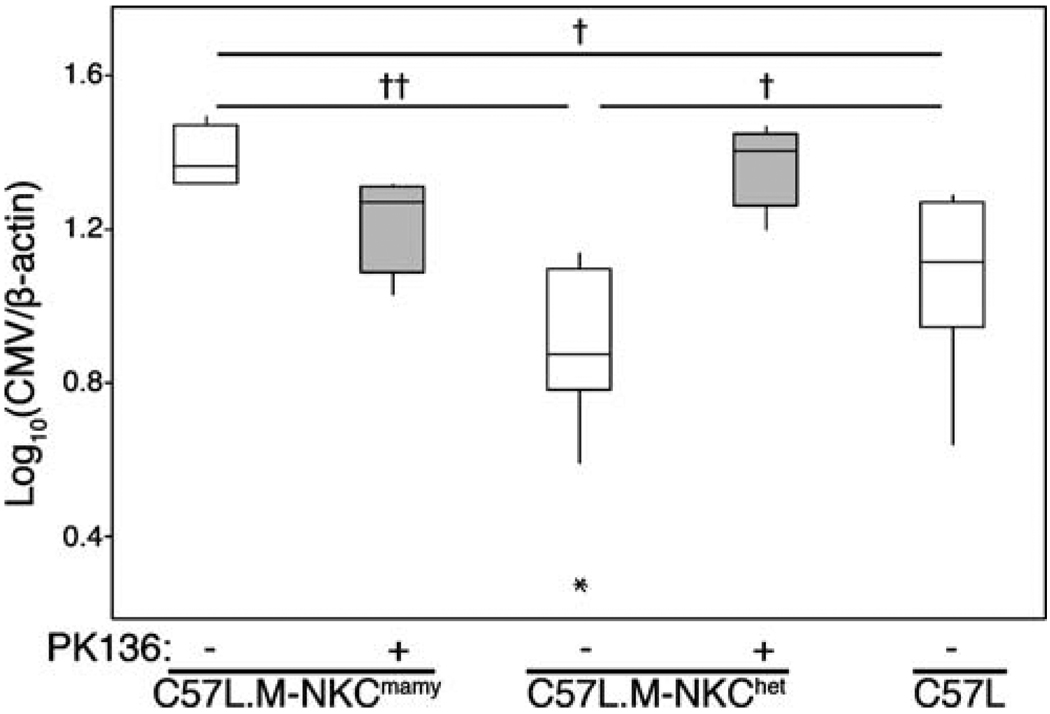
To validate its role in MCMV resistance, we crossed an NKCmamy haplotype onto the C57L background. C57L.M-NKCmamy and C57L.M-NKChet congenic mice were generated and tested with MCMV infection. We found that C57L.M-NKChet had significantly lower virus levels than C57L.M-NKCmamy and C57L (Fig. 4). Interestingly, NKChet protection was mediated through NK cells since their depletion prior to infection led to a significant increase in both spleen (p≤0.02) and liver (p≤0.05) virus levels. Thus, an NKC locus contributes MCMV resistance in this genetic system, which further underscores its importance in viral immunity.
Fig. 4.
