SUMMARY
Cancer is driven by mutation. Worldwide, tobacco smoking is the major lifestyle exposure that causes cancer, exerting carcinogenicity through >60 chemicals that bind and mutate DNA. Using massively parallel sequencing technology, we sequenced a small cell lung cancer cell line, NCI-H209, to explore the mutational burden associated with tobacco smoking. 22,910 somatic substitutions were identified, including 132 in coding exons. Multiple mutation signatures testify to the cocktail of carcinogens in tobacco smoke and their proclivities for particular bases and surrounding sequence context. Effects of transcription-coupled repair and a second, more general expression-linked repair pathway were evident. We identified a tandem duplication that duplicates exons 3-8 of CHD7 in-frame, and another two lines carrying PVT1-CHD7 fusion genes, suggesting that CHD7 may be recurrently rearranged in this disease. These findings illustrate the potential for next-generation sequencing to provide unprecedented insights into mutational processes, cellular repair pathways and gene networks associated with cancer.
INTRODUCTION
More than 1 billion people worldwide smoke tobacco1. With 20x greater risk of developing lung cancer than non-smokers and increased risk of many other tumour types, a smoker's lifestyle choice represents the most significant carcinogenic exposure confronting health services today. Tobacco smoke contains more than 60 mutagens that bind and chemically modify DNA2,3, and these brand the lung cancer genome with characteristic mutational patterns. Point mutations in, for example, TP53 and KRAS show different signatures between smokers and non-smokers with lung cancer2-4. However, such studies have been limited to a few genes, and it is unclear how representative these findings are of mutational processes across the whole genome5. In vitro assays and mouse models have been important tools for testing the mutagenicity of individual chemical constituents of tobacco smoke, but are of limited value for generalising to the complexity of smoking behaviours, systemic metabolism and cancer development in humans. Massively parallel sequencing technologies promise the capacity to paint a genome-wide portrait of mutation in human cancer. Such data will provide unprecedented insights into the relative contributions of different tobacco carcinogens to mutation in vivo, the effects of local DNA structure on mutability and the cellular defence mechanisms against exogenous mutagens.
Lung cancer is the leading cause of cancer-related deaths worldwide, developing in more than a million new patients annually6. Small cell lung cancer (SCLC), representing 15% of cases, is a distinct subtype associated with a typical clinical picture of early metastasis, initial response to chemotherapy but subsequent relapse, and a 2-year survival of <15%7. Several tumour suppressor genes are inactivated, including TP53 (80-90% of cases8), RB1 (60-90% of cases9,10) and PTEN (13% of cases11). Infrequent activating mutations have been found in PIK3CA, EGFR and KRAS (all 10% or lower; http://www.sanger.ac.uk/genetics/CGP/cosmic/), and MYC is amplified in 20% of cases.
The development of massively parallel sequencing technologies makes it feasible to catalogue all classes of somatically acquired mutation in a cancer, including base substitutions12,13, insertions and deletions (indels)12,13, copy number changes14 and genomic rearrangements14. Reports from high-coverage sequencing of two acute myeloid leukaemia genomes have been published, which have concentrated on detecting point mutations in exons and regulatory regions12,13. Here, we report the first detailed analysis of a human cancer classically associated with tobacco smoking, giving unprecedented insights into the mutational burden associated with this lifestyle choice. Such analyses signpost the advances that will be made in our understanding of the pathogenesis of cancer as we sequence hundreds to thousands of human tumours15.
RESULTS
Sequencing of a SCLC cell line
Most small cell lung cancers are not surgically resected7, meaning that cell lines are an indispensable resource for studying this disease. NCI-H209 is an immortal cell line derived from a bone marrow metastasis of a 55 year old male with SCLC, taken before chemotherapy16. The smoking history of the patient is not recorded16. However, the specimen showed histologically typical small cells with classic neuroendocrine features: >97% of such tumours are associated with tobacco-smoking17,18. An EBV-transformed lymphoblastoid line, NCI-BL209, has been generated from the patient. NCI-H209 has been extensively characterised by spectral karyotyping, capillary sequencing and high-resolution copy-number array (http://www.sanger.ac.uk/genetics/CGP/cosmic/).
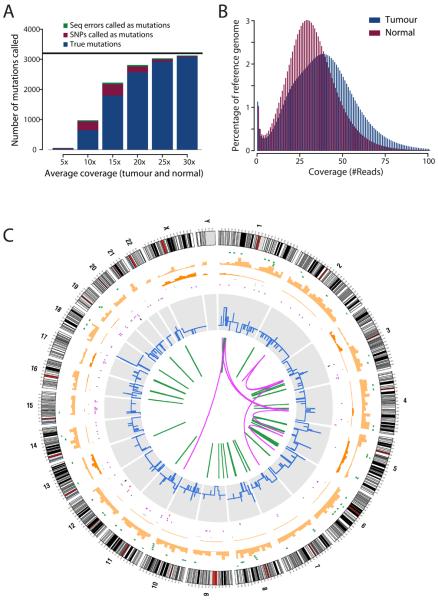
Using the SOLiD platform, we generated 25bp short-read, mate-pair shotgun sequences from the tumour and matched normal genomes. Based on detailed power calculations, we estimated that tumour and normal genomes should be sequenced to 30-fold depth to identify somatically acquired genetic variants with high sensitivity and distinguish them from both sequencing errors and germline polymorphisms (figure 1A). In total, 112Gb (39x coverage) from the tumour and 90Gb (31x) from the normal were aligned to the reference genome (figure 1B).
Figure 1.
The compendium of somatic mutations in a small cell lung cancer genome. (A) Power calculations showing the number of true somatic substitutions detected (blue) and mis-calls (SNPs called as somatic mutations, burgundy, and sequencing errors called as mutations, green) for different levels of sequence coverage. Calculations are based on a true mutation prevalence of 1/Mb (black line). (B) Histogram of the actual coverage achieved per base of the tumour (blue) and normal (burgundy) genomes. (C) Figurative representation of the catalogue of somatic mutations in the genome of NCI-H209. Chromosome ideograms are shown around the outer ring and are oriented pter-qter in a clockwise direction with centromeres indicated in red. Other tracks contain somatic alterations: validated insertions (light green rectangles); validated deletions (dark green rectangles); heterozygous (light orange bars) and homozygous (dark orange bars) substitutions shown by density per 10 megabases; coding substitutions (coloured squares; silent in grey, missense in purple, nonsense in red and splice site in black); copy number (blue lines); validated intrachromosomal rearrangements (green lines); validated interchromosomal rearrangements (purple lines).
