Abstract
Metastable and somatically heritable patterns of DNA methylation provide an important level of genomic regulation. In this article, we review methods for analyzing these genome-wide epigenetic patterns and offer a perspective on the ever-expanding literature, which we hope will be useful for investigators who are new to this area. The historical aspects that we cover will be helpful in interpreting this literature and we hope that our discussion of the newest analytical methods will stimulate future progress. We emphasize that no single approach can provide a complete view of the overall methylome, and that combinations of several modalities applied to the same sample set will give the clearest picture. Given the unexpected epigenomic patterns and new biological principles, as well as new disease markers, that have been uncovered in recent studies, it is likely that important discoveries will continue to be made using genome-wide DNA methylation profiling.
Keywords: bisulfite sequencing, CpG island, differential methylation hybridization, DNA methylation, methylation profiling, methylated CpG island recovery assay, methylated DNA immunoprecipitation, methylation-sensitive SNP chip analysis, next-generation DNA sequencing
In 1942, Waddington defined epigenetics as the development of phenotypes from genotypes [1]. Since then, the term has taken on a specific molecular meaning, mostly referring to the patterning of DNA methylation and histone modifications in chromosomes. Epigenetic patterns are largely maintained during somatic cell proliferation, and are important in diverse physiological and pathophysiological phenomena. Among the various types of epigenetic modifications of the genome, DNA methylation is certainly one of the most stable. Methylation of DNA in mammalian cells is generally restricted to the 5 position of the pyrimidine ring of cytosine residues located in CpG dinucleotides (5-methylcytosine [5-mC]). In mammalian genomes, CpG dinucleotides are somewhat depleted on average, but are found densely clustered within sequences known as CpG islands (CGIs). These CGIs are typically in the range of 0.5–2 kb in size and located within 1 kb of transcription start sites. Under normal circumstances most, although not all, CGIs are unmethylated [2–4]. In contrast to CGIs, which are usually protected from methylation, in the remainder of the mammalian genome a high percentage of CpG dinucleotides, both in unique sequences and repetitive elements, are found to be variably to densely methylated [4,5]. DNA methyltransferase enzymes are responsible for creating (i.e., de novo methylation; DNA methyltransferase [DNMT] 3a, DNMT3b) and propagating (i.e., maintenance methylation; DNMT1) tissue-specific patterns of CpG methylation in the human genome (reviewed in [6,7]). As shown in experiments with knockout mice, each of these enzymes is essential for viability of the conceptus to term. Once established, the faithful transmission of methylation patterns to daughter cells is thought to be primarily due to DNMT1 activity and, consistent with this role, the DNMT1 enzyme localizes to DNA replication foci in the nuclei of S-phase cells [8]. The other two enzymes play greater roles in de novo methylation. Interestingly, Métivier et al. reported that DNMT3a and DNMT3b can also actively demethylate 5-mC through deamination [9,10]. On the other hand, Rai et al. suggested that the 5-mC removal in zebrafish embryos is mediated through two enzymatic reactions, namely the deamination of 5-mC by deaminase (AID) and the reparation of G:T mismatch by thymine glycosylase (Mbd4) [11]. For more discussion of this important topic of DNA demethylation, we refer interested readers to a recent review article [12].
A large body of research findings has demonstrated that epigenetic aberrations in cancer cells significantly contribute to tumor initiation, invasion, metastasis and resistance to chemotherapy (reviewed in [13,14]). Silencing of tumor suppressor genes by promoter hypermethylation has been commonly observed in a number of cancers including, but by no means restricted to, colon, bladder, stomach, breast, uterine and renal carcinomas [13,15,16]. This phenomenon is functionally quite important, and in some types of cancers, including gastric carcinomas, it appears that tumor suppressor genes are inactivated more frequently by their promoter hypermethylation than by mutations [17]. Despite hypermethylation in promoter-associated CGIs, many tumors exhibit overall genomic hypomethylation, affecting nonisland sequences prominently, including repetitive elements and the pericentromeric regions [14,18]. In addition to its role in silencing tumor suppressor genes by de novo methylation in malignant cells, DNA methylation is medically important for several reasons, as will be discussed.
Epigenetic therapy with demethylating drugs
Based on the fact that some chemicals can interfere with DNA methyltransferase activity and thereby revert hypermethylated DNA to a hypomethylated state in mammalian genomes, demethylating drugs have been actively studied and validated for clinical applications [19]. Some of them, such as decitabine, or 5-aza-2′-deoxycytidine, are effective anticancer agents in some tumor types, and are now widely used on low-dose schedules, for example to prolong transfusion independence and survival in myelodysplastic syndrome.
DNA methylation & diseases involving imprinted genes
DNA methylation enforces the selective silencing of one parental allele at imprinted loci in normal human tissues. When this dose-regulating mechanism goes awry, owing to mistakes in DNA methylation early in development (‘epimutations’), the result can be any of several well-characterized epigenetic diseases, including the growth disorders Beckwith–Wiedemann syndrome and Silver–Russell syndrome, the neurodevelopmental disorders Angelman syndrome and Prader–Willi syndrome, and the endocrine disorders transient neonatal diabetes mellitus and Albright osteodystrophy. In most of these disorders, some cases are due to mutations (point mutations, microdeletions and large-scale deletions) in the relevant imprinted genes or imprinting control sequences, while other cases are due to epimutations – that is, altered methylation without any mutation in the DNA sequence. DNA methylation is also a mechanism for ‘locking-in’ the silent state of genes on the inactive X-chromosome. Based on recent publications describing unique methylation patterns in human and mouse stem cells, changes in DNA methylation play a role in the differentiation of stem cells into their committed and mature cellular progeny [20].
Epigenetic mechanisms in other diseases
DNA methylation is increasingly being studied in complex human diseases beyond cancer and the imprinting disorders. Altered CpG methylation may have an important role in propagating adverse changes in gene expression in intrauterine growth restriction and pre-eclampsia, common neuropsychiatric disorders and age-related dementias such as Alzheimer’s disease, atherosclerosis, diabetes and the metabolic syndrome, and potentially also in non-neoplastic lung diseases such as pulmonary fibrosis.
Methods for analyzing DNA methylation
General considerations in profiling DNA methylation
Here we review currently available and emerging laboratory methods for investigating DNA methylation patterns in health and disease. We emphasize cancer research, but the methods are equally applicable to other disease states. Current research in epigenetics is largely driven by novel technologies [21], and over the past decade, studies of DNA methylation have grown dramatically, and become one of the most dynamic and rapidly developing fields in molecular biology. Most approaches for large-scale methylation profiling rely on an initial genomic fractionation step followed by analysis of methylation patterns across the genome by microarray-based or sequencing-based approaches. Previously, microarrays for this purpose contained clones from libraries of CGIs [22–24]. As the field progressed, more comprehensive global approaches were accomplished using whole-genome comparative genomic hybridization tiling arrays or bacterial artificial chromosomes arrays [25–28]. To achieve global epigenetic profiling, a variety of commercial microarrays are now available. Whole-genome arrays containing overlapping oligonucleotides tiling through large areas of mammalian genomes, including but not restricted to promoter regions, are available from NimbleGen (Roche, Basel, Switzerland), Agilent (CA, USA) and Affymetrix (CA, USA). These platforms have been used successfully for analyzing DNA methylation, typically with probes made from DNA fractionated by methylation-sensitive restriction-enzyme treatment and affinity purification of methylated DNA by either methyl-binding proteins or methylated DNA immunoprecipitation (MeDIP) [24,29–31]. These approaches will likely remain a mainstay in mammalian epigenetics for some time, as they have major advantages of dense genomic coverage and high sample throughput at a reasonable cost.
