Abstract
The LIM-domain containing transcription factor, Lhx1, is involved in the regulation of early gastrulation cell movements, kidney organogenesis and other processes in vertebrate model organisms. To follow the expression of this gene in live embryos, we created transgenic zebrafish expressing enhanced green fluorescent protein (EGFP) under the control of lhx1a regulatory regions. Tg(lhx1a:EGFP) pt303 recapitulates the expression of endogenous lhx1a beginning at early gastrula stages through 72 hours of development with only few exceptions. In addition, over-expression of the Nodal ligand, ndr1, results in the concomitant expansion of the transgene and endogenous lhx1a expression. Treatment of Tg(lhx1a:EGFP) pt303 embryos with the small molecule SB-431542, an inhibitor of Nodal signaling, results in the loss of both transgene and endogenous lhx1a expression. These experiments suggest that Tg(lhx1a:EGFP) pt303 is regulated in a manner similar to endogenous lhx1a. Therefore, this reporter can be utilized not only for monitoring lhx1a expression, but also for numerous applications, including chemical genetics screening.
Keywords: lhx1a, transgene, zebrafish, kidney, gastrulation
Introduction
LIM domain-containing proteins have been implicated in numerous biological processes including cell-fate determination, tissue-specific gene expression, neuronal pathfinding, and actin organization. This motif was originally identified within LIM-homeodomain proteins (LHX), a family of proteins of which Lhx1 (formerly known as Lim1) is a member (Dawid et al. 1998). Lhx1 plays a number of roles during the course of vertebrate development, including an important early role in gastrulation. It is expressed in the organizer region (and its functional equivalents) in Xenopus, mice, and zebrafish (Shawlot and Behringer 1995; Taira et al. 1992; Toyama and Dawid 1997), and Lhx1 deficiency in both mice and Xenopus results in the loss of all cephalic structures (Hukriede et al. 2003; Kodjabachian et al. 2001; Shawlot and Behringer 1995).
Lhx1 also regulates the development of a number of organs, including the kidney. The vertebrate kidney develops from the intermediate mesoderm (IM), and Lhx1 serves as a marker of progenitor cells in this tissue (Carroll and Vize 1999; Karavanov et al. 1996; Toyama and Dawid 1997). In the mouse it continues to be expressed in both the mesonephric and metanephric kidneys where it is required for nephron formation. Loss of Lhx1 in mice results in the disruption of IM differentiation and the absence of functional kidneys (Shawlot and Behringer 1995).
There are multiple pathways that regulate Lhx1 expression during development, including Nodal, which acts in the early embryo. Members of the Nodal signaling family are TGF-β ligands that serve as inducers of both mesoderm and endoderm and also pattern the dorsal-ventral axis of the vertebrate embryo (Schier and Talbot 2005). Nodals signal through EGF-CFC co-receptors and type I and II Activin receptors. In zebrafish, over-expression of synthetic mouse nodal RNA leads to ectopic expression of lhx1a in embryos beginning at 30% epiboly (Toyama et al. 1995). In addition, a mutation in the EGF-CFC, one-eyed pinhead (oep), results in the complete loss of lhx1a expression during gastrulation (Watanabe et al. 2002).
Because of the numerous roles that Lhx1 plays throughout development, we created an EGFP transgene in zebrafish to follow lhx1a expression in vivo, and to further our understanding of its regulation during gastrula stages and pronephric development. Tg(lhx1a:EGFP)pt303expression recapitulates lhx1a expression at all stages analyzed, and modulation of the Nodal signaling pathway, either by over-expression of ndr1 or treatment with a small molecule inhibitor, confirms that this transgene is regulated in the same manner as the endogenous lhx1a gene. Therefore, Tg(lhx1a:EGFP)pt303should serve as a useful resource for investigating gastrulation and mechanisms of cell-fate specification within the kidney.