Bioinformatic algorithms were developed to identify somatically acquired genetic variation from the sequencing data (supplementary figure 1, supplementary tables 1-5), subjected to rigorous validation by PCR and capillary sequencing. We had previously identified 29 base substitutions, of which 22 (76%) were called by our algorithm from the SOLiD sequencing data (supplementary results; supplementary table 6). 79 novel coding substitutions and 354 randomly chosen genome-wide variants called by the algorithm were also tested. 77 (97%) of the coding substitutions and 333 (94%) of the random variants were confirmed as genuine somatic mutations (supplementary table 7). Neither of two known indels in coding sequence was identified. Of putative somatic indels that were called, the true-positive rate was 25% by capillary sequencing (supplementary results; supplementary table 8). Therefore, only somatic indels which were confirmed by capillary sequencing are reported here. All somatic genomic rearrangements called by anomalous read-pairs were validated by PCR and capillary sequencing across the breakpoint, as previously described14.
Repertoire of somatic mutation
Overall, 22,910 somatically acquired substitutions were identified across the NCI-H209 genome, and a further 65 indels, 334 copy number segments and 58 structural variants were confirmed (table 1, figure 1C, supplementary tables 1-5).
Table 1.
Somatically acquired genomic variants of all classes in a SCLC genome.
| Variant | Number |
|---|---|
| Somatic substitution | 22,910 |
| Coding | 132 (0.6%) |
| Nonsense | 4 |
| Non-synonymous | 92 |
| Synonymous | 36 |
| Non-coding, transcribed | 181 (0.8%) |
| Untranslated region (UTR) | 118 |
| Non-coding RNA | 63 |
| Intronic | 6464 (28%) |
| Splice site | 3 |
| Other intronic | 6461 |
| Intergenic | 16,131 (70%) |
|
| |
| Insertions and deletions | 65 |
| Coding (frameshift) | 2 (3%) |
| Intronic | 25 (38%) |
| Intergenic | 38 (58%) |
|
| |
| Genomic rearrangements | 58 |
| Deletions | 18 (31%) |
| Tandem duplications | 9 (16%) |
| Other non-inverted intrachromosomal rearrangements | 9 (16%) |
| Inverted intrachromosomal rearrangements | 15 (26%) |
| Interchromosomal rearrangements | 7 (12%) |
|
| |
| Copy number segments | 334 |
For point mutations in coding regions, we found the previously described RB1 C706F mutation, known to abrogate protein function19, and the mutation that disrupts a splice site in TP53. Combined loss of RB1 and TP53 is a characteristic feature of SCLC, confirming that NCI-H209 is genetically typical of this disease. One G>T transversion generated a premature stop codon in MLL2. We have observed clustering of truncating mutations in this gene, a histone methyltransferase, in renal cancer (manuscript submitted). Of coding variants, 92 are predicted to change amino acids, and 36 are synonymous. Since cancer is a clonal disease in which the phenotypic consequences of mutation are subject to Darwinian natural selection, accumulation of mutations conferring selective advantage on cancer subclones will manifest as an excess of non-synonymous mutations. However, the observed non-synonymous:synonymous ratio of 2.56:1 is not significantly different from that expected by chance (p=0.3), suggesting that the majority of coding variants do not confer a selective advantage to the cancer.
Due to the limited throughput of capillary sequencing, there has previously been little attempt to explore regulatory regions of the genome for potential oncogenic mutations. To address this, we extracted somatic substitutions occurring within 2kb either side of known transcription start sites, which would generally include gene promoters. Mutations were evenly distributed across the 4kb regions (supplementary figure 2A). We applied hidden Markov models to predict which substitutions might affect transcription factor binding sites. The distribution observed was no different to that seen in random, simulated sets of ‘mutations’ (supplementary figure 2B), suggesting that, analogous to substitutions in coding sequence, most of those found in regulatory regions are selectively neutral to the cancer. Nonetheless, as with coding mutations, there may be a small number which alter transcription factor binding and affect gene regulation, thus providing phenotypic variation for selection to act upon. For example, a T>G mutation 49bp upstream of the transcription start site of a gene in the RAS oncogene family, RAB42, is predicted to have significant disruptive effects on a potential binding motif for the RAS-responsive RREB1 transcription factor (p=3×10−98; supplementary figure 2C).
Taken together, these data suggest that the majority of mutations in coding and promoter regions of the NCI-H209 genome are passenger events, conferring no selective advantage to the cells. Ranking algorithms can be useful to prioritise variants for further study, but the key evidence for identifying driver mutations is recurrence in independent tumour samples, supplemented by functional studies.
Multiple mutation signatures in NCI-H209
Tobacco smoke contains more than 60 carcinogens which bind and chemically modify DNA, characteristically forming bulky adducts at purine bases (guanine and adenine)3. Adducts distort the DNA helix and, if not corrected by nucleotide excision repair (NER) or other pathways, allow non-Watson-Crick pairing during DNA replication. The physicochemical properties of the mutagen determine which adduct is formed, what repair mechanism is induced and which mis-pairing is permissible3. The substantial mutational load carried in the NCI-H209 genome allows us to discern with great statistical power several distinct mutation signatures, genomic records of the medley of mutagens deposited in the airways and lungs by tobacco smoking.
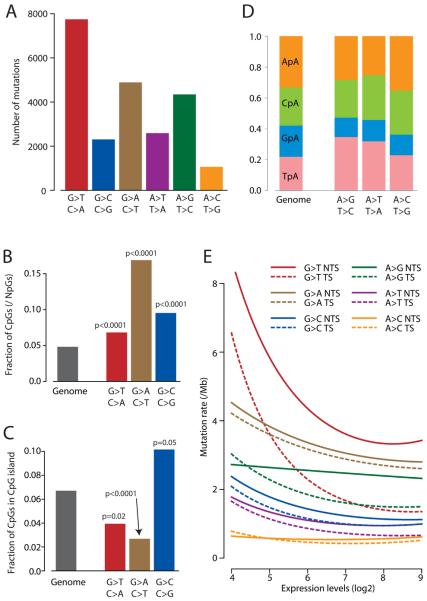
G>T/C>A transversions were the commonest change observed (34%), followed by G>A/C>T (21%) and A>G/T>C (19%) transitions (figure 2A). This distribution is remarkably similar to the pattern of substitutions observed in TP53 in SCLC cases curated from the published literature (supplementary figure 3). This implies firstly that the NCI-H209 genome is typical of SCLC, and therefore of tobacco-associated mutational profiles, and secondly that the majority of mutations were acquired in vivo, not during cell culture. G>T transversions caused by polycyclic aromatic hydrocarbons occur more frequently at methylated CpG dinucleotides in vitro and in TP5320,21. To explore this genome-wide, we compared the base preceding G>T mutations with the base before wild-type guanines in NCI-H209 (figure 2B). CpG dinucleotides were significantly enriched amongst the G>T mutation set compared to controls (odds ratio (OR), 1.5; 95% CI, 1.3-1.6; p<0.0001). We can use the fact that only 10-20% of CpG dinucleotides in CpG islands are constitutively methylated compared to 60-70% outside of CpG islands22 to assess how cytosine-methylation affects mutations at the neighbouring guanine (figure 2C). G>T mutations at CpG dinucleotides were significantly more likely to be found outside CpG islands than expected by chance (OR, 1.8; 95% CI, 1.1-2.8; p=0.02), suggesting that these transversions do indeed preferentially occur at methylated CpGs.
Figure 2.