Before discussing each type of genomic microarray and its applications, we need to describe in more detail the methods of genomic fractionation and probe preparation for identifying methylated and unmethylated DNA sequences. Choices for this purpose are: genomic DNAs can be treated with methylation-sensitive restriction endonucleases, which discriminate sequences based on methylation status; and DNA can be immunoprecipitated by antibodies that recognize methylcytosine or processed by affinity purification on methyl-binding protein beads, such that the resultant DNA is enriched for methylated sequences. Another nonfractionation approach, with different technical advantages compared with the above fractionations, is to use sodium bisulfite, which converts unmethylated cytosines into uracil residues without changing methylated cytosines. This is an important approach, as bisulfite sequencing is considered to be the gold standard for validating DNA methylation patterns.
As one of the most promising technologies for genomic analysis, next-generation (NextGen) sequencing is just beginning to take center stage as another approach in DNA methylation profiling. We will discuss this methodology and its application in studying DNA methylation in a later section. Some of the representative DNA methylation methods included in this review are illustrated in Figures 1–3, and we will explore each of them in more detail.
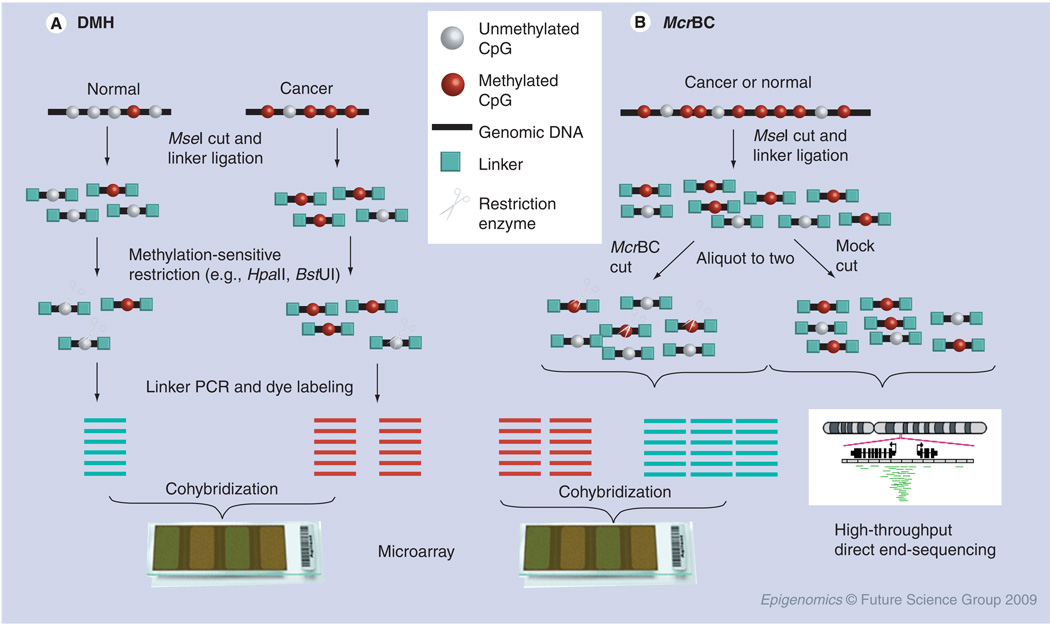
Figure 1. Methodologies based on methylation-sensitive restriction.
(A) DMH methodology; (B) McrBC methodology. DMH: Differential methylation hybridization; McrBC: A methylation-sensitive restriction enzyme that cleaves two closely spaced (55–100 bp) methylated cytosines and preferentially digests densely methylated DNA sequences.
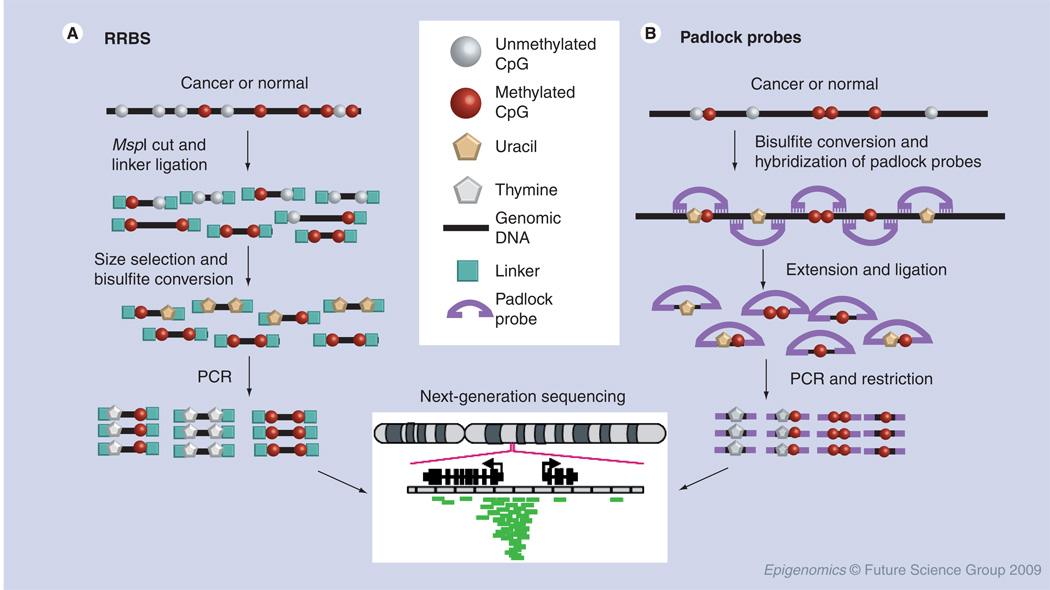
Figure 3. High-throughput bisulfite sequencing.
(A) RRBS methodology; (B) Padlock probes methodology. RRBS: Reduced representation bisulfite sequencing.
Restriction endonuclease digestion followed by microarray analysis
The site-specificity and methylation dependence of several available restriction enzymes make restriction digestion a powerful approach for genomic fractionation. In principle, these technologies permit only one of the two fractions (either methylated or unmethylated) to remain intact after the restriction digestions, and subsequently those intact probes can be labeled and hybridized to an array. By contrast, fragments that have been cut at internal sites fail to amplify and drop out from this genomic representation. For example, as shown in Figure 1A, Huang et al. developed differential methylation hybridization (DMH) arrays by combining restriction endonucleases and microarrays for high-throughput examination of the methylation status of CGIs in human genomes [22,32–34]. In these studies, genomic DNA was fragmented by restriction with MseI, a four-base cutter that cleaves bulk DNA into small fragments (<200 bp). This endonuclease recognizes sequences that rarely occur within GC-rich regions, leaving most CGIs intact. Then, the restricted fragments were ligated to synthetic linkers and further restricted with methylation-sensitive endonucleases, BstUI and/or HpaII. The BstUI was selected for the methylation analysis because more than 80% of CGIs contain BstUI sites. The BstUI-treated DNAs were used as templates for the subsequent linker-dependent PCR. Methylated DNAs resist the restriction digestions, and hence can be amplified. By contrast, the unmethylated DNAs were digested by the endonucleases and failed to be amplified. The resulting PCR products were next labeled with fluorescent dyes. In general, Cy3 dye was used for DNA from control normal patients, while Cy5 dye was used to denote DNA from the patients with cancer. Equal amounts of final amplicons from both groups were mixed well and overlaid on a DMH microarray slide upon which the human CGI library probes were printed. After stringent hybridization, weak or unbound amplicons were eliminated by extensive washing and the resulting microarray slides were subject to high-resolution fluorescence scanning using a laser beam. The ratio of Cy5 over Cy3 intensities would reflect the methylation status in the cancer group relative to that in the normal counterparts within each locus.