Results
Generation of lhx1a transgenic line
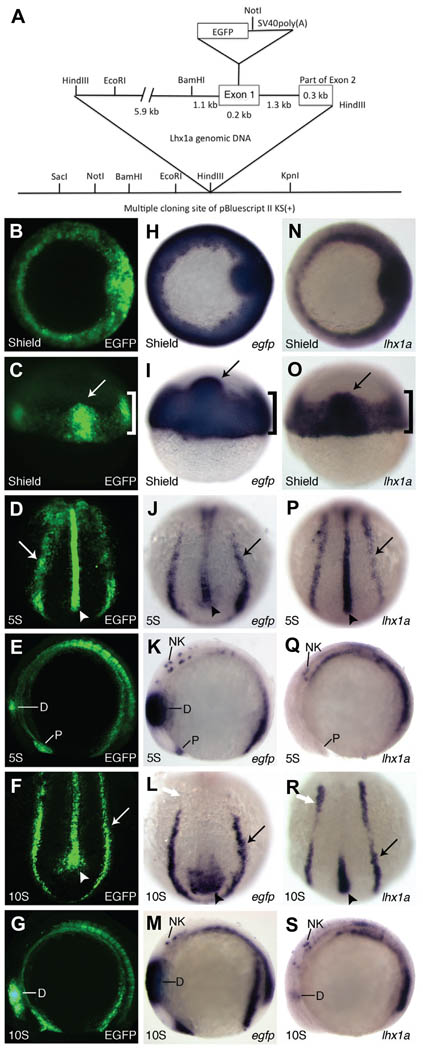
A BAC clone containing the lhx1a genomic region was identified and an 8.8 kb fragment containing 5’ untranslated sequence, exon 1, and part of exon 2 was isolated and subcloned (Fig. 1A). This fragment contains the first intron, which is important for transcriptional regulation of both the Xlim1 and lhx1a genes in Xenopus and zebrafish, respectively (Rebbert and Dawid 1997; Watanabe et al. 2002). EGFP was inserted downstream of the start codon and the lhx1a:EGFP DNA fragment was cloned into the pI-SceI meganuclease vector for generating transgenic lines (Thermes et al. 2002). The injected embryos were raised to adulthood and screened for germline carriers by EGFP expression. Three founder transgenics were identified and while levels of expression varied between lines, the expression patterns were similar. Homozygous lines were created from these founders (allele designations: pt301, pt302, pt303). Tg(lhx1a:EGFP) fish are fecund and develop normally suggesting that the insertion of this transgene does not cause any deleterious effects. For all subsequent experiments, we used Tg(lhx1a:EGFP)pt303, the line with the strongest expression.
Fig. 1. Construct design and early Tg(lhx1a:EGFP)pt303 expression.
(A) Schematic of lhx1a:EGFP construct. (B–S) Tg(lhx1a:EGFP)pt303 embryos. (B–G) Tg(lhx1a:EGFP)pt303 expression. (H–M) In situ hybridization for egfp. (N–S) In situ hybridization for lhx1a. (B,H,N) Shield stage embryos, animal view. (C,I,O) Shield stage embryos, dorsal view. Arrow marks the shield and bracket denotes the marginal cells. (D,J,P) 5-somite stage embryos, dorsal-posterior view. Arrow points to intermediate mesoderm and arrowhead marks the notochord. (E,K,Q) 5-somite stage embryos, lateral view. Anterior is to the left. (F,L,R) 10-somite stage embryos, dorsal-posterior view. Arrow points to the intermediate mesoderm and arrowhead marks the notochord. White arrow denotes anterior expression domain of lhx1a in the intermediate mesoderm. (G,M,S) 10-somite stage embryos, lateral view. Anterior is to the left. Abbreviations: D, diencephalon; NK, neural keel; P, polster.
Early expression of Tg(lhx1a:EGFP)pt303
EGFP protein was first analyzed at shield stage where it is localized to the marginal cells (Fig. 1 B,C, bracket) and the shield (Fig. 1 B,C, arrow), the fish equivalent of Spemann’s Organizer in Xenopus and the mouse node. By 5-somites, EGFP protein is seen predominantly in the notochord (Fig. 1D, arrowhead) and bilaterally in the intermediate mesoderm (IM) (Fig. 1D, arrow), the tissue that will ultimately give rise to the kidney. There is also expression in the polster and the diencephalon (Fig. 1E). By 10-somites, EGFP protein remains in the most posterior region of the notochord as well as the IM (Fig. 1F, arrowhead and arrow, respectively). There is still strong expression in the diencephalon (Fig. 1G).
At the RNA level, egfp expression recapitulates what is seen with the protein, although some structures can be seen in finer detail through in situ hybridization. Similar to the protein, egfp mRNA is expressed in the marginal cells and the shield (Fig. 1 H,I, bracket and arrow, respectively). At 5-somites, expression is seen throughout the notochord and the IM (Fig. 1J, arrowhead and arrow, respectively). It is present in the polster and neural keel as well as in the diencephalon where expression is aberrantly strong (Fig. 1K). By 10-somites, down-regulation of egfp expression is beginning in the IM and the notochord (Fig. 1L, arrow and arrowhead, respectively). Expression is still present in the neural keel and remains strong in the diencephalon (Fig. 1M) at this stage.