The mutation profile of NCI-H209. (A) Numbers of mutations in each of the 6 possible mutation classes. (B) Fraction of the three classes of guanine mutations occurring at CpG dinucleotides in NCI-H209, with p values reflecting the comparison with the expected fraction across the genome (grey). (C) Fraction of guanine mutations at CpGs which are found in CpG islands for each of the three classes of mutation. P values reflect comparison with the genome-wide fraction (grey) of CpGs found in CpG islands (and hence more likely to be constitutively unmethylated) versus outside CpG islands (high rates of constitutive methylation). (D) Distribution of the four NpA dinucleotides for each of the three classes of adenine mutation in NCI-H209, compared to the expected distribution across the genome (left). (E) Fitted curves showing the effects of gene expression and strand bias on mutation prevalence for the six classes of adenine and guanine mutation in NCI-H209. The y axis is expressed as mutations per Mb of at-risk nucleotides, namely mutations/1,000,000 Gs for G>T.
We next assessed the base preceding the guanine for G>A and G>C mutations (figure 2B). For G>A transitions, striking enrichment of CpG dinucleotides was observed in the mutation set compared to wild-type guanines in the genome (OR, 4.0; 95% CI, 3.7-4.3; p<0.0001), and these showed a strong propensity to occur outside CpG islands (OR, 2.6; 95% CI, 1.6-4.1; p<0.0001). This is consistent with the well-described phenomenon of spontaneous deamination of methylated cytosine to uracil, read as thymine. Although G>C transversions showed a similar enrichment for CpG context (OR, 2.2; 95% CI, 1.9-2.5; p<0.0001), these were significantly more likely to occur within CpG islands (OR, 0.6; 95% CI, 0.4-1.0; p=0.05), suggesting that the carcinogen responsible targets unmethylated CpG dinucleotides. In keeping with previous reports23,24, we found that the guanine base in G>C transversions was more frequently followed by an adenine than expected by chance (OR, 1.4; 95% CI, 1.3-1.5; p<0.0001).
For mutations involving adenines, fewer substitutions of all classes were seen at GpA dinucleotides than expected by chance (p<0.0001; figure 2D), and A>T and A>G occurred significantly more frequently at TpA than expected (p<0.0001). Among somatically acquired indels, single base-pair insertions were more likely to be gains of A or T nucleotides than C or G (8:1). Curiously, single base deletions favoured loss of C/G nucleotides, rather than A/T (26:12), and there was a propensity for the C/G deletions to occur at CC or GG dimers or longer (18/26). In contrast to the frequency of indels at runs of A or T nucleotides, deletions at C or G tracts are not well described, and our findings may reflect a distinct mutation signature.
Thus, the sequence context of the ~23,000 mutations in the NCI-H209 genome provides tremendous power to identify multiple distinctive mutation signatures, not evident from targeted resequencing studies of limited genomic regions.
Imprint of two DNA repair pathways
Several pathways can repair DNA lesions caused by exogenous carcinogens. Bulky adducts on purines are the predominant form of DNA damage induced by tobacco carcinogens, and can be sufficiently disruptive to impede RNA polymerase when they occur on the transcribed strand of genes. Stalled RNA polymerases can recruit the nucleotide excision repair machinery, leading to excision of the altered nucleotide, preventing mutation. In studies of TP53 mutations in lung cancer, G>T transversions occur more frequently on the non-transcribed strand2,5, suggesting that many of the same lesions occurring on the transcribed strand are correctly identified and removed by the cell. We found that guanine and adenine substitutions are generally less frequent on the transcribed than the non-transcribed strand (supplementary figure 4), confirming that purines appear to be the major target of carcinogens in tobacco smoke.
We next correlated mutation prevalence to gene expression (figure 2E). For a given level of gene expression, the effects of transcription-coupled repair are revealed by the significant separation of curves for mutations on the transcribed and non-transcribed strands. We found evidence for significant transcription-coupled repair for G>T transversions (p<0.0001), as well as A>G (p=0.003) and A>T (p=0.03), possibly G>C (p=0.08), but not G>A (p=0.3) or A>C (p=0.8) mutations. Thus, the extent of transcription-coupled repair differs for the various classes of mutation, presumably reflecting differences in the ability of the transcription-coupled repair machinery to recognise and/or repair different adduct lesions.
For most mutations, there appears to be another novel expression-linked repair pathway that operates on both strands and is at least as numerically important as transcription-coupled repair. Thus, significantly lower mutation prevalence, on both transcribed and non-transcribed strands, was observed in more highly expressed genes for G>T (p<0.0001), G>A (p<0.0001), G>C (p<0.0001) and A>T (p<0.0001). Again, there are some interesting differences across mutation classes in the relative contributions of the two repair pathways. For A>G mutations, only transcribed strand mutations decreased with higher gene expression, suggesting that transcription-coupled repair is the more important pathway for preventing such events. In contrast, G>A mutations occurred equally on transcribed and non-transcribed strands, but mutations on both strands were significantly reduced in more highly expressed genes, suggesting that the novel expression-linked repair pathway is more important than transcription-coupled repair here.
Taken together, these data imply that at least two separate DNA repair pathways have been enlisted for protection of the NCI-H209 genome, notwithstanding the difficulties in extrapolating cell line expression levels to in vivo expression during cancer progression. The fact that the two pathways have operated with differing efficacy across the six classes of mutation implies that the lesions have distinct physicochemical effects on DNA structure, with variable recognition and excision by the genome surveillance machinery.
Genomic rearrangements and copy number
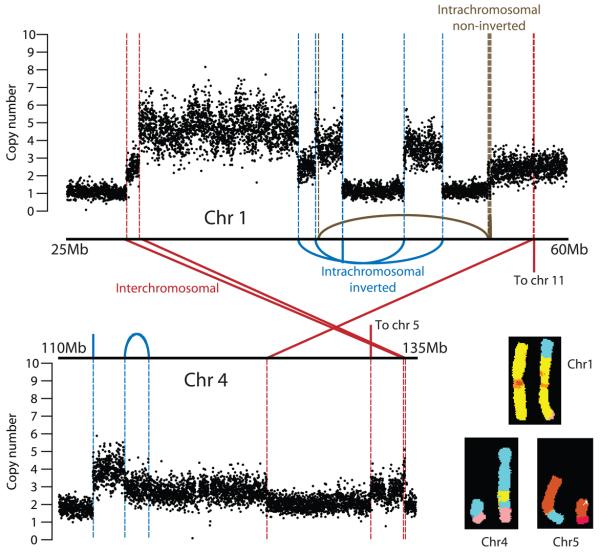
We identified 58 somatically acquired genomic rearrangements in the NCI-H209 genome. These include 18 (31%) deletions and 9 (16%) tandem duplications. The majority of rearrangements, however, cannot be ascribed to classical structural variant patterns, due to the considerably greater complexity of somatically acquired rearrangements compared to germline events. This is exemplified by a set of rearrangements incorporating regions from chromosomes 1p32-36 and 4q25-28 (figure 3). Here, most of the intrachromosomal rearrangements are in inverted orientation, but cannot be classical inversions since they demarcate copy number changes and do not have reciprocal breakpoints. By similar reasoning, most interchromosomal rearrangements also appear to be unbalanced. Other clusters of unbalanced rearrangements were found in NCI-H209, including chromosomes 3q and 5q, and we have seen this phenomenon in many other solid tumour genomes.