To judge the potential false discoveries in this method, a methylation-insensitive isoschizomer of HpaII, namely MspI, can be employed in a control reaction. This methodology was applied for profiling promoter methylation in 8091 human genes [35]. Fukasawa et al. employed a similar approach and carried out promoter methylation studies in human lung cancer [36]. In their studies, the methylated HpaII-resistant DNA fragments and MspI-cleaved products were amplified and labeled with Cy3 and Cy5, respectively, and then were cohybridized to microarrays containing promoters of 288 cancer-related genes [36]. By using the similar restriction fractionation approach, HpaII tiny fragment enrichment by ligation-mediated PCR assay (HELP) uses MspI representations as an internal control [37]. Because MspI is the methylation-insensitive isoschizomer of HpaII, it would cut every possible HpaII restriction site across the genome. By comparing the two profiles generated by each enzyme, Khulan et al. performed both intra- and inter-genomic DNA-methylation analyses in 6.2 Mb of the mouse genome, and identified 223 novel tissue-specific differentially methylated regions [37].
A limitation in utilizing BstUI and HpaII to assess methylation status is that these enzymes identify only a limited fraction of genome CpG sites. To improve sensitivity, Nouzova et al. added a reverse approach by using genomic representations made by digesting with McrBC, an unusual restriction enzyme that cleaves methylated, rather than unmethylated, DNA (Figure 1B). McrBC cuts between two closely spaced (55–100 bp) methylated cytosines in the context (G/A)Cmet, and therefore preferentially digests densely methylated regions of DNA, such as abnormally methylated CGIs in cancer cells and normally methylated repetitive and intragenic sequences in most cells and tissues. As an example of this method, a greater number of hypermethylated loci were identified in an acute promyelocytic leukemia cell line than in normal peripheral blood mononuclear cells, and the sensitivity of the analysis was greater compared with the conventional approach using only HpaII and BstUI [38]. Irizarry et al. developed comprehensive high-throughput arrays for relative methylation (CHARM) from the McrBC assay by improving statistical procedures and the array design algorithm [39]. By using the CHARM assay to detect genome-wide DNA methylation in colon cancers, this group reported hypermethylated CpG sites in ‘CGI shores’, which were defined as the regions approximately 2 kb away from a CGI [40]. This interesting finding is consistent with the DNA methylation ‘spreading theory’, which describes that de novo methylation may begin at the flanking CpG sites and progressively invade into the core of the island [41]. As will be described and illustrated in Figure 1B, genomic fractionation into methylated (HpaII-resistant) and unmethylated (McrBC-resistant) fractions can also be exploited for mapping DNA methylation by conventional and NextGen sequencing [5].
Recently, a methodology to enrich the unmethylated DNA with the involvement of multiple enzyme-mediated restrictions was developed [31]. In this approach, genomic DNA was subjected to a cocktail of methylation-sensitive restriction endonucleases (HpaII, Hin6I, AciI, Hin1I and Hpych4IV). This combinatorial approach provides a better coverage (up to 41%) of all CpG dinucleotides in mammalian genomes [31]. After the multiple restrictions of genomic DNA, a double-stranded adaptor was ligated to the CpG overhangs. At this point, the relatively short and amplifiable DNA fragments were predominantly derived from unmethylated regions and were susceptible to the subsequent PCR amplifications. However, if any residual fragments that harbor methylated cytosines remained, they could be further eliminated by a second step of methylation-specific restriction using McrBC prior to the PCR. After the two-step restrictions and subsequent PCR, amplicons were labeled with different fluorescent cyanine (Cy5 or Cy3) dyes for either sample or reference DNA, mixed and cohybridized to oligonucleotide microarrays [31]. In another variation of DMH, enriching unmethylated DNA, the total genomic DNA was first restricted with HpaII, ligated to special linkers prior to subsequent PCR amplifications, and then the resultant amplicons were hybridized to arrays of promoter sequences [42]. This approach relied on the occurrence of two methylation-sensitive restriction sites in close proximity. In this case, if the restriction sites were both unmethylated, they were susceptible to HpaII and could be ligated to linkers followed by PCR amplification. By contrast, if the sites at either or both ends were methylated in the genome then restriction cuts were prevented and longer fragments were generated that were poorly susceptible to PCR amplification.
As DMH methods rely on PCR amplification prior to microarray analysis, Chen et al. conducted a test to determine at which cycles of PCR the amplification is discriminative, and found that PCR should be carried out for less than 29 cycles to prevent overamplification of the partially restricted DNA fragments and yet yield sufficient PCR products for intact DNAs [43]. Besides PCR cycles, another potential bias associated with PCR amplification of GC-rich sequences, including CGIs, is the choice of polymerase enzyme. Pike et al. compared the efficiency of three ‘GC-improved’ DNA polymerases (AccuPrime™ DNA polymerase [Invitrogen, CA, USA], ThermalAce™ [Invitrogen], DNA polymerase and GC-RICH PCR enzyme [Invitrogen]) to the classical Taq polymerase and found advantages to using these higher efficiency enzymes [44].
Of historical importance, another restriction enzyme-based methodology not involving microarrays is restriction landmark genomic scanning (RLGS), a two-dimensional gel electrophoresis approach combining restriction enzyme polymorphism and DNA methylation-sensitive sites for genome-wide analysis of DNA methylation and expression. This method entails the restriction digestion of genomic DNA with methylation-sensitive restriction enzymes (NotI or AscI) followed by radiolabeling of the restriction fragments and two-dimensional gel electrophoresis; this method yields a pattern of spots on autoradiograms representing unmethylated sites in the genome of the experimental sample being analyzed [45]. Comparison of the patterns of spots found with two different samples has uncovered important differentially methylated genes in diverse cancer types, ranging from leukemias to lung cancers [46–48].
Profiling allele-specific methylation on microarrays
Allele-specific epigenetic modifications are the hallmark of imprinted loci. This type of allelic asymmetry is increasingly recognized as occurring at a subset of nonimprinted loci as well. While simple DMH does not yield information on allele-specific methylation, restriction enzyme-based approaches followed by microarray analysis have now been developed to produce such allele-specific data. In a single nucleotide polymorphism (SNP) chip-based method, called methylation-sensitive SNP chip analysis (MSNP), Yuan et al. and Kerkel et al. used restriction endonuclease-based methylation profiling on Affymetrix SNP arrays to determine net methylation and allele-specific methylation genome-wide [49,50]. In the initial proof-of-principle study using 50K SNP arrays, genomic DNA was first cleaved by XbaI in the presence or absence of HpaII. The resulting DNA fragments were ligated to adaptor linkers, followed by PCR amplification, labeling and hybridization to the SNP arrays, which contain oligonucleotides matching both alleles of a large number of SNPs, distributed at roughly equal intervals along the human genome. The resultant allele-specific SNP hybridization intensities derived from XbaI genomic representations were compared with those from the XbaI/HpaII representation (and control representations with XbaI/MspI). As will be discussed, MSNP can now be applied to 250 K and 1M SNP arrays that use NspI or StyI as the generic restriction enzymes, but the principle remains the same. With regard to DNA methylation, the SNPs on the arrays fall into three general categories. The first is class 1 SNPs, fragments that lack internal HpaII sites, yield genetic information (copy number aberrations and loss of heterozygosity) and serve as invariant internal controls in the DNA methylation analysis. Class 2 SNPs are within fragments that contain HpaII sites at positions other than the SNP itself. In fact, these SNPs are the informative ones for assessing both net (average of the two alleles) and allele-specific DNA methylation. Class 3 SNPs, the rarest category, fall within a CCGG sequence and therefore create or destroy a HpaII restriction site, or have adjacent polymorphic HpaII sites, based on the SNP database. These SNPs are not reliable for assessing DNA methylation but, like the class 1 SNPs, they are useful for assessing loss of heterozygosity and DNA copy number.