MA/My NKC allele contributes to virus control in NKChet H-2b mice. Box and whisker plots for virus levels determined by QPCR 3.5 d.p.i. of PBS (white) PK136-treated (gray) C57L.M-NKCmamy and C57L.M-NKChet as well as PBS-treated C57L.M-NKCc57l(n=4–10 per group) mice with 1 × 105 PFU SGV. Significance as determined by Wilcoxon test is indicated (††p≤0.001, †p≤0.05). No significant protective effect was observed in the liver
Genetic determinants of MCMV resistance in the livers of infected F2 mice
Fine differences in spleen and liver MCMV immunity have been detected (Scalzo et al. 1990; Tay and Welsh 1997), though IFN-γ and perforin contributed through NK cells were needed in both organs (Loh et al. 2005; Sumaria et al. 2009). In the current work, virus levels were significantly lower in spleens than in livers of infected H-2k homozygous and heterozygous F2 offspring (Fig. 1). In contrast, virus levels were lower in livers than in spleens of infected C57L and H-2b F2 offspring (Fig. 1) and this differential genetic and immune control of MCMV replication in spleens and livers of infected mice is consistent with our previous work (Xie et al. 2007, 2009). The MHC region of chromosome 17 had a dramatic influence on liver immunity (Table 6), accounting for 36.9% of the trait variance while non-MHC loci had limited effects in this organ. It is interesting to note that no chromosome 6 linkage was observed in our genome-wide or conditional analysis nor statistically significant differences noted between virus levels in C57L mice with various NKC haplotypes (data not shown). Thus, the MHC-independent protection via the NKC was organ-specific and MHC-mediated control is critical in the liver.
Table 6.
Haley–Knott regression analysis of F2 intercross cohort, liver (n=230)
| Chr | Position (cM) | Nearest marker | LOD | % Variance | p |
|---|---|---|---|---|---|
| 8 | 62.9 | SCOR8.85.14 | 1.43 | 2.8 | 0.90 |
| 11 | 6 | D11Mit2 | 2.00 | 3.9 | 0.50 |
| 12 | 13 | D12Mit285 | 1.63 | 3.2 | 0.78 |
| 17 | 1 | D17Mit16 | 23.00 | 36.9 | <0.001 |
| 19 | 2 | D19Mit68 | 1.45 | 2.9 | 0.8894 |
Discussion
In this study, two non-MHC MCMV-resistance loci have been mapped using classical genetic strategies. A suggestive NKC-linked QTL (MA/My allele) on chromosome 6 which gave enhanced protection in F2 animals has been validated. The protective effect of D6Wum42mamy is surprising given that C57L and MA/My NKC haplotypes are highly related with the exception of the proximal NKC (Brown et al. 2001b). Previously unappreciated, a D6Wum42mamy protective effect in F2 offspring and D6Wum42het in our NKC congenic strains has revealed that the NKC can provide protection independent of H-2k. In addition, the model indicated that MA/My NKC alleles afforded additive protection when in combination with H-2k MHC. Both of these findings distinguish this interaction from epistasis, as both gene regions provide virus control independently and additive protection when in combination.
The genetic analysis and biological confirmation for NKC involvement in virus resistance using NKC congenic animals suggest that NKC polymorphism can directly impact NK-mediated virus immunity. This is an unexpected finding given that distinct NKC-Ly49 haplotypes are thought to exist in mice based on genetic probing with microsatellite and single-nucleotide polymorphism (SNP) markers (Brown et al. 2001b; Lee et al. 2001b; Scalzo et al. 2005) that correspond well with particular Southern restriction fragment length polymorphism (RFLP) groupings for clustered Ly49 genes (reviewed in Brown and Scalzo 2008). As we have previously shown that MA/My and C57L NKC-Ly49 haplotypes are related (Brown et al. 2001b), we hypothesized that Ly49 receptors displayed by NK cells of either strain are likely to be very similar in structure (Brown et al. 2001b). Despite this, we found that their Ly49G2 receptors actually differ by a single amino acid in the extracellular domain (Xie et al. 2009). Subtle sequence variation detected within Ly49g might hint that the haplotypes actually differ; however, additional sequencing of the NKC-Ly49 gene clusters is needed to answer this question.
A recent study has shown that differences in the ligandbinding domain of the Ly49G2 receptor determines its capacity to interact with an MHC class I Dk ligand (Silver et al. 2002). Thus, even modest variation in Ly49 or other NKC-encoded receptors can have a significant impact on ligand binding, which could then also influence NK cell effector functions, including those pertaining to virus immunity. This is more interesting in light of recent studies which have shown that select human KIR and HLAcombined genotypes influenced susceptibility to chronic viral infection (Alter et al. 2007; Carrington and Martin 2006; Khakoo et al. 2004; Martin et al. 2002, 2007; Parham 2005). An important feature of the genetic system studied here is that it provides a new model for investigating how NKC and MHC genetic variation can affect NK-mediated virus immunity.