Figure 3.
Localised complexes of somatically acquired genomic rearrangements in NCI-H209. Copy number plots across regions on chromosomes 1 and 4 are shown. Inverted intrachromosmal rearrangements (blue), non-inverted intrachromosomal rearrangements (brown) and interchromosomal rearrangements (red) are shown in relation to copy number changes. The inset shows the representative chromosomes on spectral karyotyping. There are three breakpoints between chromosome 1 (yellow) and 4 (light blue), and a translocation between chromosomes 4 and 5 (tan).
Chromosomal rearrangements can juxtapose two genes: if they are in the same orientation with an intact open reading frame, an oncogenic fusion gene may result. In NCI-H209, a predicted in-frame fusion gene was created by a 240kb deletion on chr16, adjoining the first two exons of CREBBP with the 3′ portion of BTBD12, a gene involved in repair of dsDNA breaks25,26. Interestingly, in acute myeloid leukaemia, CREBBP is recurrently fused with MYST327. RT-PCR showed that the predicted CREBBP-BTBD12 fusion transcript is expressed in NCI-H209, but not in 55 other SCLC cell lines. The significance of the predicted fusion gene with respect to cancer development is therefore unclear.
CHD7 rearrangements in SCLC cell lines
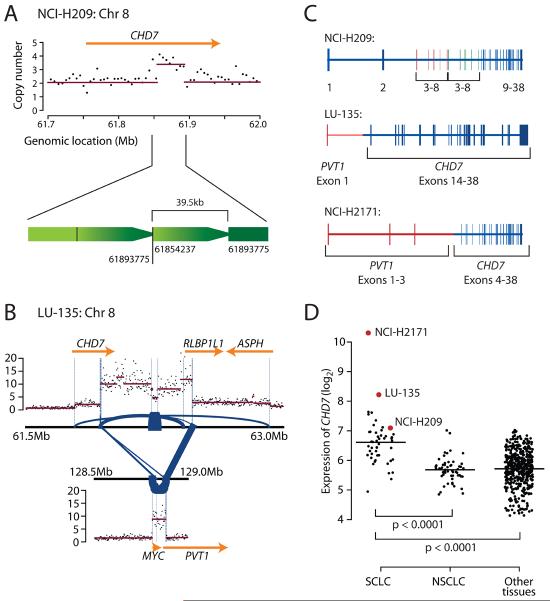
Intrachromosomal rearrangements can also result in internal rearrangements of genes, through loss or duplication of exons. A 39kb tandem duplication was found in CHD7, predicted to lead to in-frame duplication of exons 3-8 (figure 4A). We previously identified a massively amplified and highly expressed fusion gene comprising exons 1-3 of PVT1, a non-coding RNA gene immediately downstream of MYC, and exons 4-38 of CHD7 in another SCLC cell line, NCI-H217114. This raises the possibility that CHD7 rearrangements may be recurrent in SCLC. Using multiplex ligation-dependent probe amplification, we identified a further SCLC cell line (LU-135) with internal exon copy number alterations, among 63 lines screened (supplementary figure 5). LU-135 was therefore studied by mate-pair sequencing (figure 4B). This demonstrated that, as for NCI-H2171, the CHD7 amplicon was linked to MYC amplification. One breakpoint predicted the existence of a fusion gene between exon 1 of PVT1 and exons 14-38 of CHD7 (figure 4C), and by RT-PCR across the breakpoint, this transcript is expressed. In keeping with genomic amplification and active expression of the PVT1 locus, NCI-H2171 and LU-135 show particularly elevated levels of CHD7 transcripts (figure 4D). SCLC cell lines on average show a log2 greater expression of CHD7 than both non-small cell lung cancer lines and other tumour types (p<0.0001).
Figure 4.
CHD7 rearrangements in SCLC cell lines. (A) A somatically acquired 39.5kb tandem duplication is found in NCI-H209. (B) The LU-135 cell line shows co-amplification of the 3′ portion of CHD7 together with MYC and the 5′ portion of PVT1. Blue lines show locations of genomic rearrangements observed in the amplicons, with the thickness of the line proportional to the number of reads spanning the breakpoint. (C) Transcripts resulting from CHD7 rearrangements are an in-frame duplication of exons 3-8 in NCI-H209 and two amplified PVT1-CHD7 fusion genes in NCI-H2171 and LU-135. (D) CHD7 is over-expressed in SCLC compared to both non-small cell lung cancer and other tumour types. LU-135 and NCI-H2171 show massive over-expression of CHD7 in keeping with the genomic amplification present in these cell lines.
Thus, CHD7 is rearranged in three SCLC cell lines. Two carry a PVT1-CHD7 fusion gene in the setting of MYC amplification. PVT1 is a non-coding gene immediately downstream of MYC, and may itself be a transcriptional target of the MYC protein28. Insertion of CHD7 into this locus with subsequent amplification gives the double hit of increased gene copy number and regulatory elements for a co-amplified transcription factor, explaining the massive over-expression seen in these cell lines. PVT1 is recurrently rearranged in variant Burkitt lymphoma translocations29, and may be oncogenic30. The NCI-H209 rearrangement is predicted to duplicate one of the two chromodomains. CHD7 is a chromatin remodeller, promoting enhancer-mediated transcription through association with histone H3K4-methylation31. Histone modifiers have been implicated as cancer genes32, and a family member, CHD5, may function as a tumour suppressor gene33. Recurrent rearrangements of CHD7 in SCLC would be an interesting extension of this theme if functional studies and genomic analyses of primary samples confirm our data.
DISCUSSION
The compendium of somatic alterations in a cancer genome is shaped by multiple intrinsic and extrinsic processes, including exposure to mutagens, selective pressures active in the tissue microenvironment, genomic instability and DNA repair pathways15. The advent of massively parallel sequencing portends an era in which unbiased, genome-wide mutation screens allow the consequences of these processes to be discerned and decoded. Even in this single lung cancer genome, we can identify several distinctive point mutation patterns, reflecting the cocktail of carcinogens present in cigarette smoke, as well as signatures of the partially successful attempts of the cell's surveillance machinery to repair DNA damage. The complete catalogue of somatically acquired mutations in a given cancer harbours the subset of variants that drive the neoplastic phenotype, and a likely candidate, CHD7 rearrangement, has emerged from the NCI-H209 genome.