In contrast to CGI arrays, SNP arrays and full-genome tiling arrays query diverse locations in the genome that are intergenic, intragenic and promoter CGI-associated. As noted previously, while much prior research in cancer epigenetics has focused on gaining methylation in CGIs, there are advantages to surveying not only islands, but also nonisland sequences. In contrast to islands, the vast majority of which are nonmethylated, nonisland sequences frequently have substantial baseline CpG methylation, so both gains and losses of DNA methylation can be evaluated as biomarkers. Moreover, nonisland CpG sequences may have important regulatory functions that will only be revealed when these sequences begin to be studied using MSNP and related genomic profiling methods such as HELP, CHARM assays and MeDIP on tiling arrays, as well as ultra-high-throughput sequencing.
New biological principles can emerge specifically from studying allele-specific epigenetic modifications by MSNP and related methods. For example, Kerkel et al. carried out a large study of allele-specific methylation in normal human hematopoietic and placental tissues. This study uncovered a strong genetic/epigenetic dependence at multiple loci with strong linkage between SNP genotypes and the propensity of a given allele (CpGs near the SNP) to become methylated [50]. This category of allele-specific methylation, being sequence-dependent, is distinct from genomic imprinting, in which the allele that becomes methylated is determined by its parental origin and not by its sequence. A method very similar to MSNP was utilized to examine allele-specific methylation on the human inactive and active X-chromosomes in females. This study revealed an interesting phenomenon of opposite patterns of methylation in CGIs versus gene bodies (intragenic sequences), such that the CGIs were found to be hypermethylated on the inactive X chromosome while the gene bodies were relatively hypermethylated on the active X chromosome [51].
Bisulfite conversion-based assays utilizing microarray platforms
To characterize methylation patterns at base-pair resolution, bisulfite conversion of DNA followed by sequencing is considered to be the gold-standard approach [52]. Bisulfite conversion-based approaches can be the first step in characterizing DNA methylation both on microarrays and by NextGen sequencing. Bisulfite treatment converts unmethylated cytosines to uracil, such that U is read as T after PCR amplification and sequencing. This conversion does not affect methylated cytosines, which remain C in the sequence. PCR amplicons generated after bisulfite conversion of genomic DNA can be hybridized to microarrays containing methylation-specific oligonucleotides (MSO; comprise 19–23 nucleotides) to query DNA methylation status [53]. As MSO probes discriminated methylated from unmethylated cytosines within a given CG-rich sequence, the quantitative differences in hybridization, which are assessed by the fluorescent intensity, can indicate the methylation status of a particular locus. For example, a set of 12 MSOs was designed to test 15 CpG sites within the CGI in the first exon of the ERα gene [53]. As each probe can interrogate interrogate 2–4 CpG sites in the CGI the methylation status of the ERα gene was identified to be strikingly different among breast cancer cell lines. Likewise, further studies applying MSO were able to classify various human tumor types by methylation patterns [54,55] and classified a specific subtype of non-Hodgkin’s lymphoma [56] based on the differential methylation profiles of several gene promoters In addition, a modification of MSO technology illustrating the methylation status of the promoter region of the MGMT gene was developed to examine colorectal cancer [57]. An increasingly popular bisulfite-based high-throughput DNA-methylation profiling platform, commercially available from Illumina, utilizes bead arrays to obtain a quantitative measure of the percentage methylation at each CpG site [58]. In the current version of this approach, genomic DNA samples are bisulfite-converted, and the bead array assay utilizes hybridization and primer extension to query the methylation status of cytosines in specific CpG dinucleotides. The information content of the bead arrays is limited only by the number of specific primers attached to the beads. The proof-of-principle study for this basic approach examined human lung cancers at 1536 specific CpG sites in 371 genes, thereby deriving panels of cancer-specific epigenetic markers [58]. For several years, the coverage offered by this system (Illumina Infinium methylation assays) has increased substantially to more than 20,000 CpGs in promoter regions of more than 14,000 genes.
MeDIP & methyl-binding domain affinity chromatography followed by microarray-based profiling
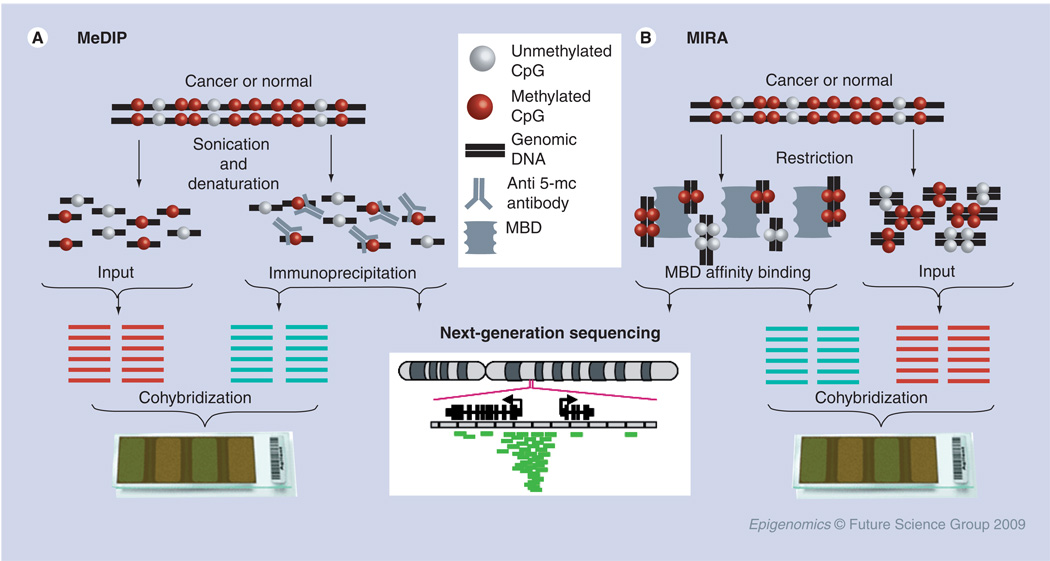
Genomic fractionation using restriction enzymes has limitations as previously noted. For example, only 3.9% of all CpG dinucleotides in human nonrepetitive regions are recognizable by HpaII [59]. Although the conversion of unmethylated cytosines with bisulfite provides a sensitive alternative, it cannot yet be flexibly or comprehensively applied to whole-genome screenings, as even the Illumina Infinium assays only query a restricted number of CpG sites. A third general type of high-throughput approach in methylation analysis applies MeDIP or affinity chromatography over a methyl-binding domain (MBD) linked to beads, followed by probe preparation and microarray hybridization (Figure 2). MeDIP utilizes nonspecific fragmentation of the genomic DNA followed by anti-5mC antibodies to enrich for methylated DNA fragments. The resultant immunoprecipitated DNA, enriched in hypermethylated sequences, and total genomic DNA (input) are labeled with fluorescent dyes Cy5 and Cy3, respectively, and cohybridized onto microarray chips (Figure 2A). The ratio of fluorescent intensity (Cy5 to Cy3) indicates the methylation status at each particular gene. MeDIP is thus a valuable general fractionation approach, compatible with any genomic microarray platform to query the level of methylation in genomic sequences. In their proof-of-principle study, Weber et al. analyzed the genome-wide methylation between male and female fibroblasts using MeDIP coupled with a comparative genomic hybridization microarray that contains bacterial artificial chromosome clones with an average tiling resolution of 80 kb [24]. In addition to demonstrating methylation differences related to cellular transformation, their results, which included but were not restricted to CGIs, revealed an interesting spatial pattern of methylation on the X chromosome, such that the inactive X chromosome was found to be hypermethylated overall at only a subset of gene-rich regions at the telomeric end and, unexpectedly, was hypomethylated overall relative to its active counterpart. One of the crucial factors in this assay is the sensitivity of the anti-5-methylcytosine antibody. Moreover, the MeDIP method is most sensitive to densely methylated sequences, as DNA fragments with many contiguous methylated CpGs are more efficiently precipitated. Approximately 200 differentially methylated genes were identified in a SW48 colon cancer cell line using MeDIP coupled with a 12k CpG island microarray [24], which were substantially fewer than the previous examination [48]. Keshet et al. coupled MeDIP to a promoter array (nearly 13,000 human gene promoters) and have identified several common motifs of promoters that were significantly methylated in various human cancer cell lines [60].
