Abstract
Adipose tissue modulates whole body metabolism and insulin sensitivity by controlling circulating lipid levels and producing molecules that can regulate fatty acid metabolism in such tissues as muscle and liver. We have developed RNA interference (RNAi) screens to identify genes in cultured adipocytes that regulate insulin signalling and key metabolic pathways. These short interfering RNA (siRNA)-based screens identified the transcriptional corepressor receptor interacting protein 140 (RIP140) (J Clin Invest 116: 125, 2006) and the mitogen-activated protein kinase (MAP4k4) (Proc Natl Acad Sci USA 103: 2087Proc Natl Acad Sci USA 103: 2006) as negative regulators of insulin-responsive hexose uptake and oxidative metabolism. Gene expression profiling revealed that RIP140 depletion upregulates the expression of clusters of genes in the pathways of glucose uptake, glycolysis, tricarboxylic acid cycle, fatty acid oxidation, mitochondrial biogenesis and oxidative phosphorylation. RIP140-null mice resist weight gain on a high-fat diet and display enhanced glucose tolerance. MAP4k4 depletion in adipocytes increases many of the RIP140-sensitive genes, increases adipogenesis and mediates some actions of tumour necrosis factor-α (TNF-α). Remarkably, another hit in our RNAi screens was fat specific protein 27 (FSP27), a highly expressed isoform of Cidea. We discovered that FSP27 unexpectedly associates specifically with lipid droplets and regulates fat storage. We conclude that RIP140, MAP4k4 and the novel lipid droplet protein FSP27 are powerful regulators of adipose tissue metabolism and are potential therapeutic targets for controlling metabolic disease. The discovery of these novel proteins validates the power of RNAi screening for discovery of new therapeutic approaches to type 2 diabetes and obesity.
Keywords: Cidec, fatty acids, glucose metabolism, insulin resistance, triglycerides, type 2 diabetes
Type 2 diabetes is associated with obesity and with the inability of pancreatic beta cells to secrete sufficient insulin to maintain normal blood glucose levels (Ahren 2005, Kahn et al. 2006, Marchetti et al. 2006, Salsali & Nathan 2006). However, mild forms of type 2 diabetes in obese patients are often associated with circulating insulin concentrations that are actually higher than observed in normal subjects. This paradox was resolved by the finding that peripheral tissues such as muscle and liver in such type 2 diabetic subjects are relatively resistant to the actions of insulin, prompting beta cells to secrete higher than normal levels of insulin. Insulin resistance in muscle of obese subjects, in particular, usually appears prior to the ultimate failure of beta cells (Petersen & Shulman 2002, Ahren 2005). This insulin resistance in skeletal muscle in turn is thought to result from defects in the signalling pathways whereby insulin enhances translocation of intracellular glucose transporters (GLUT4) to the cell surface membrane, while the total expression of the GLUT4 protein is the same in normal and diabetic subjects (Huang & Czech 2007). Interestingly, such syndromes as obesity and lipdystrophy are thought to confer insulin resistance to skeletal muscle, even in subjects who are not diabetic (Sovik et al. 1996). When the beta cells of such subjects exhibit impaired insulin secretion, type 2 diabetes results. Thus, understanding the molecular connections between obese or lipodystrophic conditions and the induction of impaired insulin signalling in skeletal muscle is a major goal in the field.
Over the past several years it has become apparent that adipose tissue in humans, mice and rats plays a key role in regulating insulin signalling in skeletal muscle. The basis for this conclusion includes experiments showing that adipose-specific deletions in key genes lead to systemic glucose intolerance and insulin resistance. For example, conditional ablation of GLUT4 in adipose tissue in mice causes impaired insulin signalling in skeletal muscle, glucose intolerance and even diabetes in some of the animals (Li et al. 2000, Rossetti et al. 1997, Stenbit et al. 1997). Conversely, selective expression of GLUT4 (Shepherd et al. 1993, Tozzo et al. 1995) at high levels in adipose tissue greatly improves insulin sensitivity in mice. Knockout of the gene encoding the adipose-specific protein adiponectin also leads to decreased insulin sensitivity in muscle and impaired glucose tolerance in vivo (Kubota et al. 2002, Pajvani & Scherer 2003, Haluzik et al. 2004, Civitarese et al. 2006), although one study failed to observe this effect (Ma et al. 2002). It has also been noted that adipose tissue-specific deficiency of peroxisome proliferator-activated receptor-γ (PPAR-γ) protects against high fat diet-induced insulin resistance (Jones et al. 2005). Furthermore, transplantation of adipose tissue from normal mice into lipodystrophic mice prevents the muscle insulin resistance and glucose intolerance of this condition (Gavrilova et al. 2000). Taken together, these and other data provide compelling evidence in support of the concept that adipose tissue metabolism strongly influences whole body glucose tolerance and skeletal muscle insulin sensitivity.