Tobacco smoke deposits many hundreds of chemicals in the airways and lungs. Each carcinogen-associated mutation represents the consequence of three processes: chemical modification of a purine by a mutagen, failure to repair the lesion by genome surveillance pathways and incorrect nucleotide incorporation opposite the distorted base during DNA replication. G>T transversions are the commonest substitution in NCI-H209, mutations previously linked to polycyclic aromatic hydrocarbons3 and acrolein21 in tobacco smoke. We found enrichment of G>T mutations at CpG dinucleotides, especially outside CpG islands, supporting in vitro evidence that these carcinogens preferentially bind methyl-CpG dinucleotides20,21. Polycyclic aromatic hydrocarbons containing a cyclopentane ring have been associated with G>C transversions34. We found them enriched at CpG dinucleotides, but, in contrast to G>T and G>A mutations, our data suggest that unmethylated-CpGs are the target here, underscoring the remarkable statistical power genome-wide mutation screens give for delineating mutation spectra.
We can also infer signatures of DNA repair in cancer genomes. Transcription-coupled repair is induced by stalling of RNA polymerase at bulky adducts on the transcribed strand, and we saw evidence for this process in NCI-H209. We have also discovered the imprint of a novel and more general form of expression-linked repair, through which mutation frequency is reduced on both strands in highly expressed genes. The expression-linked decrease in mutation frequency may reflect global genomic nucleotide excision repair, in which distorting adducts are corrected genome-wide35. Why this pathway should be more effective in highly transcribed regions is unclear. One possibility is that ssDNA formed on both strands during transcription facilitates recognition of the adduct, and there is some evidence that components of the NER pathway can recognize adducts in ssDNA36. Strikingly, some mutation types were repaired almost exclusively by transcription-coupled repair (A>G), some showed evidence for only the more general expression-linked repair (G>A), while others had features of both mechanisms (G>T, A>T). Such differences are presumably determined by the physicochemical properties of the multitudinous adducts induced by carcinogens present in tobacco smoke.
On average, lung cancer develops after 50 pack-years of smoking37. Candidate gene resequencing studies suggest that the mutation prevalence in NCI-H209 is similar to that of primary lung cancers38,39. If the majority of mutations derive from the mélange of mutagens present in tobacco smoke, the clone of cells that ultimately becomes cancerous would acquire, over its lifetime, an average of one mutation for every 15 cigarettes smoked. If this is the case in a localised cluster of cells, then the number of mutations acquired across the whole bronchial tree from even one cigarette must be substantial. The data presented here demonstrate the power of whole genome sequencing to disentangle the many complex mutational signatures found in cancers induced by tobacco smoke.
METHOD SUMMARY
Massively parallel, shotgun sequencing of genomic DNA from NCI-H209 and the matched EBV-transformed B-cell line was performed on the SOLiD platform (Life Technologies, Foster City, CA) to a target >30-fold depth. Mate-pair 25bp reads were mapped back to the NCBI36 reference genome in 2-base encoded colour space. Bespoke bioinformatic algorithms were developed to identify all classes of somatically acquired genetic variant from the sequencing data. PCR and capillary sequencing on DNA from both tumour and normal lines was performed to confirm all putative somatically acquired genomic rearrangements and indels, together with random samples of coding and non-coding substitutions. To identify potential regulatory consequences of point mutations, substitutions in promoter regions were compared against random samples of simulated variants for effects on predicted transcription-factor binding sites. The sequence context of mutation positions was compared against randomly sampled genomic positions which had sufficient sequence coverage by chi-squared tests. Gene expression levels for NCI-H209 were determined on the Affymetrix U133A microarray, and analysed for mutation prevalence by Poisson regression.
METHODS
Power calculations
To estimate the levels of coverage required for accurate detection of somatically acquired base substitutions, we make the following assumptions:
Tumour DNA is approximately diploid, with 3.2Gb per cell.
All reads can be mapped back correctly to the genome, even if they have a mutation, SNP or sequencing error.
Mutations and SNPs are heterozygous (homozygous variants are much easier to detect accurately).
Mutation prevalence 1/Mb; SNP prevalence 1/kb; sequencing error rate averaged 1/100 bases, with moderate degrees of systematic error (allowing different bases in the genome to have different error rates); overdispersion 2.0 (ratio of the variance of coverage as reads/base exceeds the mean reads/base).
- We use the following strategy for calling a mutation at a given base, which is covered by CT reads in the tumour DNA and CN reads in the normal DNA:
- Of the CT reads in tumour DNA, the substitution is seen in a greater number of reads than the other two possible substitutions, with at least as many reads as a particular threshold, κ(CT).
- Of the CN reads in the normal DNA, the number of reads containing a particular substitution must be fewer than a threshold, κ(CN).
The probability of calling a true mutation and the probabilities of mis-calls of sequencing errors and SNPs as mutations are calculated for each level of coverage of tumour and normal genomes, based on the κ(CT) and κ(CN) thresholds above, a binomial distribution of alleles (for heterozygous variants) and sequencing errors following a beta-binomial distribution. The distribution of coverages for the genome is then modeled by a negative binomial distribution to account for overdispersion, and the overall true and false positive rates summed.
Sequencing
Genomic DNA was used to construct mate pair libraries of different insert sizes, The smallest insert libraries were from 600bp-800bp and the largest were 3-4kb. Library preparation, emulsion PCR, slide preparation and sequencing were all performed according to the manufacturer's protocol (Applied Biosystems SOLiD Library Preparation Protocol). Sequencing generated 25bp tags in colour space from each end of the DNA fragment. Primary data analysis including image analysis and base-calling was carried out with the corona pipeline (Applied Biosystems). Alignment of reads to the NCBI36 reference genome (translated into colour space) was performed with corona-lite (version 0.32), allowing 2 mismatches in mapping and up to 4 in the pairing step. Duplicate read pairs with identical coordinates were removed, and only uniquely mapping reads were used for analysis.
Substitution detection
The corona-lite pipeline (version 0.32) was used to generate a preliminary list of variant bases from the uniquely aligning reads. We used the optimal thresholds defined in point 5 of the power calculations above (based on a mutation prevalence of 8/Mb, as estimated from capillary sequence data in COSMIC) to determine whether there was sufficient evidence for calling a somatic substitution or not at each base in this preliminary list. Resulting tumour-specific substitutions were further filtered to remove (i) those residing in regions of LOH in the normal cell line, (ii) those potentially due to misalignment in segmental duplications and near sequence gaps, (iii) those corresponding to polymorphic positions in dbSNP, (iv) those potentially due to misalignment or miscalls as they are adjacent to SNPs or within 5 bp of insertions and deletions and (v) those where all supporting reads contained the putative variant in the first or last 5bp of the read (to reduce effects of misalignment across indels). Substitutions were annotated using Ensembl version 52.
The bioinformatic algorithm initially compares the tumour genome against the published reference genome to identify variants, before comparing it against the EBV-transformed line to determine whether the variant is somatically acquired or germline. Thus, a mutation in the B-cell line induced by EBV transformation will not be falsely detected as a variant in the SCLC line. However, one potential artefact resulting from changes induced by EBV transformation is in regions where there is loss of heterozygosity – this could potentially lead to false mutation calls in the tumour because the EBV line contains only one allele at heterozygous SNP positions. For this reason, we excluded regions of LOH in the EBV line from the mutation-calling algorithm (filter criterion (i) above).