Evidence for this was revealed by comparison of MCMV-resistance traits in F2 and NKC congenic animals without H-2k protection. In the case of H-2b F2 animals, only those that were homozygous for D6Wum42mamy experienced a protective effect. However, in NKC congenic animals without H-2k resistance, we found that NKChet mice restrained MCMV significantly better than their NKCmamy or NKCc57l littermates. This finding suggests that still other unknown genetic modifiers may have affected MCMV-resistance traits in individual F2 animals. While we have not rigorously examined numbers of NK cells in F2 animals, we have found similar numbers of splenic NK cells at steady state in C57L, MA/My, and reciprocal H-2 and NKC congenic animals (data not shown). Thus, a distinct NKChet advantage in virus resistance might occur via increased NK Ly49 receptor polymorphism which could broaden NK receptor diversity for a given NK cell and possibly its cumulative binding capacity for self-MHC class I ligands and chances for selection to the pool of educated NK cells (Johansson et al. 2009). Interestingly, recent data have shown that KIR AB haplotype heterozygosity is associated with enhanced responsiveness of human CD56dim NK cells to cytokine stimulation (Korbel et al. 2009) and with protection from HIV infection (Jennes et al. 2006).
Among human populations of the world, selective pressures, including resistance to infection and reproductive competency, have apparently influenced retention of KIR A and B haplotypes specialized for either function, respectively (Parham 2008). The AA KIR haplotype has been associated with preeclampsia and recurrent miscarriage (RM) when the fetus possesses at least one C2 HLA haplotype, which has been proposed to occur via inhibition of uterine NK cells (Hiby et al. 2004, 2008). Perhaps such inhibition of uterine NK cells in MLF1 mothers contributed to the significant underrepresentation of H-2k/b MLF2 offspring and the striking low frequency of animals with a combined D17Mit16c57l D19Mit68c57l genotype. In the current work, uterine NK cells in close proximity to fetal H-2k/b trophoblasts in the decidua could be strongly inhibited due to engagement of NK inhibitory receptors in MLF1 mothers, whose NK cells become licensed on H-2k and H-2b class I proteins. If so, NK-dependent trophoblast invasion, spiral artery remodeling, and establishment of the fetal–maternal interface may be hampered, thereby leading to a decreased blood supply to the developing fetus and loss. NKChet encoded NK receptor diversity in congenic mice may provide a model for understanding the basis for how selective pressure is at work to shape NK cell functions and contributions to individuals and populations.
Another important discovery predicted by our genetic analysis, a non-MHC, non-NKC MCMV-resistance QTL has been mapped on chromosome 19. Curiously, D19Mit68c57l was associated with MCMV resistance; thus the QTL is understood to be transgressive since it confers protection, but was contributed by the susceptible strain. More interestingly, its greatest effect was observed among the group of mice without H-2k resistance (i.e., D17Mit16c57l), as revealed by stratifying for both markers. Despite a merely suggestive linkage, D19Mit68c57l D17Mit16c57l F2 animals were significantly underrepresented (i.e., observed frequency= 1.45%) in the cohort. Thus, its overall effect may have been underestimated in the current study. A trend toward greater resistance was also seen in D19Mit68c57l D17Mit16mamy F2 offspring. Furthermore, we have shown that H-2k MCMV resistance is significantly greater in the C57L background than in the MA/My background (Xie et al. 2007), which has implicated still-unknown genetic modifiers. A prime candidate for additional MCMV-resistance effects is this D19Mit68c57l-linked QTL. The region flanking D19Mit68 contains several interesting candidates known to be involved in virus defense including Unc93b1 (Tabeta et al. 2006), NF-κB subunit p65 (RelA), and PTK1 (Mlk3). Validation and refined mapping for a chromosome 19 QTL is in progress.