The efficacy of this transgene lies in its ability to recapitulate endogenous lhx1a expression. For the majority of tissues examined, this appears to be the case. At shield stage, there is strong expression of endogenous lhx1a in the shield and the margin (Fig. 1 N,O, arrow and bracket, respectively). By 5-somites, expression is predominantly seen in the notochord and IM, although weak expression can also be seen in the polster and neural keel (Fig. 1P, arrowhead and arrow, respectively and 1Q). At 10-somites, lhx1a expression is down-regulated in the notochord and throughout most of the IM (Fig. 1R, arrowhead and arrow, respectively). While diencephalon expression is first reported at this stage, lhx1a expression remains in the neural keel (Fig. 1S).
The anterior IM expression of lhx1a (Fig. 1R, white arrow) ultimately contributes to proximal fates in the pronephric kidney, and presumably corresponds with the expression seen in the proximal tubule at 24 hours post fertilization (hpf) (Toyama and Dawid 1997). This expression domain is not found in the transgene (Fig. 1L, white arrow), although EGFP protein seems to perdure (Fig. 1F), suggesting variations between endogenous lhx1a expression and Tg(lhx1a:EGFP)pt303. Overall, the expression of Tg(lhx1a:EGFP)pt303 agrees with the endogenous expression pattern of lhx1a. The main differences observed are earlier and stronger transgene expression in the diencephalon (Toyama and Dawid 1997; Toyama et al. 1995) and an absence of anterior IM expression at the 10-somite stage.
Tg(lhx1a:EGFP)pt303 expression at 24 hpf through early larval stages
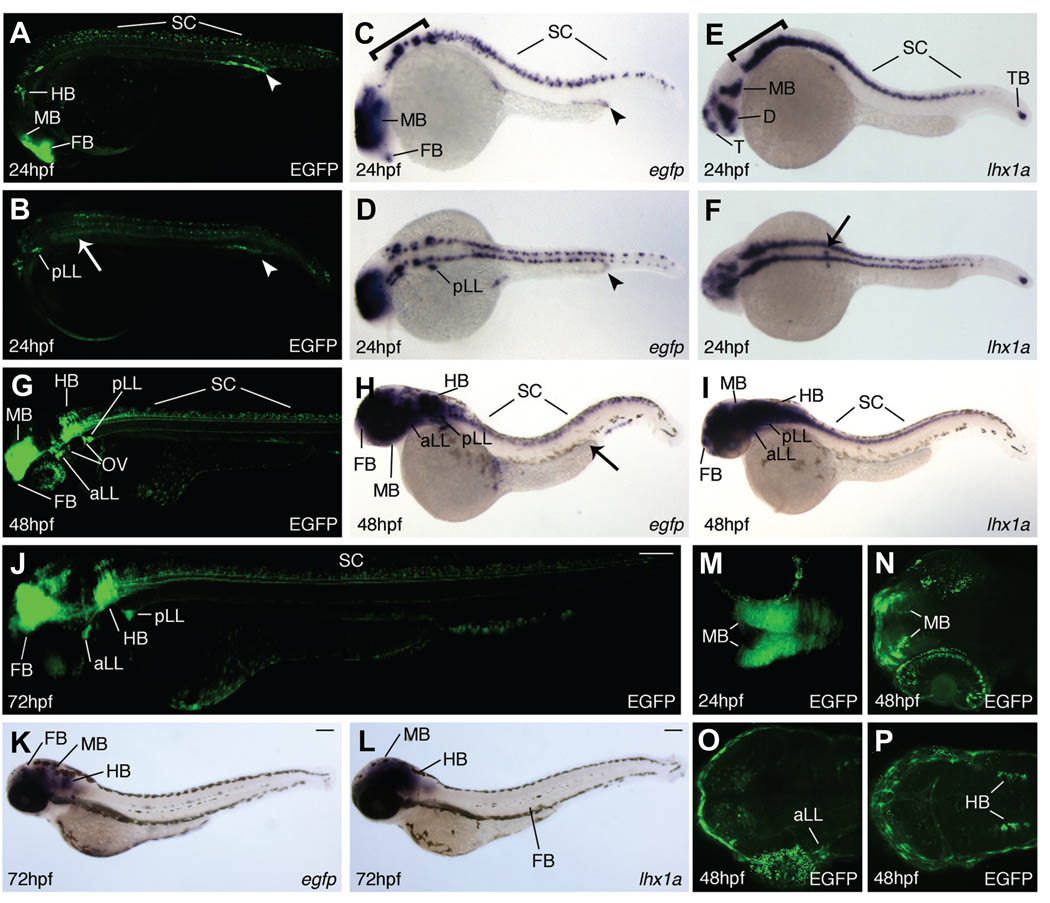
Tg(lhx1a:EGFP)pt303 embryos at 24 hpf have a dynamic expression pattern. EGFP protein is localized to the forebrain, midbrain, and hindbrain (Fig. 2A). It is also expressed in the neurons that run the length of the spinal column (Fig. 2A). A dorsal view of a transgenic embryo at 24hpf also shows expression in the posterior lateral line ganglia (Fig. 2B). In the kidney, EGFP protein is present in the most distal region of the pronephric tubule and duct (Fig. 2 A,B, arrowhead). The expression domains of egfp mRNA coincide with those seen at the protein level. Expression is seen in the forebrain, midbrain, and hindbrain as well as in the neurons within the spinal cord (Fig. 2C). Strong expression is found within the posterior lateral line ganglia while kidney expression is limited to the distal tubule and duct (Fig. 2 C,D, arrowhead).