Based upon the above considerations, we reasoned that adipose tissue was a suitable target tissue to search for novel proteins that regulate adipocyte metabolism directly and perhaps whole body metabolism indirectly. Identification of such proteins might provide useful insight into the molecular basis of insulin signalling or other aspects of adipocyte function, and may lead to potent targets for therapeutic agents. We thus developed a strategy for such identifications based upon a two step process (Powelka et al. 2006, Tang et al. 2006): (1) Selection of candidate genes/proteins from the entire database of mouse and human genes, based on high, selective expression in adipocytes and their regulation by changes in metabolic states, and (2) Screening the function of these candidate genes by RNA interference (RNAi)-based gene silencing in cultured adipocytes, using insulin signalling to glucose transport as a reporter assay. We developed methods for silencing genes with pools of short interfering RNA (siRNA) oligonucleotides in cultured 3T3-L1 adipocytes (Jiang et al. 2003) as well as in primary mouse and human adipocytes (Puri et al. 2007). Following successful functional analysis of several candidate genes with this method (Jiang et al. 2003, Mitra et al. 2004, Zhou et al. 2004, Powelka et al. 2006, Tang et al. 2006), we were able to miniaturize the procedure and perform gene silencing assays in a higher throughput mode (30–50 genes per week). These experiments have led to the identification of several genes that encode proteins with novel functions in regulating adipocyte biology. Three of these proteins we have discovered by this approach are briefly summarized in this review, and may be potential targets for development of therapeutic agents in future studies.
RIP140 as a negative regulator of oxidative metabolism
Receptor interacting protein 140 (RIP140) was one of a set of genes we identified in a subtractive cDNA library designed to isolate transcripts enriched in RNA from insulin-responsive adipocytes and muscle and thus potentially involved in the pathway of regulated glucose uptake common to these cell types. Upon siRNA-mediated knockdown of expression in our miniaturized screen, RIP140 was the most prominent of several targets in this set which resulted in an enhancement of insulin-stimulated deoxyglucose uptake (Powelka et al. 2006). This enhancement of insulin-responsive deoxyglucose uptake appears to be largely the consequence of an increase in GLUT4 expression in these cells.
Although the upregulation of RIP140 expression during adipogenesis had been previously noted (Soukas et al. 2001), at that time investigations into its role in adipocyte biology had not been reported. RIP140 was originally identified as a transcriptional repressor found in a complex with oestrogen receptor (Cavailles et al. 1995). Subsequent reports have suggested that RIP140 can interact with a wide spectrum of nuclear receptors including members of the PPAR family (L’Horset et al. 1996, Treuter et al. 1998, Tazawa et al. 2003), and thus is likely to play a role in modulating the transcriptional programme activated by these factors. RIP140 is similar to other nuclear receptor cofactors in that it serves as a scaffolding protein that couples the receptor to transcription modulatory activities (Fig. 1). RIP140 itself contains nine nuclear receptor-interacting LXXLL motifs, and in turn, four repressor domains which can recruit histone deacetylases or C-terminal binding protein (Christian et al. 2004). RIP140 was the first example of a repressor reported to associate with several ligand-bound nuclear receptors (L’Horset et al. 1996). This finding was contrary to the notion that part of the mechanism of nuclear receptor activation by ligands is dismissal of basal repressors associated with the apo-receptor (Gurevich et al. 2007).
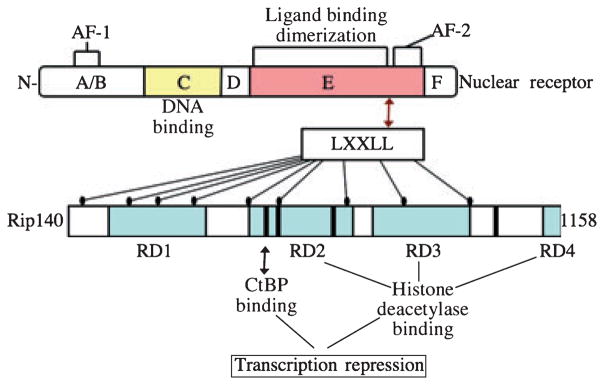
Figure 1.
Domain structure of receptor interacting protein 140 (RIP140). Interaction of the RIP140 protein with nuclear receptors is mediated by nine LXXLL motifs capable of interacting with the ligand binding domain of the receptor. In turn, via four independent repressor domains (RD1-4) RIP140 can recruit histone deacetylases as well as C-terminal binding protein (CtBP) whose activities repress transcription from the promoter bound by the nuclear receptor.