Figure 2. Affinity purification of hypermethylated DNA.
(A) MeDIP methodology; (B) MIRA methodology. MBD: Methyl-binding domain; MeDIP: Methylated DNA immunoprecipitation; MIRA: Methylated CGI recovery assay.
Unlike conventional MeDIP, which employs a monoclonal antibody against 5-mC in the context of single-stranded DNA, MBD-based affinity purification is an alternative approach to enrich hypermethylated DNA in the genome. An example is methylated CGI recovery assay (MIRA), which utilizes the very high affinity of the MBD2/MBD 3-like 1 complex to purify methylated DNA (Figure 2B). MIRA is not sequence-dependent, and does not require a denaturation step to make the DNA single stranded. MIRA coupled with microarray-based analyses provides high-resolution genome-wide methylation profiling. By this approach, Rauch et al. identified a tumor suppressor gene, DLEC1, as well as 11 homeodomain-family genes that were frequently methylated in primary human lung cancers [61]. It is worth noting that the MIRA approach has now been commercialized by Life Sciences Technologies (Invitrogen). Another laboratory also pursued this affinity approach, in conjunction with a second step of partial strand melting, followed by direct sequencing of the methylated DNA, for successfully identifying methylated CGIs in human lung cancers [62]. A similar strategy, using MBD2 fused to the Fc fragment of human IgG1 by protein A-sepharose, was recently developed [63]. Finally, in an interesting variant of the well known ChIP-on-chip approach for studying chromatin proteins, Ballestar et al. globally examined the distribution of MBD2 as a surrogate marker for densely clustered methylated CpGs in the genome of breast cancer cells [64].
As noted above, in considering the interpretation of microarray experiments using these methods for preparing the probes, it is important to keep in mind that both the MBDs and the anti-5-methyl-C antibody are specifically recognizing only the most densely methylated DNA sequences and are not pulling down DNA that is methylated, even at all available CpGs, in regions of the genome that have a low CpG content. Such regions are better examined by restriction endonuclease-based approaches. It is worthwhile to note that several studies described improved analytic algorithms for interpreting genome-wide methylation data generated by MeDIP experiments [65,66].
Thus, no single approach, with the possible exception of extensive and deep sequencing of bisulfite-converted DNA (which is still slightly beyond current practical capabilities for large sets of biological samples) can give a truly complete picture of the overall epigenome. In summary, combinations of several modalities applied to the same sample set will give the clearest picture.
NextGen sequencing & its application in analyzing DNA methylation
The availability of NextGen sequencing, that is, a high-throughput sequencing technique, offers much higher coverage per run and relatively lower cost for genome-wide sequencing than previous technologies. Therefore, it provides a more cost-effective platform for large-scale methylation detection. As summarized in Table 1, the most widely used high-throughput sequencing platforms on the market are the 454 Genome sequencer (Roche), Solexa technology (Illumina, CA, USA) and the SOLiD platform (Applied Biosystems Inc., CA, USA). NextGen sequencing technology is in an exponential development stage. Some recent review papers have provided a more comprehensive view of this technology [67,68]. Owing to limited space, a brief introduction to these technologies and their applications in the methylome will be discussed here.
Table 1.
Overview of next-generation DNA sequencing technologies.
| Platform | PCR mechanism | Synthesis method | Read-length (bp) | Parallel reads |
|---|---|---|---|---|
| 454 | Emulsion PCR | Pyrosequencing | 200–500 | 1 million |
| Solexa | Bridge PCR | Reversible terminators | up to 75 | 150 million |
| SOLiD | Emulsion PCR | Ligation | up to 50 | 400 million |
The 454 system was the first platform available for NextGen sequencing. According to the principle of real-time pyrophosphate DNA sequencing method, DNA fragments are ligated to adaptors and subjected to emulsion PCR with water-in-oil microreactors. The sequencing signals are collected through the fluorescence generated from luciferin substrate in the sequencing-by-synthesis (SBS) reaction. The system could conduct more than 1 million individual reads at lengths of up to 500 bp. Illumina developed another high-throughput sequencing platform, Solexa genome analyzer, by using a parallelized sequencing approach. The principle of the analyzer is based on bridge PCR, in which the forward and reverse primers are attached to a solid surface so that the adaptor-ligated DNA fragments can be annealed on the surface and subjected to PCR amplification. The system takes advantage of SBS technology with reversible fluorescently labeled terminators, allowing detection of each single synthesized base in a real-time fashion. Almost 150 million reads could be analyzed within 6 h with an accuracy of 99%. Although up to 75 bp of length could be detected in paired-end sequencing, an average of 36 bp would provide much higher accuracy because of signal decay and dephasing. The third platform, the SOLiD system, is based on a sequencing-by-ligation technique. The system propagates sequencing on the template DNA fragments by ligating a pool of fluorescently labeled octamers that contain random oligonucleotide combinations. Each cycle of hybridization and ligation was proceeded by cleavage of the 3′ end of the ligated octomer and addition of the next fluorescent probe. A total of 400 million sequence tags can be generated per run and the length of each read can reach 50 bp.
By taking advantage of high-throughput sequencing technology, an obvious ultimate objective is to achieve cost-effective complete-genome bisulfite sequencing, which will reveal the methylation status of every CpG dinucleotide. A tour-de-force whole-genome bisulfite sequencing study of Arabidopsis thaliana, a widely used plant genetic model, was recently carried out by the Jacobsen laboratory by using the Solexa platform [69]. Their paper contains an interesting comparison of the relative accuracy of array-based and bisulfite sequencing-based methods and, not surprisingly, the sequencing-based approach was more accurate. Another advantage of the sequencing-based analysis was the ability to score methylation in most types of repetitive sequences, which are less easily probed by microarray-based methods.
Unfortunately, whole-human genome bisulfite sequencing currently remains just out of reach because of the larger genome size and relatively small portion of methylated cytosine in mammalian genomes, compared with Arabidopsis thaliana. Therefore, new approaches have been developed to either enrich high-CpG-density sequences or target specific CpG sites through padlock (molecular inversion) probes. In a pilot study, Taylor et al. used a MDB affinity column to enrich methylated DNA from either normal peripheral blood leukocytes or several types of human lymphomas. The 454 NextGen sequencing platform was used for high-throughput sequencing of bisulfite PCR amplicons [70]. Although this approach circumvents and improves the previously rate-limiting step of cloning the bisulfite PCR products, the total sequencing reads remain less than 1 million. To further extend NextGen sequencing technology to the genome-wide scale, Meissner et al. reported a reduced representation bisulfite sequencing method (Figure 3A), in which 90% of CpG islands in the mouse genome could be covered through MspI fractionation. DNA fragments were then subjected to bisulfite conversion and high-throughput sequencing by the Solexa system [71]. Bestor and associates are utilizing the SOLiD sequencing platform to conduct direct end-sequencing of restriction enzyme-digested and size-fractionated genomic DNA – an approach that they previously validated by using high-volume conventional sequencing [5].