Fig. 2. Expression of Tg(lhx1a:EGFP)pt303 during later development.
All animals are oriented such that anterior is to the left. (A–P) Tg(lhx1a:EGFP)pt303 embryos and larvae. ( A , B , G , J , M – P ) Tg(lhx1a:EGFP)pt303 expression. (C,D,H,K) In situ hybridization for egfp. (E,F,I,L) In situ hybridization for lhx1a. (A–F) 24 hpf embryos. Arrow marks position of pronephric proximal tubule and arrowhead delineates distal tubule and duct. (A,C,E) Lateral view, bracket denotes hindbrain. (B,D,F) Dorsal view. (G–I) 48 hpf larvae, arrow in H marks distal tubule and duct in the kidney. (J–L) 72 hpf larvae. (J) Montage of two images to display entire larvae. (M–P) Longitudinal optical sections of transgenic animals. (M) 24hpf embryo. (N–P) 48hpf larva. Abbreviations: aLL, anterior lateral line ganglia; FB, forebrain; HB, hindbrain; MB, midbrain; OV, otic vesicle; pLL, posterior lateral line ganglia; SC, spinal column; TB, tailbud.
Endogenous lhx1a is found in the forebrain, specifically in the telencephalon and diencephalon, as well as in the midbrain and hindbrain (Fig. 2E, bracket marks hindbrain). It is also seen in the spinal cord neurons (Fig. 2E). Outside of the CNS, expression of lhx1a is restricted to the most proximal region of the pronephric tubule and to the tailbud (Fig. 2 E,F, arrow). Neither of these expression domains are conserved in Tg(lhx1a:EGFP)pt303embryos at 24 hpf. Loss of proximal tubule expression (Fig. 2 C,D compared to 2 E,F) is consistent with the loss of anterior IM expression seen as early as 10 somites (Fig. 1L compared to 1R, white arrows). In addition, missing transgene expression in the tailbud (Fig. 2 C,D compared to 2 E,F) may be due to defuse, reduced egfp expression in the most posterior region of the notochord at 10 somites (Fig. 1L compared to 1R, arrowheads). These data suggest that additional regulatory elements may be necessary to completely mimic the endogenous expression pattern of lhx1a.
By 48 hpf, strong expression of Tg(lhx1a:EGFP)pt303 at both the protein and RNA level is seen in the CNS (Fig. 2 G,H). Expression is maintained in the posterior lateral line ganglia and is now apparent in the anterior lateral line ganglia (Fig. 2 G,H). Expression is also seen in the otic vesicle, which is a conserved expression domain in Xenopus laevis (Karavanov et al. 1996), but is difficult to detect at the mRNA level (Fig. 2 G,H). However, transgene expression is no longer readily apparent in the kidney (Fig. 2H, arrow). This expression pattern recapitulates what is seen for endogenous lhx1a at this stage (Fig. 2I). By 72 hpf, EGFP protein and egfp mRNA are still present in the brain (Fig. 2 J,K) but only the protein remains in spinal column neurons (Fig. 2J). Since endogenous lhx1a is no longer expressed in the neurons of the spinal column (Fig. 2L), this may represent perdurance of GFP protein in this tissue.