LOH in the normal cell line was determined by analysis of the EBV-transformed B-cell line on an Affymetrix SNP6 array, using hidden Markov model algorithms for identification of allele-specific copy number.
Insertion & deletion detection
Small insertions (up to 3 bp) and deletions (up to 11 bp) were called using corona-lite (version 0.4). Indels found in the tumour and not in the normal were further filtered to require (i) minimum three supporting tumour reads, (ii) minimum one read on each strand, (iii) no LOH in the normal, (iv) maximum 100X coverage (to remove regions of misalignment) and (v) minimum 30X normal coverage (to reduce the number of germline indels in the set).
Structural variant detection
Abnormal read pairs that mapped to the genome with MAQ at an unexpected distance or orientation were identified, grouped and filtered as described14. To define somatic variants, at least 8 independent read pairs were required in the tumour, and a maximum number in the normal were allowed as defined based on the same optimal thresholds used in substitution calling.
Confirmation by capillary sequencing
A set of 79 novel coding mutations and 354 randomly chosen, genome-wide substitutions predicted by the algorithm, and 262 small insertions and deletions, were taken forward for independent validation by capillary sequencing across the region of the mutation. Structural variants were confirmed by PCR across the breakpoint and capillary sequencing the breakpoint. All confirmations were done in both the tumour and normal to determine if the variants were somatic or germline.
Copy number determination
Reads were counted in windows across the genome, corrected for genome uniqueness as described14. Circular binary segmentation was used to segment the data, using a Bioconductor package originally designed for array-CGH data40. The adaptation of the algorithm for shotgun sequencing data has been described in detail elsewhere14.
Substitution analysis
For mutation context, the bases ±10 bp from the mutation were extracted and the number of each base counted. Context of equivalent changes (eg. C>T and G>A) were combined. The background context was determined from 200,000 randomly sampled bases from the genome at positions in which sufficient coverage of the tumour and normal genomes was achieved. Strand bias was calculated based on annotating each mutation as to whether it fell on the transcribed or untranscribed strand in Ensembl 52. Gene expression data were derived from the Affymetrix U133A array. Chi-squared testing was performed to detect departures of observed mutation context from that expected in the covered genome. Poisson regression was used for the analysis of the effects of gene expression on mutation prevalence, incorporating the number of at-risk bases in each gene footprint as the offset, allowing quadratic terms for the relationship between expression and mutation prevalence, and using a dummy variable for transcribed vs non-transcribed strand mutations. The image of the entire NCI-H209 genome was produced using Circos41.
Analysis of promoter mutations
The promoter region of each gene was defined as the 2000bp each side of the transcription start site (TSS) for its representative transcript. Some of the promoters defined this way overlapped, giving 63.9 Mbp of unique sequence. We mapped the substitutions to the gene with the nearest TSS, and confirmed that the reference bases provided matched the corresponding bases in the reference genome. We investigated the impact of the somatic substitutions on transcription factor binding to the promoter regions using a Sunflower model42 based on the JASPAR databases's43 set of vertebrate transcription factors. The database contains two matrices for the transcription factor Lhx3: one based on human data, and one from mouse. The mouse factor was excluded and the remaining 100 matrices used to build the model with an unbound prior probability of 0.99; leaving 1 × 10−4 for each factor. The G+C content was close to 50% for the promoters for both lines, so we used a at distribution for the unbound emission probabilities (A = C = G = T = 0.25).
The reference sequences for each promoter, plus 300bp of padding sequence on each end (4600 bp per promoter in total) were downloaded from Ensembl 53. The padding sequence is necessary for two reasons. First, the posterior probability decoding of Sunflower is unreliable near the beginning and end of a sequence42. Second, if a transcription factor binding site overlaps the beginning or end of the sequence, it cannot be correctly modeled as only part of its state loop will be traversed.
For the set of observed substitutions, we generated the variant promoters from the reference sequences. We also randomly sampled the unobserved substitutions according to the mutation distributions of the observed substitutions. As a further control, these were restricted to the same mutation type per promoter as the observed substitutions. For example, if a promoter had a C>T substitution at position −160, the set of possible unobserved substitutions for that promoter would be all of the other possible C>T and G>A substitutions. If a promoter had multiple observed substitutions, any of those types were allowed in the sampling. Variant sequences for 1000 sets of random unobserved substitutions were generated. The set size was the same as the observed set size, for a total of 315,000 unobserved substitutions. Using the pairwise comparison functionality of Sunflower and the 100-factor JASPAR model, we compared the variant sequences to the reference sequences, obtaining a relative entropy value for each. To ensure that the results were related to real binding sites and not random noise, we randomly shuffled the columns of all the JASPAR position weight matrices (PWMs) to create new sets of matrices. From 100 such sets, we built Sunflower models using the same parameters as for the real JASPAR data, and compared the variant promoter sequences to their references for the observed substitutions.
CHD7 analysis
Multiplex ligation-dependent probe amplification (MLPA) for CHD7 was performed using the commercially available assay (MRC-Holland b.v., Amsterdam, Netherlands) according to manufacturer's instructions. Identification of rearrangements in LU-135 was performed using a 3-4kb mate-pair library sequenced on the SOLiD platform, as described above. Rearrangements were confirmed by PCR and capillary sequencing across the breakpoint. RT-PCR across aberrant exon junctions was used to assess expression of the PVT1-CHD7 fusion transcripts (NCI-H2171 and LU-135) and the internal duplication of exons 3-8 (NCI-H209). RT-PCR products were capillary sequenced to confirm their veracity. Gene expression data were derived from the Affymetrix U133A array for the core cell lines listed on our website (http://www.sanger.ac.uk/genetics/CGP/CellLines/).
Supplementary Material
Supplementary figure 1. Outline of bioinformatic algorithms for identification of somatically acquired genetic variants of all classes. T, tumour genome; N, normal genome; seq, sequencing; var, variant; κ(CT), threshold for number of reads containing variant allele required in tumour genome sequence data, for given level of coverage at that base; κ(CN), maximum number of reads containing variant allele in normal genome sequence data at that base.
Supplementary figure 2. Analysis of somatic substitutions falling in regions predicted to contain the promoters of protein-coding genes in the Ensembl database. (A) Number of substitutions falling in 100bp bins for the 2kb either side of gene transcription start sites in the Ensembl database. (B) Comparison of the predicted effects on transcription factor binding sites of observed mutations (red) and random sets of mutations (blue). Relative entropy denotes the magnitude of the predicted effect of the variant on the likelihood of transcription factor binding. (C) Example of a mutation predicted to obliterate a binding site for the RAS-responsive transcription factor, RREB1. The consensus binding sequence for RREB1 is (CCCCACCA / TGGTGGGG), and a putative element is seen 49bp downstream of the transcription start site of the gene RAB42. A T>G somatic substitution in NCI-H209 is predicted to reduce the potential for RREB1 to bind to the site with high confidence (p=3×10−98).