A second approach to selectively sequence the CpG-rich methylome is to target specific CpG sites across the genome. By taking advantage of the pilot study from The Encyclopedia of DNA Elements (ENCODE) project, which provided detailed DNA sequence information for 1% of the human genome [72], two research groups simultaneously reported the utilization of padlock probes to target specific CpG sites across the genome [73,74]. The probes were first synthesized by programmable DNA microarrays based on targeted DNA sequences. As shown in Figure 3B, padlock probes were designed to target both the 5′ and 3′ ends of specific CpG sites in bisulfite-converted DNA, so that the CpG sites fall in the gap between two probes. The methylation status of CpG sites was captured by single-ligation amplification reaction. The beauty of this technique is that the library of tens of thousands of multiplexed padlock probes are amplified in one single tube. The captured targeting CpG sites in padlock loops were then subjected to NextGen sequencing. Although only 66,000 CpG sites, accounting for approximately 0.25% of total CpG sites in human genome, were sequenced in the studies, the assay is ready to expand the representation across genome by following the increasing coverage of the ENCODE project.
New biological generalities are emerging from these studies examining both CGIs and nonisland sequences; for example, Meissner et al. found that most developmental changes in CpG methylation occur outside of promoter regions [71], and Ball et al. found a trend for gene bodies to be hypermethylated as a function of active transcription [73]. Rapid technical progress and new discoveries can be anticipated in this exciting area.
Other approaches for profiling DNA methylation
Profiling on expression microarrays combined with drug-induced demethylation
This approach relies on the functional criterion of transcriptional activation to discover genes that have been epigenetically silenced in cancer cells. Demethylating drugs, notably 5-aza-2′-deoxycytidine (5′), can reactivate the expression of genes that have been silenced via promoter hypermethylation by combining 5′ treatment with standard mRNA expression profiling on oligonucleotide or cDNA microarrays. This method is straightforward to generate lists of genes that fit this criterion [75]. The approach is popular and effective; however, its caveats are that it often requires the use of immortalized cell lines, which frequently contain artifactual gains of methylation at many loci [76], and that some of the activated loci might be turning on as an indirect consequence of exposure to the drug and not directly related to methylation of their promoter regions. Thus, after candidate genes are selected from microarray analysis, it is necessary to analyze their methylation patterns in cells before and after treatment by other assays, such as bisulfite sequencing or methylation-specific PCR. Meanwhile, these methylation findings in vitro can be validated in primary tumors.
Profiling methylation in archival formalin-fixed paraffin-embedded samples
Methods that can give access to the huge archival collections of well-characterized normal and disease (cancer, other) samples stored in pathology departments of all major medical centers are obviously highly desirable. Furthermore, some key questions, notably defining the epigenetic changes that occur at the earliest stages of tumor initiation, such as in situ carcinomas and dysplasias, can only be addressed in microdissected material, which is often obtained from formalin-fixed paraffin-embedded (FFPE) specimens. The obvious limitations are limited amounts of extractable DNA and substantial degradation of this DNA under the harsh conditions necessary for extracting it from the formaldehyde cross-linked tissues. Nonetheless, restriction digestion/PCR-based protocols have been developed that work well with this material and can be coupled with custom microarrays for medium-throughput applications [77]. Bisulfite conversion/PCR-based protocols are also effective in this setting, as was emphasized in the original report by Herman and coworkers describing the widely used methylation-sensitive PCR (MSP) method [78], and this approach continues to be used in many medium-throughput studies [79,80]. It will be interesting to see how ultra-high-throughput sequencing may open up new vistas in the analysis of DNA methylation in archival FFPE material.
Moderate-throughput quantitative methylation analysis by quantitative-MSP (MethyLight)
When given a detailed knowledge of the pattern of CpG methylation at a given locus in diseased versus normal tissues, it is generally possible to design methylation-sensitive PCR primers and an internal doubly labeled fluorescent TaqMan® probe (Applied Biosystem Inc.). This possibility allows measurements of the degree of methylation by real-time PCR of bisulfite-converted genomic DNA. This method, which has been validated in several studies by Laird and colleagues, has an experimentally proven sensitivity capable of detecting one methylated DNA molecule in a background of 10,000 unmethylated ones [81,82]. Thus, quantitative MSP, with basic robotics or with multichannel pipetting by hand, is suitable for use in small laboratories both for medium-throughput profiling of a group of candidate genes in a group of experimental samples (~30 genes in ~100 samples) and for diagnostic and screening studies involving the screening of patient samples (blood, urine, sputum and other cytology preparations) for small numbers of tumor cells [83].
Mass spectrometry (Sequenom MassArray)
Using the mass spectrometry-based platform manufactured by Sequenom (CA, USA), methylation patterns can be unambiguously established quantitatively and at single-base-pair resolution [84]. This medium- to high-throughput approach, which relies on bisulfite conversion followed by PCR amplification of specific loci and site-specific cleavage of an in vitro-transcribed RNA copy of the converted DNA prior to mass spectrometry, has been successfully utilized to profile DNA methylation in human non-small-cell lung cancers [85], as well as breast tumors [86]. It has the advantages of high sample throughput and single-base-pair resolution determination of CpG methylation patterns.
Screening large collections of experimental samples for methylation status of specific genes using methylation target arrays
Huang and coworkers have developed a strategy for methylation profiling that reverses the usual approach of hybridizing an array of genes with a probe made from a single experimental sample. Instead, each address on the methylation target arrays (MTA) microarray contains the amplified methylated fraction of the genome from a single sample (tumor, normal, other disease state or tissue). The custom array, containing a large number of experimental samples, is then hybridized with a probe for a specific gene (promoter CGI) or other locus of interest. Based on the hybridization intensity at each address on the array, it is possible to score each sample for the presence or absence of methylation of that sequence. In design, the MTA array is thus analogous to tissue microarrays, which are used to assess immunopositivity for specific protein antigens in large series of human tumors and normal tissues. In the MTA method, the probes to be spotted on the custom microarrays are synthesized by PCR of linker-ligated CGI restriction fragments from the experimental samples of interest that have first been digested with methylation-sensitive restriction enzymes. In their proof-of-principle study, Chen et al. used MTA to determine the frequencies of hypermethylation of a group of ten genes in 93 primary breast cancers [43].
Conclusion & future perspective
Epigenomic studies have made remarkable progress toward revealing a detailed molecular picture of site-specific DNA methylation in mammalian and plant genomes. Because much of the relevant technology has come from the field of cancer epigenetics, we have given some emphasis to this area here. However, we have also touched on some of the broader applications in other human diseases, and basic biology.
This review discussed the epigenetic and epigenomic technologies used to discover gene silencing in human carcinogenesis. High-throughput analyses can be utilized in at least three ways. First, they provide tools for monitoring the ‘cancer-specific’ epigenetic markers during cancer early detection and prognosis. In fact, epigenetic therapies are now approved for various hematological malignancies and are currently in clinical trials for solid tumors. The high-throughput epigenetic assessments could be used for evaluating the efficacy of a therapy regimen. Second, these advanced technologies are likely to evolve molecular mechanisms that in part account for tumorigenicity; in this regard, such knowledge can be incorporated to develop new therapeutic strategies. Third, NextGen sequencing technologies are in strong demand for defining human epigenomes at single-base resolution, which cannot be achieved by conventional methods [87,88]. In summary, since most human cancers now appear to be associated with aberrant gene expression in part due to epigenetic alterations, the contribution of high-throughput epigenetic studies cannot be overstated.
This field has seen explosive growth, and although many references are listed here, not every relevant study is cited. Instead, an attempt has been made to cover some of the methods that may be most useful for individual laboratories that are interested in methylation profiling as a tool for asking targeted biological and clinical questions (as summarized in Table 2). A number of companies, not a comprehensive list, are named as manufacturers of platforms and reagents, but this selection is based entirely on references to the corresponding scientific publications and no commercial endorsements are implied.
Table 2.