To analyze the expression of Tg(lhx1a:EGFP)pt303 in finer detail, we performed two-photon confocal microscopy on 24 hpf embryos and 48 hpf larvae, focusing on the CNS. At 24 hpf, transgene expression can be seen in both hemispheres of the midbrain (Fig. 2M) and this strong midbrain expression remains at 48 hpf (Fig. 2N). Sectioning deeper into the larva, expression in the anterior lateral line ganglia (Fig. 2O) and the hindbrain (Fig. 2P) becomes apparent. This transgene expression coincides with lhx1a expression previously shown in histological sections (Toyama and Dawid 1997). Overall, these data suggest that the expression of Tg(lhx1a:EGFP)pt303 recapitulates that of the endogenous lhx1a gene with only a few exceptions, and support the usefulness of this transgene for monitoring lhx1a during development in live embryos.
Responsiveness of Tg(lhx1a:EGFP)pt303 to modulation of Nodal signaling
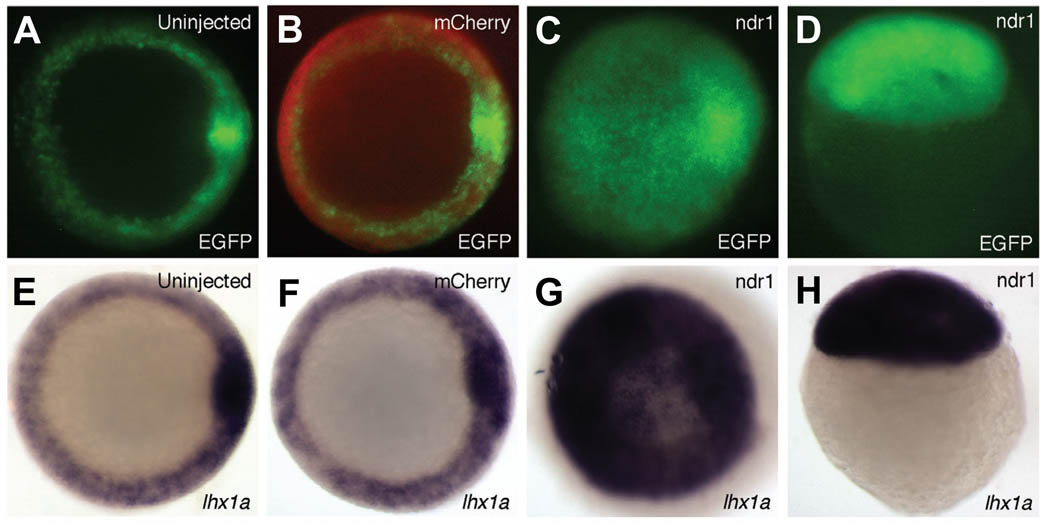
To test whether Tg(lhx1a:EGFP)pt303 is responsive to signaling pathways known to regulate endogenous lhx1a expression, we over-expressed ndr1, one of the zebrafish ligands of the Nodal pathway. It was previously shown that injection of low doses of ndr1 resulted in ectopic expression of lhx1a in shield-stage embryos (Rebagliati et al. 1998). As compared to controls, injection of Tg(lhx1a:EGFP)pt303 embryos with 25pg ndr1 mRNA led to an expansion of both transgene and endogenous lhx1a expression (Fig. 3). These data suggest that Tg(lhx1a:EGFP)pt303, similar to the endogenous lhx1a gene, is responsive to over-expression of Nodal and is therefore regulated by this pathway.
Fig. 3. Responsiveness of Tg(lhx1a:EGFP)pt303 to perturbations in Nodal signaling.
(A–H) Shield stage Tg(lhx1a:EGFP)pt303 embryos. (A–C,E–G) Animal view. (D,H) Lateral view. (A,E) Uninjected control embryos. (B,F) Embryos injected with 125pg mCherry RNA. (C,D,G,H) Embryos injected with 100pg mCherry RNA and 25pg ndr1 RNA. (A–D) Tg(lhx1a:EGFP)pt303 expression. (B) Overlay of mCherry expression (red) displays an embryo that was successfully injected. (E–H) In situ hybridization for lhx1a.