Our data support a model whereby distinct genetic factors may affect virus control in different organs, with H-2 playing a predominant role. Interestingly, we consistently observed in H-2b mice that MCMV levels are significantly lower in liver than spleen. Because NKT cells are abundant in this organ, and because they can rapidly produce important cytokines including IFN-γ, they too might contribute some protection. Indeed, it has been shown that NKT cells are rapidly activated after MCMV infection and that stimulation of NKT cells during MCMV infection can augment NK cell-mediated virus control in spleen and liver of BALB/c and C57BL/6 mice (Wesley et al. 2008; van Dommelen et al. 2003). Nonetheless, a direct role for NKT cell-mediated MCMV resistance has not been demonstrated. In accord with this, we have not found a significant alteration in MCMV levels in spleen or liver after depletion of T cells, including NKT cells, via anti-CD4 and anti-CD8 treatment (Xie et al. 2009).
In summary, distinct NKC and chromosome 19 (non-MHC) loci independently affect MCMV resistance, in addition to MHC protection, in this defined model genetic system. Elucidating the additive interplay among non-MHC and MHC adds to our understanding of how NK cell receptor and MHC ligand polymorphism, and their interactions can enhance viral immunity.
Acknowledgments
We thank Corinne Abalos and Susan Alejandra Sainz for technical assistance. This work was supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease Grant R01 AI050072. M.D.S was supported by the NIH Interdisciplinary Training Program in Immunology (5T32 AI07496) and the NIH Biotechnology Training Program (T32 GM08715).
Contributor Information
Michael D. Stadnisky, Department of Microbiology, University of Virginia School of Medicine, Charlottesville, VA 22908, USA
Ani Manichaikul, Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA 22908, USA; Department of Public Health Sciences, Division of Biostatistics and Epidemiology, University of Virginia School of Medicine, Charlottesville, VA 22908, USA.
Alyssa G. Lundgren, Department of Medicine, Division of Nephrology, University of Virginia School of Medicine, Charlottesville, VA 22908, USA
Michael G. Brown, Department of Microbiology, University of Virginia School of Medicine, Charlottesville, VA 22908, USA Department of Medicine, Division of Nephrology, University of Virginia School of Medicine, Charlottesville, VA 22908, USA; Beirne B. Carter Center for Immunology Research, University of Virginia School of Medicine, Charlottesville, VA 22908, USA.
References
- Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- Beutler B, Georgel P, Rutschmann S, Jiang Z, Croker B, Crozat K. Genetic analysis of innate resistance to mouse cytomegalovirus (MCMV) Brief Funct Genomic Proteomic. 2005;4:203–213. doi: 10.1093/bfgp/4.3.203. [DOI] [PubMed] [Google Scholar]
- Broman KW, Speed TP. A model selection approach for the identification of quantitative trait loci in experimental crosses. J R Statist Soc B. 2003;64:641–656. doi: 10.1534/genetics.108.094565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Brown MG, Scalzo AA. NK gene complex dynamics and selection for NK cell receptors. Semin Immunol. 2008;20:361–368. doi: 10.1016/j.smim.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MG, Dokun AO, Heusel JW, Smith HR, Beckman DL, Blattenberger EA, Dubbelde CE, Stone LR, Scalzo AA, Yokoyama WM. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001a;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- Brown MG, Scalzo AA, Stone LR, Clark PY, Du Y, Palanca B, Yokoyama WM. Natural killer gene complex (Nkc) allelic variability in inbred mice: evidence for Nkc haplotypes. Immunogenetics. 2001b;53:584–591. doi: 10.1007/s002510100365. [DOI] [PubMed] [Google Scholar]
- Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- Chalmer JE, Mackenzie JS, Stanley NF. Resistance to murine cytomegalovirus linked to the major histocompatibility complex of the mouse. J Gen Virol. 1977;37:107–114. doi: 10.1099/0022-1317-37-1-107. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KA, Devora G, Lai WC, O'Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers MP, Kielczewska A, Loredo-Osti JC, Adam SG, Makrigiannis AP, Lemieux S, Pham T, Lodoen MB, Morgan K, Lanier LL, Vidal SM. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat Genet. 2005;37:593–599. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dighe A, Rodriguez M, Sabastian P, Xie X, McVoy M, Brown MG. Requisite H2k role in NK cell-mediated resistance in acute murine cytomegalovirus-infected MA/My mice. J Immunol. 2005;175:6820–6828. doi: 10.4049/jimmunol.175.10.6820. [DOI] [PubMed] [Google Scholar]
- Grundy JE, Mackenzie JS, Stanley NF. Influence of H-2 and non-H-2 genes on resistance to murine cytomegalovirus infection. Infect Immun. 1981;32:277–286. doi: 10.1128/iai.32.1.277-286.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Heeney JL, Dalgleish AG, Weiss RA. Origins of HIV and the evolution of resistance to AIDS. Science. 2006;313:462–466. doi: 10.1126/science.1123016. [DOI] [PubMed] [Google Scholar]
- Hiby SE, Walker JJ, O'Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum Reprod. 2008;23:972–976. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- Jennes W, Verheyden S, Demanet C, Adje-Toure CA, Vuylsteke B, Nkengasong JN, Kestens L. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol. 2006;177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- Johansson S, Salmon-Divon M, Johansson MH, Pickman Y, Brodin P, Karre K, Mehr R, Hoglund P. Probing natural killer cell education by Ly49 receptor expression analysis and computational modelling in single MHC class I mice. PLoS ONE. 2009;4:e6046. doi: 10.1371/journal.pone.0006046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O'Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- Korbel DS, Norman PJ, Newman KC, Horowitz A, Gendzekhadze K, Parham P, Riley EM. Killer Ig-like receptor (KIR) genotype predicts the capacity of human KIR-positive CD56dim NK cells to respond to pathogen-associated signals. J Immunol. 2009;182:6426–6434. doi: 10.4049/jimmunol.0804224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Landolfo S, Gariglio M, Gribaudo G, Lembo D. The human cytomegalovirus. Pharmacol Ther. 2003;98:269–297. doi: 10.1016/s0163-7258(03)00034-2. [DOI] [PubMed] [Google Scholar]
- Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet. 2001a;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- Lee SH, Gitas J, Zafer A, Lepage P, Hudson TJ, Belouchi A, Vidal SM. Haplotype mapping indicates two independent origins for the Cmv1s susceptibility allele to cytomegalovirus infection and refines its localization within the Ly49 cluster. Immunogenetics. 2001b;53:501–505. doi: 10.1007/s002510100359. [DOI] [PubMed] [Google Scholar]
- Loh J, Chu DT, O'Guin AK, Yokoyama WM, Virgin HW., IV Natural killer cells utilize both perforin and gamma interferon to regulate murine cytomegalovirus infection in the spleen and liver. J Virol. 2005;79:661–667. doi: 10.1128/JVI.79.1.661-667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange JS. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. 2002;4:1545–1558. doi: 10.1016/s1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol. 2008;20:311–316. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Sabastian P, Clark P, Brown MG. Cmv1- independent antiviral role of NK cells revealed in murine cytomegalovirus-infected New Zealand White mice. J Immunol. 2004;173:6312–6318. doi: 10.4049/jimmunol.173.10.6312. [DOI] [PubMed] [Google Scholar]