Summary of some major methods for profiling DNA methylation.
| Methodology | Examples | Advantages | Major limitations | Ref. |
|---|---|---|---|---|
| DNA microarray based on methylation-sensitive restriction |
DMH | The first isoschizomers-based (BstUI and HpaII) microarray for analysis of hypermethylated CGIs in cancer genome |
BstUI restriction sites mainly distribute in CGIs with a poor coverage for CpG sites in other genomic regions |
[22,32–34] |
| HELP | Less bias toward CGIs; better coverage on genomic regions with low CpG density |
Not for detection of DNA hypermethylation in genome |
[37] | |
| McrBC | Higher restriction sensitivity than that of other methylation-sensitive enzymes; preferentially digests densely methylated genomic regions, such as CGIs and repetitive sequences |
Compromised annotation for the genomic locations in data analysis |
[38] | |
| CHARM (based on McrBC) |
With the improvement of statistical procedures and array design algorithm; less bias toward CGI; better location annotation and highly quantitative. |
Relatively lower resolution than that in sequencing-based technology |
[39,40] | |
| Allele-specific methylation on microarray | MSNP | Detection of allele-specific DNA methylation at imprinted and nonimprinted loci; measurement of copy number aberrations and loss of heterozygosity at the same time |
May lose DNA methylation information for the genomic region that are not covered by SNP microarray platform |
[49,50] |
| DNA microarray based on affinity purification |
MeDIP and MIRA |
Efficient technologies for detection of highly methylated CpG sites in the genome, such as CGIs and repetitive sequences |
Less sensitive to genomic regions containing low evel of DNA methylation |
[24,60,61] |
| Next-generation sequencing | RRBS;Padlock probes; MeDIP-Seq; MIRA-Seq; Direct end sequencing |
RRBS and padlock approaches are high-throughput and genome-wide bisulfite sequencing platforms; all methods based on next-generation sequencing provide much higher resolution for detecting DNA methylation |
The cost remains a major concern for the whole-genome sequencing at this stage |
[71,73,74] |
| Other technologies | MethyLight™ and Sequenom MassArray |
Both methods are suitable for quantitative methylation profiling at multiple loci with large sample size |
Only cover limited loci | [81–86] |
CGI: CpG island; CHARM: Comprehensive high-throughput arrays for relative methylation; CpG: Cytosine–phosphate–guanine; DMH: Differential methylation hybridization; HELP: HpaII tiny fragment enrichment by ligation-mediated PCR; MeDIP: Methylated DNA immunoprecipitation; MIRA: Methylated-CpG island recovery assay; MSNP: Methylation sensitive SNP chip analysis; RRBS: Reduced representation bisulfite sequencing.
Executive summary
The prototype of methylome profiling is based on methylation-sensitive restriction, and typical examples are differential methylation hybridization, HpaII tiny fragment enrichment by ligation-mediated PCR, McrBC and comprehensive high-throughput arrays for relative methylation assays.
Methylated DNA immunoprecipitation and methylated CpG island recovery assay are representatives of affinity purification for methylated DNA.
Methylation-sensitive SNP chip analysis can detect genome-wide allele-specific methylation at imprinted and nonimprinted loci.
Next-generation sequencing is one of the most promising technologies to achieve deeper coverage and higher resolution for DNA methylation analysis.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript
Bibliography
Papers of special note have been highlighted as:
▪of interest
▪▪ of considerable interest
- 1.Waddinton C. The epigenotype. Endeavor 1. 1942:18–24. [Google Scholar]
- 2.Antequera F. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 2003;60(8):1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Illingworth R, Kerr A, Desousa D, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6(1):E22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 5.Rollins RA, Haghighi F, Edwards JR, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16(2):157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 8.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71(5):865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 9.Kangaspeska S, Stride B, Metivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452(7183):112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 10.Metivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 11.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133(7):1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 15.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 17.Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell. 2004;5(2):121–125. doi: 10.1016/s1535-6108(04)00033-9. [DOI] [PubMed] [Google Scholar]
- 18.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 19.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 2006;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 20.Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2(2):123–134. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Plass C. Cancer epigenomics. Hum. Mol. Genet. 2002;11(20):2479–2488. doi: 10.1093/hmg/11.20.2479. [DOI] [PubMed] [Google Scholar]
- 22. Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum. Mol. Genet. 1999;8(3):459–470. doi: 10.1093/hmg/8.3.459. ▪▪ First array-based high-throughput study on CpG island hypermethylation in cancers by using differential methylation hybridization.
- 23.Heisler LE, Torti D, Boutros PC, et al. CpG island microarray probe sequences derived from a physical library are representative of CpG islands annotated on the human genome. Nucleic Acids Res. 2005;33(9):2952–2961. doi: 10.1093/nar/gki582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. ▪▪ Methylated DNA immunoprecipitation (MeDIP) was first reported in this study.
- 25.Ching TT, Maunakea AK, Jun P, et al. Epigenome analyses using bac microarrays identify evolutionary conservation of tissue-specific methylation of shank3. Nat. Genet. 2005;37(6):645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- 26.Cowell JK, Hawthorn L. The application of microarray technology to the analysis of the cancer genome. Curr. Mol. Med. 2007;7(1):103–120. doi: 10.2174/156652407779940387. [DOI] [PubMed] [Google Scholar]
- 27.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 2004;36(3):299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 28.Lippman Z, Gendrel AV, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat. Methods. 2005;2(3):219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 29.Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc. Natl Acad. Sci. USA. 2008;105(1):252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi H, Nagae G, Tsutsumi S, et al. High-resolution mapping of DNA methylation in human genome using oligonucleotide tiling array. Hum. Genet. 2007;120(5):701–711. doi: 10.1007/s00439-006-0254-6. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher A, Kapranov P, Kaminsky Z, et al. Microarray-based DNA methylation profiling: technology and applications. Nucleic Acids Res. 2006;34(2):528–542. doi: 10.1093/nar/gkj461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan PS, Chen CM, Shi H, et al. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61(23):8375–8380. [PubMed] [Google Scholar]
- 33.Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Huang TH. Applications of CpG island microarrays for high-throughput analysis of DNA methylation. J. Nutr. 2002;132(8 Suppl.):S2430–S2434. doi: 10.1093/jn/132.8.2430S. [DOI] [PubMed] [Google Scholar]
- 34.Wei SH, Chen CM, Strathdee G, et al. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin.Cancer Res. 2002;8(7):2246–2252. [PubMed] [Google Scholar]
- 35.Hatada I, Fukasawa M, Kimura M, et al. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25(21):3059–3064. doi: 10.1038/sj.onc.1209331. [DOI] [PubMed] [Google Scholar]
- 36.Fukasawa M, Kimura M, Morita S, et al. Microarray analysis of promoter methylation in lung cancers. J. Hum. Genet. 2006;51(4):368–374. doi: 10.1007/s10038-005-0355-4. [DOI] [PubMed] [Google Scholar]
- 37.Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the help assay. Genome Res. 2006;16(8):1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nouzova M, Holtan N, Oshiro MM, et al. Epigenomic changes during leukemia cell differentiation: analysis of histone acetylation and cytosine methylation using CpG island microarrays. J. Pharmacol. Exp. Ther. 2004;311(3):968–981. doi: 10.1124/jpet.104.072488. [DOI] [PubMed] [Google Scholar]
- 39. Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780–790. doi: 10.1101/gr.7301508. ▪▪ Compared three major methylation profiling methods - MeDIP, HELP and McrBC assays. Also developed the CHARM method by improving the McrBC assay through new algorithms and array design.