Supplementary figure 3. Comparison of the mutation spectrum between (A) 245 published substitutions in TP53 in SCLC cases recorded by the IARC database (http://www-p53.iarc.fr/) and (B) 22,910 genome-wide somatic substitutions in NCI-H209.
Supplementary figure 4. Strand bias in mutation prevalence across transcribed regions of the NCI-H209 genome. The numbers of mutations of each of the 6 classes of guanine and adenine mutations are shown, split by whether they occur on the transcribed (TS) or non-transcribed strand (NTS), as defined by the Ensembl transcript database.
Supplementary figure 5. Multiplex ligation-dependent probe amplification (MLPA) of CHD7 in SCLC cell lines. Each histogram shows the normalised signal intensity for probes in exon order across the gene, followed by probes from control genomic regions. The first histogram is a representative example from a cell line without evidence for exon copy number variation. The following three plots represent the three cell lines in which rearrangements of CHD7 were identified.
Supplementary table 1. Genome-wide list of 22,910 somatic substitutions identified by bioinformatic analysis of sequencing data from NCI-H209. Chromosome and base-pair are shown (by the NCBI36 genome build), together with the reference base at that position, the predicted variant, the zygosity and the sequence context (10bp either side of and including the variant base).
Supplementary table 2. Genome-wide list of 65 somatically acquired small insertions and deletions identified by bioinformatic analysis of short-read sequencing data from NCI-H209, all subsequently confirmed as genuine, somatic changes by capillary sequencing. Chromosome and base-pair (in a range where necessary) are shown according to the NCBI36 genome build, together with the zygosity, the predicted variant, status and the sequence context (10bp either side of and including the variant base).
Supplementary table 3. List of 58 somatically acquired genomic rearrangements in NCI-H209. All structural variants have been confirmed by PCR across the breakpoint, with bidirectional sequencing confirming the segments involved. All but two have had the breakpoint annotated to base-pair resolution: for the other two, we provide a range of genomic positions encompassing the breakpoints. Length and sequence of either microhomology or non-templated sequence at the junction are shown.
Supplementary table 4. List of copy number segments across the genome for NCI-H209. The copy number changes associated with identified somatically acquired genomic rearrangements are marked.
Supplementary table 5. Table of somatically acquired substitutions and indels in coding sequence. The mutations, genomic positions and genes are shown. Predicted protein consequences are shown based on the Ensembl transcript quoted. The validation status refers to whether the variant has been confirmed by capillary sequencing or whether no confirmatory sequencing was attempted.
Supplementary table 6. Sensitivity of bioinformatic algorithm for identifying known somatic single-base substitutions in NCI-H209. From capillary sequencing of ~4000 genes (5Mb), we previously identified 29 somatic single-base substitutions. Of these, 22 (76%) were correctly identified independently from the SOLiD sequencing data.
Supplementary table 7. Specificity of bioinformatic algorithm for identifying somatic single-base substitutions in NCI-H209. A set of 79 novel coding substitutions and 354 randomly chosen genome-wide variants called by the algorithm were tested by capillary sequencing to ascertain veracity. Of these, 77 (97%) of the coding substitutions and 333 (94%) of the random variants were confirmed as genuine somatic mutations. The true and false positives are shown, together with possible explanations, where identified, for the false positives.
Supplementary table 8. Specificity of bioinformatic algorithm for identifying somatic indels in NCI-H209. A set of 262 novel indel calls made by the algorithm were assessed by capillary sequencing, of which 65 (25%) were confirmed as genuine somatic indels.
ACKNOWLEDGEMENTS
This work was supported by the Wellcome Trust (grant reference 077012/Z/05/Z). PJC is a Kay Kendall Leukaemia Fund Intermediate Clinical Fellow. IV is supported by the Human Frontiers Science Programme. JM and AG are supported by an NCI grant (NCI P50CA70907).
Footnotes
AUTHOR CONTRIBUTIONS
EDP undertook the development and implementation of bioinformatic algorithms for analysis of the sequencing data, assisted by Meynert, DJ, DB, KWL, CG, GO, LS, LC, MJ, CL, Marshall, Menzies, AB, JWT, Mangion, YAS, SFM, HEP, EFT, GLC, CCL, EB, MDR KJM and PJC. PJS, SOM and DJM were responsible for generating libraries and running sequencers, together with downstream validation analyses, assisted by MLL, IV, SNZ, HRD, LJM, CL and SE. Minna and AG generated the cell lines. EDP, MRS, PAF and PJC directed the research and wrote the manuscript, which all authors have approved.
AUTHOR INFORMATION
Genome sequence data have been deposited at the European Genome-Phenome Archive (EGA, http://www.ebi.ac.uk/ega/) which is hosted by the EBI, under accession number EGAS00000000051. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. SOM, Mangion, YAS, SFM, HEP, EFT, GLC, CCL, MDR, KJM are employees of Life Technologies, the manufacturers of the SOLiD platform. The other authors have no conflicts of interest to report. Correspondence and requests for materials should be addressed to pc8@sanger.ac.uk.