- Rodriguez MR, Lungren A, Sabastian P, Li QGC, Brown MG. A Cmv2 QTL on chromosome X affects MCMV resistance in New Zealand male mice. Mamm Genome. 2009;20:414–423. doi: 10.1007/s00335-009-9203-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalzo AA, Fitzgerald NA, Simmons A, La Vista AB, Shellam GR. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J Exp Med. 1990;171:1469–1483. doi: 10.1084/jem.171.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalzo AA, Fitzgerald NA, Wallace CR, Gibbons AE, Smart YC, Burton RC, Shellam GR. The effect of the Cmv-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J Immunol. 1992;149:581–589. [PubMed] [Google Scholar]
- Scalzo AA, Lyons PA, Fitzgerald NA, Forbes CA, Yokoyama WM, Shellam GR. Genetic mapping of Cmv1 in the region of mouse chromosome 6 encoding the NK gene complex-associated loci Ly49 and musNKR-P1. Genomics. 1995;27:435–441. doi: 10.1006/geno.1995.1074. [DOI] [PubMed] [Google Scholar]
- Scalzo AA, Manzur M, Forbes CA, Brown MG, Shellam GR. NK gene complex haplotype variability and host resistance alleles to murine cytomegalovirus in wild mouse populations. Immunol Cell Biol. 2005;83:144–149. doi: 10.1111/j.1440-1711.2005.01311.x. [DOI] [PubMed] [Google Scholar]
- Scalzo AA, Corbett AJ, Rawlinson WD, Scott GM, Degli-Esposti MA. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol Cell Biol. 2007;85:46–54. doi: 10.1038/sj.icb.7100013. [DOI] [PubMed] [Google Scholar]
- Silver ET, Lavender KJ, Gong DE, Hazes B, Kane KP. Allelic variation in the ectodomain of the inhibitory Ly-49G2 receptor alters its specificity for allogeneic and xenogeneic ligands. J Immunol. 2002;169:4752–4760. doi: 10.4049/jimmunol.169.9.4752. [DOI] [PubMed] [Google Scholar]
- Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, Scalzo AA, Fremont DH, Yokoyama WM. Recognition of a virusencoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumaria N, van Dommelen SL, Andoniou CE, Smyth MJ, Scalzo AA, Degli-Esposti MA. The roles of interferon-gamma and perforin in antiviral immunity in mice that differ in genetically determined NK-cell-mediated antiviral activity. Immunol Cell Biol. 2009;87:559–566. doi: 10.1038/icb.2009.41. [DOI] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- Tay CH, Welsh RM. Distinct organ-dependent mechanisms for the control of murine cytomegalovirus infection by natural killer cells. J Virol. 1997;71:267–275. doi: 10.1128/jvi.71.1.267-275.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dommelen SL, Tabarias HA, Smyth MJ, Degli-Esposti MA. Activation of natural killer (NK) T cells during murine cytomegalovirus infection enhances the antiviral response mediated by NK cells. J Virol. 2003;77:1877–1884. doi: 10.1128/JVI.77.3.1877-1884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat RL, Clark PY, Brown MG. Quantitative measurement of infectious murine cytomegalovirus genomes in real-time PCR. J Virol Methods. 2003;112:107–113. doi: 10.1016/s0166-0934(03)00197-6. [DOI] [PubMed] [Google Scholar]
- Wesley JD, Tessmer MS, Chaukos D, Brossay L. NK cell-like behavior of Vα14i NK T cells during MCMV infection. PLoS Pathog. 2008;4:e1000106. doi: 10.1371/journal.ppat.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Dighe A, Clark P, Sabastian P, Buss S, Brown MG. Deficient major histocompatibility complex-linked innate murine cytomegalovirus immunity in MA/My.L-H2b mice and viral downregulation of H-2k class I proteins. J Virol. 2007;81:229–236. doi: 10.1128/JVI.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Stadnisky MD, Brown MG. MHC class I Dk locus and Ly49G2+ NK cells confer H-2k resistance to murine cytomegalovirus. J Immunol. 2009;182:7163–7171. doi: 10.4049/jimmunol.0803933. [DOI] [PMC free article] [PubMed] [Google Scholar]