- 40.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stirzaker C, Song JZ, Davidson B, Clark SJ. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004;64(11):3871–3877. doi: 10.1158/0008-5472.CAN-03-3690. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Yu Q, Cho AH, et al. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia. 2005;7(8):748–760. doi: 10.1593/neo.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CM, Chen HL, Hsiau TH, et al. Methylation target array for rapid analysis of CpG island hypermethylation in multiple tissue genomes. Am. J. Pathol. 2003;163(1):37–45. doi: 10.1016/S0002-9440(10)63628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pike BL, Groshen S, Hsu YH, et al. Comparisons of PCR-based genome amplification systems using CpG island microarrays. Hum. Mutat. 2006;27(6):589–596. doi: 10.1002/humu.20329. [DOI] [PubMed] [Google Scholar]
- 45.Smiraglia DJ, Plass C. The study of aberrant methylation in cancer via restriction landmark genomic scanning. Oncogene. 2002;21(35):5414–5426. doi: 10.1038/sj.onc.1205608. [DOI] [PubMed] [Google Scholar]
- 46.Liu TH, Raval A, Chen SS, Matkovic JJ, Byrd JC, Plass C. CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Cancer Res. 2006;66(2):653–658. doi: 10.1158/0008-5472.CAN-05-3712. [DOI] [PubMed] [Google Scholar]
- 47.Park J, Brena RM, Gruidl M, et al. CpG island hypermethylation profiling of lung cancer using restriction landmark genomic scanning (RLGS) analysis. Cancer Biomark. 2005;1(2–3):193–200. doi: 10.3233/cbm-2005-12-307. [DOI] [PubMed] [Google Scholar]
- 48.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24(2):132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 49. Yuan E, Haghighi F, White S, et al. A single nucleotide polymorphism chip-based method for combined genetic and epigenetic profiling: validation in decitabine therapy and tumor/normal comparisons. Cancer Res. 2006;66(7):3433–3451. doi: 10.1158/0008-5472.CAN-05-3739. ▪▪ By using Affymetrix single nucleotide polymorphism arrays, the authors reported the first methylation-sensitive single nucleotide polymorphism chip analysis study, a genome-wide allele-specific methylation profiling.
- 50.Kerkel K, Spadola A, Yuan E, et al. Genomic surveys by methylation-sensitive snp analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40(7):904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 51.Hellman A, Chess A. Gene body-specific methylation on the active × chromosome. Science. 2007;315(5815):1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 52.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22(15):2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12(1):158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aihara T, Takatsuka Y, Itoh K, et al. Phase II study of concurrent administration of doxorubicin and docetaxel as first-line chemotherapy for metastatic breast cancer. Oncology. 2003;64:124–130. doi: 10.1159/000067771. [DOI] [PubMed] [Google Scholar]
- 55.Adorjan P, Distler J, Lipscher E, et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 2002;30(5):E21. doi: 10.1093/nar/30.5.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi H, Maier S, Nimmrich I, et al. Oligonucleotide-based microarray for DNA methylation analysis: principles and applications. J. Cell Biochem. 2003;88(1):138–143. doi: 10.1002/jcb.10313. [DOI] [PubMed] [Google Scholar]
- 57.Kimura N, Nagasaka T, Murakami J, et al. Methylation profiles of genes utilizing newly developed CpG island methylation microarray on colorectal cancer patients. Nucleic Acids Res. 2005;33(5):E46. doi: 10.1093/nar/gni046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16(3):383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat. Rev. Genet. 2004;5(6):446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 60.Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38(2):149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 61. Rauch T, Li H, Wu X, Pfeifer GP. Mira-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66(16):7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. ▪▪ The MIRA assay was first reported in this paper. This is an alternative approach based on affinity purification of hypermethylated DNA in the genome.
- 62.Shiraishi M, Sekiguchi A, Terry MJ, et al. A comprehensive catalog of CpG islands methylated in human lung adenocarcinomas for the identification of tumor suppressor genes. Oncogene. 2002;21(23):3804–3813. doi: 10.1038/sj.onc.1205454. [DOI] [PubMed] [Google Scholar]
- 63.Gebhard C, Schwarzfischer L, Pham TH, et al. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66(12):6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- 64.Ballestar E, Paz MF, Valle L, et al. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22(23):6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Down TA, Rakyan VK, Turner DJ, et al. A bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26(7):779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pelizzola M, Koga Y, Urban AE, et al. Medme: An experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18(10):1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ansorge WJ. Next-generation DNA sequencing techniques. N. Biotechnol. 2009;25(4):195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 68.Shendure JA, Porreca GJ, Church GM. Overview of DNA sequencing strategies. Curr. Protoc. Mol. Biol. 2008;Chapter 7(Unit 7):1. doi: 10.1002/0471142727.mb0701s81. [DOI] [PubMed] [Google Scholar]
- 69. Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. ▪▪ First complete genome bisulfite sequencing study on Arabidopsis, which contains smaller genome size and higher density of methylated cytosine than mammalian genomes.
- 70.Taylor KH, Kramer RS, Davis JW, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67(18):8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 71. Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. ▪▪ Reported reduced representation bisulfite sequencing. method and conducted high-throughput bisulfite sequencing on those selected genomic regions. The methylation information of 90% of all cytosine–phosphate–guanine (CpG) islands in the mouse genome could be covered by this method.
- 72.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27(4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Deng J, Shoemaker R, Xie B, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27(4):353–360. doi: 10.1038/nbt.1530. ▪▪ Both [73] and [74] reported genome-wide targeting and sequencing of CpG sites by using padlock probes and NextGen sequencing technologies. The authors provide a promising strategy to circumvent costly prohibitive whole-genome bisulfite sequencing.
- 75.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 2002;31(2):141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 76.Smiraglia DJ, Rush LJ, Fruhwald MC, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum. Mol. Genet. 2001;10(13):1413–1419. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- 77.Melnikov AA, Scholtens DM, Wiley EL, Khan SA, Levenson VV. Array-based multiplex analysis of DNA methylation in breast cancer tissues. J. Mol. Diagn. 2008;10(1):93–101. doi: 10.2353/jmoldx.2008.070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.To KF, Leung WK, Lee TL, et al. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int. J. Cancer. 2002;102(6):623–628. doi: 10.1002/ijc.10783. [DOI] [PubMed] [Google Scholar]
- 80.Shen L, Catalano PJ, Benson AB, 3rd, O’Dwyer P, Hamilton SR, Issa JP. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin.Cancer Res. 2007;13(20):6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cottrell SE, Laird PW. Sensitive detection of DNA methylation. Ann. NY Acad. Sci. 2003;983:120–130. doi: 10.1111/j.1749-6632.2003.tb05967.x. [DOI] [PubMed] [Google Scholar]
- 82.Eads CA, Danenberg KD, Kawakami K, et al. Methylight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28(8):E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kagan J, Srivastava S, Barker PE, Belinsky SA, Cairns P. Towards clinical application of methylated DNA sequences as cancer biomarkers: a joint NCI’s EDRN and NIST workshop on standards, methods, assays, reagents and tools. Cancer Res. 2007;67(10):4545–4549. doi: 10.1158/0008-5472.CAN-06-2888. [DOI] [PubMed] [Google Scholar]
- 84.Ehrich M, Nelson MR, Stanssens P, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl Acad. Sci. USA. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ehrich M, Field JK, Liloglou T, et al. Cytosine methylation profiles as a molecular marker in non-small cell lung cancer. Cancer Res. 2006;66(22):10911–10918. doi: 10.1158/0008-5472.CAN-06-0400. [DOI] [PubMed] [Google Scholar]
- 86.Lin H-JL, Zuo T, Lin C-H, et al. Breast cancer-associated fibroblasts confer akt1-mediated epigenetic silencing of cystatin m in epithelial cells. Cancer Res. 2008;68(24):10257–10266. doi: 10.1158/0008-5472.CAN-08-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones P, Martienssen R. A blueprint for a human epigenome project: the AACR human epigenome workshop. Cancer Res. 2005;65:11241–11246. doi: 10.1158/0008-5472.CAN-05-3865. [DOI] [PubMed] [Google Scholar]
- 88.American Association for Cancer Research Human Epigenome Task Force; European Union, Network of Excellence. Scientific Advisory Board: Moving ahead with an international human epigenome project. Nature. 2008;454(7205):711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]