REFERENCES
- 1.Jha P. Avoidable global cancer deaths and total deaths from smoking. Nat Rev Cancer. 2009;9:655–64. doi: 10.1038/nrc2703. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem Res Toxicol. 2008;21:160–71. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeifer GP, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–51. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 4.DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res. 2004;567:447–74. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Rodin SN, Rodin AS. Origins and selection of p53 mutations in lung carcinogenesis. Semin Cancer Biol. 2005;15:103–12. doi: 10.1016/j.semcancer.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Toh CK. The changing epidemiology of lung cancer. Methods Mol Biol. 2009;472:397–411. doi: 10.1007/978-1-60327-492-0_19. [DOI] [PubMed] [Google Scholar]
- 7.Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc. 2008;83:355–67. doi: 10.4065/83.3.355. [DOI] [PubMed] [Google Scholar]
- 8.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 9.Horowitz JM, et al. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci U S A. 1990;87:2775–9. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori N, et al. Variable mutations of the RB gene in small-cell lung carcinoma. Oncogene. 1990;5:1713–7. [PubMed] [Google Scholar]
- 11.Yokomizo A, et al. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene. 1998;17:475–9. doi: 10.1038/sj.onc.1201956. [DOI] [PubMed] [Google Scholar]
- 12.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mardis ER, et al. Recurring Mutations Found by Sequencing an Acute Myeloid Leukemia Genome. N Engl J Med. 2009 doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carney DN, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–23. [PubMed] [Google Scholar]
- 17.Barbone F, Bovenzi M, Cavallieri F, Stanta G. Cigarette smoking and histologic type of lung cancer in men. Chest. 1997;112:1474–9. doi: 10.1378/chest.112.6.1474. [DOI] [PubMed] [Google Scholar]
- 18.Lubin JH, Blot WJ. Assessment of lung cancer risk factors by histologic category. J Natl Cancer Inst. 1984;73:383–9. doi: 10.1093/jnci/73.2.383. [DOI] [PubMed] [Google Scholar]
- 19.Kaye FJ, Kratzke RA, Gerster JL, Horowitz JM. A single amino acid substitution results in a retinoblastoma protein defective in phosphorylation and oncoprotein binding. Proc Natl Acad Sci U S A. 1990;87:6922–6. doi: 10.1073/pnas.87.17.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denissenko MF, Chen JX, Tang MS, Pfeifer GP. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc Natl Acad Sci U S A. 1997;94:3893–8. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Z, Hu W, Hu Y, Tang MS. Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc Natl Acad Sci U S A. 2006;103:15404–9. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fekairi S, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svendsen JM, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozman M, et al. Type I MOZ/CBP (MYST3/CREBBP) is the most common chimeric transcript in acute myeloid leukemia with t(8;16)(p11;p13) translocation. Genes Chromosomes Cancer. 2004;40:140–5. doi: 10.1002/gcc.20022. [DOI] [PubMed] [Google Scholar]
- 28.Carramusa L, et al. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol. 2007;213:511–8. doi: 10.1002/jcp.21133. [DOI] [PubMed] [Google Scholar]
- 29.Zeidler R, et al. Breakpoints of Burkitt's lymphoma t(8;22) translocations map within a distance of 300 kb downstream of MYC. Genes Chromosomes Cancer. 1994;9:282–7. doi: 10.1002/gcc.2870090408. [DOI] [PubMed] [Google Scholar]
- 30.Guan Y, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–55. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 31.Schnetz MP, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagchi A, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–75. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 34.Jackson MA, Lea I, Rashid A, Peddada SD, Dunnick JK. Genetic alterations in cancer knowledge system: analysis of gene mutations in mouse and human liver and lung tumors. Toxicol Sci. 2006;90:400–18. doi: 10.1093/toxsci/kfj101. [DOI] [PubMed] [Google Scholar]
- 35.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, et al. Interactions of human replication protein A with single-stranded DNA adducts. Biochem J. 2005;385:519–26. doi: 10.1042/BJ20041151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lubin JH, et al. Cigarette smoking and cancer risk: modeling total exposure and intensity. Am J Epidemiol. 2007;166:479–89. doi: 10.1093/aje/kwm089. [DOI] [PubMed] [Google Scholar]
- 38.Davies H, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–5. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 39.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 41.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffmann MM, Birney E. An effective model for natural selection in promoters. 2009 doi: 10.1101/gr.096719.109. Manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vlieghe D, et al. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–7. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Outline of bioinformatic algorithms for identification of somatically acquired genetic variants of all classes. T, tumour genome; N, normal genome; seq, sequencing; var, variant; κ(CT), threshold for number of reads containing variant allele required in tumour genome sequence data, for given level of coverage at that base; κ(CN), maximum number of reads containing variant allele in normal genome sequence data at that base.
Supplementary figure 2. Analysis of somatic substitutions falling in regions predicted to contain the promoters of protein-coding genes in the Ensembl database. (A) Number of substitutions falling in 100bp bins for the 2kb either side of gene transcription start sites in the Ensembl database. (B) Comparison of the predicted effects on transcription factor binding sites of observed mutations (red) and random sets of mutations (blue). Relative entropy denotes the magnitude of the predicted effect of the variant on the likelihood of transcription factor binding. (C) Example of a mutation predicted to obliterate a binding site for the RAS-responsive transcription factor, RREB1. The consensus binding sequence for RREB1 is (CCCCACCA / TGGTGGGG), and a putative element is seen 49bp downstream of the transcription start site of the gene RAB42. A T>G somatic substitution in NCI-H209 is predicted to reduce the potential for RREB1 to bind to the site with high confidence (p=3×10−98).
Supplementary figure 3. Comparison of the mutation spectrum between (A) 245 published substitutions in TP53 in SCLC cases recorded by the IARC database (http://www-p53.iarc.fr/) and (B) 22,910 genome-wide somatic substitutions in NCI-H209.
Supplementary figure 4. Strand bias in mutation prevalence across transcribed regions of the NCI-H209 genome. The numbers of mutations of each of the 6 classes of guanine and adenine mutations are shown, split by whether they occur on the transcribed (TS) or non-transcribed strand (NTS), as defined by the Ensembl transcript database.
Supplementary figure 5. Multiplex ligation-dependent probe amplification (MLPA) of CHD7 in SCLC cell lines. Each histogram shows the normalised signal intensity for probes in exon order across the gene, followed by probes from control genomic regions. The first histogram is a representative example from a cell line without evidence for exon copy number variation. The following three plots represent the three cell lines in which rearrangements of CHD7 were identified.
Supplementary table 1. Genome-wide list of 22,910 somatic substitutions identified by bioinformatic analysis of sequencing data from NCI-H209. Chromosome and base-pair are shown (by the NCBI36 genome build), together with the reference base at that position, the predicted variant, the zygosity and the sequence context (10bp either side of and including the variant base).
Supplementary table 2. Genome-wide list of 65 somatically acquired small insertions and deletions identified by bioinformatic analysis of short-read sequencing data from NCI-H209, all subsequently confirmed as genuine, somatic changes by capillary sequencing. Chromosome and base-pair (in a range where necessary) are shown according to the NCBI36 genome build, together with the zygosity, the predicted variant, status and the sequence context (10bp either side of and including the variant base).
Supplementary table 3. List of 58 somatically acquired genomic rearrangements in NCI-H209. All structural variants have been confirmed by PCR across the breakpoint, with bidirectional sequencing confirming the segments involved. All but two have had the breakpoint annotated to base-pair resolution: for the other two, we provide a range of genomic positions encompassing the breakpoints. Length and sequence of either microhomology or non-templated sequence at the junction are shown.
Supplementary table 4. List of copy number segments across the genome for NCI-H209. The copy number changes associated with identified somatically acquired genomic rearrangements are marked.
Supplementary table 5. Table of somatically acquired substitutions and indels in coding sequence. The mutations, genomic positions and genes are shown. Predicted protein consequences are shown based on the Ensembl transcript quoted. The validation status refers to whether the variant has been confirmed by capillary sequencing or whether no confirmatory sequencing was attempted.
Supplementary table 6. Sensitivity of bioinformatic algorithm for identifying known somatic single-base substitutions in NCI-H209. From capillary sequencing of ~4000 genes (5Mb), we previously identified 29 somatic single-base substitutions. Of these, 22 (76%) were correctly identified independently from the SOLiD sequencing data.
Supplementary table 7. Specificity of bioinformatic algorithm for identifying somatic single-base substitutions in NCI-H209. A set of 79 novel coding substitutions and 354 randomly chosen genome-wide variants called by the algorithm were tested by capillary sequencing to ascertain veracity. Of these, 77 (97%) of the coding substitutions and 333 (94%) of the random variants were confirmed as genuine somatic mutations. The true and false positives are shown, together with possible explanations, where identified, for the false positives.
Supplementary table 8. Specificity of bioinformatic algorithm for identifying somatic indels in NCI-H209. A set of 262 novel indel calls made by the algorithm were assessed by capillary sequencing, of which 65 (25%) were confirmed as genuine somatic indels.