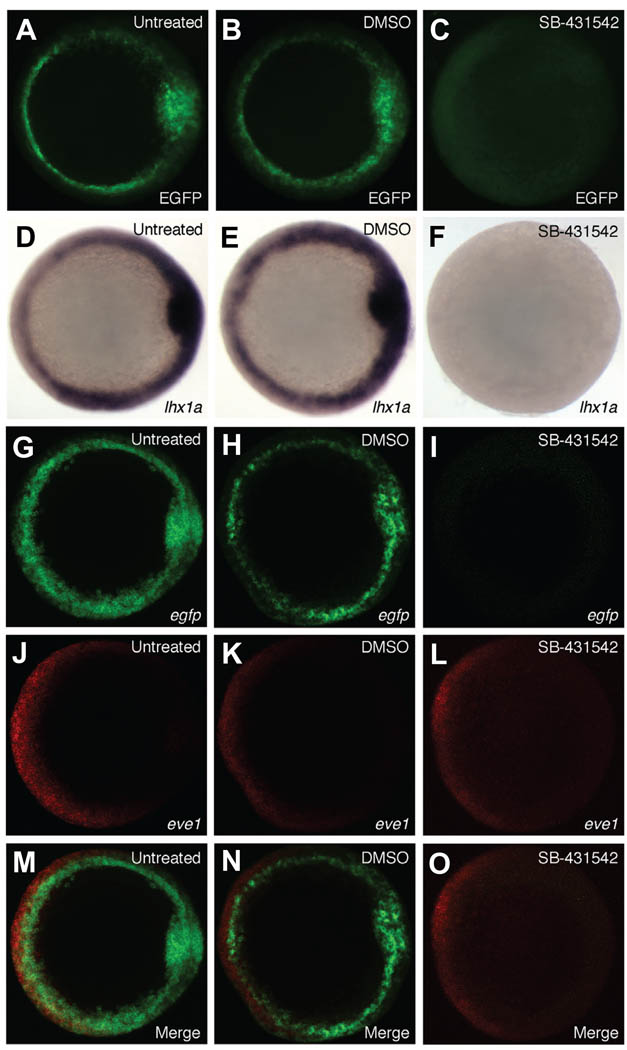
In a complimentary experiment, we utilized a small molecule to inhibit the Nodal pathway. SB-431542 blocks the activity of the type I activin receptor-like kinases (ALK) ALK4, ALK5, and ALK7 in mammalian cell culture by preventing phosphorylation of downstream effector molecules Smad2 and Smad3 (Inman et al. 2002). This compound inhibits Nodal signaling in zebrafish embryos (Hagos and Dougan 2007), and at high doses phenocopies sqt;cyc double mutants, which eliminate early Nodal signals in the fish (Feldman et al. 1998; Hagos and Dougan 2007). As compared to controls, treatment of Tg(lhx1a:EGFP)pt303 embryos with 800µM SB-431542 led to a complete loss of transgene and endogenous lhx1a expression (Fig. 4 A–F). Eve1, a marker of the ventral side of the embryo, served as a negative control to show that treatment with SB-431542 does not disrupt all endogenous signaling events in the early embryo. As compared to controls, eve1 expression is still present in SB-431542 treated embryos (Fig. 4 G–O). Taken together, these data suggest that the effect seen with SB-431542 is specific, and that Tg(lhx1a:EGFP)pt303, like endogenous lhx1a, is regulated by Nodal signaling and is capable of responding to perturbations in this pathway.
Fig. 4. Modulation of Tg(lhx1a:EGFP)pt303 expression by SB-431542.
(A–O) Shield stage Tg(lhx1a:EGFP)pt303 embryos, animal view. (A,D,G,J,M) Untreated control embryos. (B,E,H,K,N) Embryos treated with 0.8% DMSO. (C,F,I,L,O) Embryos treated with 800 µM SB-431542. (A–C) Tg(lhx1a:EGFP)pt303 expression. (D–F) In situ hybridization for lhx1a. (GO) Fluorescent in situ hybridization. (G–I) egfp. (J–L) eve1. (M–O) Merged images.
Discussion
Here we present evidence that the transgenic line, Tg(lhx1a:EGFP)pt303, recapitulates the endogenous expression of lhx1a and is regulated in the same manner during early development in the zebrafish embryo. Strong expression of this transgene begins during gastrula stages. In addition, Tg(lhx1a:EGFP)pt303 is detected in the pronephric kidney during early somitogenesis, which to date is the earliest published expression of a zebrafish transgenic reporter in the nephric field. Comparison with previous data suggests that this nephric expression of Tg(lhx1a:EGFP)pt303 may serve as a marker of kidney progenitor cells (Serluca and Fishman 2001). There are some inconsistencies between the transgene and the endogenous expression pattern of lhx1a. Specifically, proximal tubule expression in the kidney and tailbud expression in the axial mesoderm are absent while diencephalon expression is aberrantly strong in the transgene. It is likely that regulatory regions are absent from this transgene, and this results in the discrepancies that we see between transgene and endogenous expression.
Our data demonstrate that Tg(lhx1a:EGFP)pt303 responds to alterations in the Nodal pathway, suggesting that it is regulated in a manner similar to the endogenous gene. Lhx1a plays a number of important roles throughout development, and this transgene will serve as a useful tool not only for gaining insight into gene regulation but also for trying to understand some of the earliest steps of cell-fate specification. Finally, since zebrafish are well-suited for small molecule screening (Vogt et al. 2009), this transgene could be used to screen for compounds that affect early gastrulation or kidney progenitor cells.
Materials and Methods
Zebrafish husbandry
Embryos expressing the Tg(lhx1a:EGFP) transgene were obtained from incrossing homozygous adults. Embryos were maintained at 28.5°C and were staged according to Kimmel et al. (Kimmel et al. 1995).
Tg(lhx1a:EGFP) construct design
An 8.8kb genomic region of the lhx1a locus was obtained from BAC 184f16 by HindIII digest and sub-cloned into pBluescript II KS(+). The upstream 5.9kb BamHI fragment was removed from this vector to facilitate the introduction of an AscI site, at base pair 12 downstream of the initiation codon, into the remaining 2.9kb fragment. This allowed for the insertion of EGFP in-frame with the first three amino acids of Lhx1a. Sequencing confirmed that no spurious mutations were created by site-directed mutagenesis. EGFP was amplified from the pEGFP-1 vector using primers with MluI sites at their 5’ ends, an enzyme compatible with AscI. Following insertion of EGFP into the construct, the 5.9kb BamHI fragment was returned to generate the original 8.8kb genomic fragment in pBluescript II KS(+). The entire lhx1a:EGFP fragment was then cloned into the pI-SceI vector by HindIII digest. 25pg of lhx1a:EGFP/pI-SceI plasmid DNA was injected into 1-cell stage embryos along with the I-SceI restriction enzyme (NEB) (Thermes et al. 2002). These injected embryos were raised to adulthood and screened for EGFP expression in known lhx1a expression domains. Three independent transgenic lines were isolated; Tg(lhx1a:EGFP)pt301, Tg(lhx1a:EGFP)pt302, and Tg(lhx1a:EGFP)pt303. All three lines displayed expression in the same domains, with slight variability in overall expression levels.
In situ hybridization
Embryos were fixed in 4% paraformaldehyde (PFA) and processed for whole mount in situ hybridization as described (Toyama and Dawid 1997) or for fluorescent in situ hybridization (FISH) (Schoenebeck et al. 2007), with the following modifications. Embryos were incubated in Cy3-tyramide or fluorescein-tyramide solution for one hour. RNA probes for lhx1a, egfp, and eve1 were used for this study.
Zebrafish mRNA microinjection
mCherry/pCS2+ was linearized with SacII and ndr1/pCS2+ was linearized with NotI. The mMessage mMachine SP6 kit (Ambion) was used to transcribe capped RNA for both genes as per manufacture’s instructions. Tg(lhx1a:EGFP)pt303 embryos were injected with either 125pg mCherry mRNA or 100pg mCherry and 25pg ndr1 mRNA at the 1-cell stage. All embryos that expressed mCherry at shield stage were either fixed in 4% PFA or were analyzed for transgene expression.
Chemical treatment of transgenic embryos
SB-431542 (4- [4-(1,3 benzodioxol-5-yl)-5-(2-pyridinyl-1H-imidazol-2-yl]benzamide) (Sigma) was stored as a 100mM stock in DMSO at - 20°C. Tg(lhx1a:EGFP)pt303 embryos at equivalent stages were arrayed into 12-well plates (maximum of 25/well) and were treated with 1.5mL of E3, 0.8% DMSO or 1.5mL of E3, 0.8% DMSO, 800 µM SB-431542 between the 256 and 512-cell stage (Hagos and Dougan 2007). Drug or vehicle was removed at shield stage by washing five times with E3 media, and embryos were either fixed in 4% PFA or were analyzed for transgene expression.
Zebrafish Imaging
Tg(lhx1a:EGFP)pt303 embryos (shield through 24 hpf) were held in place on glass bottom culture dishes (MatTek Corporation) with 1% low melting point agarose. Larvae (48 and 72 hpf) were immobilized with tricaine and placed on similar dishes. Tg(lhx1a:EGFP)pt303expression was imaged with either a Zeiss 510 Meta or a Leica TCS SP5 confocal microscope. Z-stacks were taken for each image and projections were made using Image J. For Tg(lhx1a:EGFP)pt303 embryos treated with SB-431542 or injected with ndr1 and/or mCherry RNA, images were taken with a Leica M205 FA epifluorescent microscope. Images were further analyzed with Adobe Photoshop CS3.
For optical sectioning, Tg(lhx1a:EGFP)pt303 embryos and larvae (24hpf and 48hpf, respectively) were fixed in 4% PFA for 1 hour at room temperature. They were then positioned in a depression slide containing 3% methylcellulose and imaged with an Olympus Fluoview FV1000 MPE system. Images were analyzed in the same manner as above.
Acknowledgements
We would like to thank Lance Davidson for help with confocal microscopy; Dave Grainy and Gretchen Blasko for fish care; Wade Znosko for critical reading of the manuscript; David Kozlowski for identification of anatomical structures; Nathan Bahary for the mCherry expression plasmid; Mike Rebagliati for the ndr1 expression plasmid and Michael Tsang for suggestions and advice. This work has been funded by the National Institutes of Health, USA: R01DK069403, R01HD053287, P30DK079307 and the NICHD Intramural Program.
References
- Carroll TJ, Vize PD. Synergism between Pax-8 and lim-1 in embryonic kidney development. Dev Biol. 1999;214:46–59. doi: 10.1006/dbio.1999.9414. [DOI] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature. 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- Hagos EG, Dougan ST. Time-dependent patterning of the mesoderm and endoderm by Nodal signals in zebrafish. BMC Dev Biol. 2007;7:22. doi: 10.1186/1471-213X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukriede NA, Tsang TE, Habas R, Khoo PL, Steiner K, Weeks DL, Tam PP, Dawid IB. Conserved requirement of Lim1 function for cell movements during gastrulation. Dev Cell. 2003;4:83–94. doi: 10.1016/s1534-5807(02)00398-2. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Karavanov AA, Saint-Jeannet JP, Karavanova I, Taira M, Dawid IB. The LIM homeodomain protein Lim-1 is widely expressed in neural, neural crest and mesoderm derivatives in vertebrate development. Int J Dev Biol. 1996;40:453–461. [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kodjabachian L, Karavanov AA, Hikasa H, Hukriede NA, Aoki T, Taira M, Dawid IB. A study of Xlim1 function in the Spemann-Mangold organizer. Int J Dev Biol. 2001;45:209–218. [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Dev Biol. 1998;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- Rebbert ML, Dawid IB. Transcriptional regulation of the Xlim-1 gene by activin is mediated by an element in intron I. Proc Natl Acad Sci USA. 1997;94:9717–9722. doi: 10.1073/pnas.94.18.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, YeloN D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serluca FC, Fishman MC. Pre-pattern in the pronephric kidney field of zebrafish. Development. 2001;128:2233–2241. doi: 10.1242/dev.128.12.2233. [DOI] [PubMed] [Google Scholar]
- Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- Taira M, Jamrich M, Good PJ, Dawid IB. The LIM domain-containing homeo box gene Xlim-1 is expressed specifically in the organizer region of Xenopus gastrula embryos. Genes Dev. 1992;6:356–366. doi: 10.1101/gad.6.3.356. [DOI] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- Toyama R, Dawid IB. lim6, a novel LIM homeobox gene in the zebrafish: comparison of its expression pattern with lim1. Dev Dyn. 1997;209:406–417. doi: 10.1002/(SICI)1097-0177(199708)209:4<406::AID-AJA8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Toyama R, O’connell ML, Wright CV, Kuehn MR, Dawid IB. Nodal induces ectopic goosecoid and lim1 expression and axis duplication in zebrafish. Development. 1995;121:383–391. doi: 10.1242/dev.121.2.383. [DOI] [PubMed] [Google Scholar]
- Vogt A, Cholewinski A, Shen X, Nelson SG, Lazo JS, Tsang M, Hukriede NA. Automated image-based phenotypic analysis in zebrafish embryos. Dev Dyn. 2009;238:656–663. doi: 10.1002/dvdy.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Rebbert ML, Andreazzoli M, Takahashi N, Toyama R, Zimmerman S, Whitman M, Dawid IB. Regulation of the Lim-1 gene is mediated through conserved FAST-1/FoxH1 sites in the first intron. Dev Dyn. 2002;225:448–456. doi: 10.1002/dvdy.10176. [DOI] [PubMed] [Google Scholar]