These observations combined with the well established roles of a variety of nuclear receptors, most prominently PPAR-γ (Lehrke & Lazar 2005), in adipocyte biology prompted us to investigate the effects of RIP140 on gene transcription in these cells. Thus we performed Affymetrix GeneChip profiling of gene expression in cultured 3T3-L1 adipocytes depleted of RIP140 with siRNA vs. control adipocytes transfected with scrambled siRNA (Powelka et al. 2006). Analysis of pathways of genes affected by RIP140 depletion revealed a remarkable concentration of significantly upregulated genes in the pathways of oxidative metabolism of glucose and fatty acids: glycolysis, tricarboxylic acid cycle and fatty acid oxidation as well as mitochondrial biogenesis and oxidative phosphorylation (Fig. 2). The majority of annotated genes in each of these pathways were significantly upregulated in response to RIP140 silencing, with most of the remainder not significantly changed, while only 4 of 142 genes in these categories were significantly downregulated (Powelka et al. 2006). Thus RIP140 seems to broadly suppress pathways of oxidative energy production in adipocytes. These changes in gene expression upon RIP140 depletion are associated with phenotypic changes including increased mitochondrial mass, increased oxygen consumption and increased glucose conversion to CO2. In addition, depletion of RIP140 results in enhanced expression of several markers of brown adipocytes (Ucp1, Cidea and Cpt1b) consistent with the higher capacity of brown fat for oxidative fuel utilization in thermogenesis.
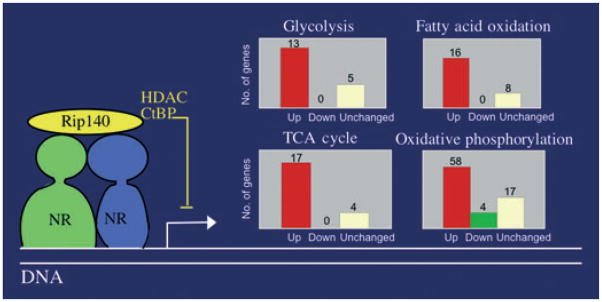
Figure 2.
Function of receptor interacting protein 140 (RIP140) in adipocytes. Via interaction with nuclear receptors (here depicted as a heterodimer such as PPAR-RXR) bound to specific recognition sites in promoters, RIP140 recruits transcription repressor activities [histone deacetylases (HDAC) and C-terminal binding protein (CtBP)] to promoter complexes thereby inhibiting transcription. Major categories of genes repressed by RIP140 function in adipocytes include those in the glycolysis, tricarboxylic acid (TCA) cycle, fatty acid oxidation and oxidative phosphorylation pathways. Inset charts depict numbers of genes whose expression is changed upon RIP140 depletion: upregulated (red bars); unchanged (white bars); downregulated (green bar).
While these studies of cellular effects of RIP140 were underway in our laboratory, Parker and colleagues, who had initially identified RIP140 in association with an oestrogen receptor (Cavailles et al. 1995), reported that mice with a whole body knockout of the RIP140 gene were lean compared with littermates, with body fat stores reduced by 70% despite near-normal food intake (Leonardsson et al. 2004). Transcriptional profiles of these animals as well as adipocytes derived from RIP140-deficient mouse embryonic fibroblasts were similar to those observed in 3T3-L1 cells (Christian et al. 2005, Powelka et al. 2006). This lean phenotype of the RIP140 knockout mouse is therefore associated with enhanced oxidative energy metabolism in the whole animal as well as in cultured cell systems. Additional studies of RIP140−/− mice demonstrated that weight gain even on a high fat diet was reduced compared with controls (Powelka et al. 2006). Furthermore, RIP140−/− animals maintained normal glucose and insulin tolerance while control animals that gained more weight developed impaired glucose disposal, as expected.
Currently, the exact mechanisms for the leaner, insulin-sensitive phenotype of high-fat challenged, RIP140−/− animals are undefined. The whole body knockout could result in contributions from insulin-sensitive organs other than fat such as muscle or liver, or even altered regulatory signals from brain or endocrine organs, given the likely participation of RIP140 in regulatory systems involving many different nuclear receptors. This question should soon be addressed with conditional knockout models; however, metabolic changes in adipose tissue are the most obvious and direct mechanism for alterations in fat deposition. Enhanced oxidative metabolism would certainly favour fatty acid oxidation, as would partial conversion to a brown adipocyte phenotype suggested by expression of brown fat marker genes Cidea and Ucp1 in RIP140-depleted cells. The expression of these markers remains low compared with endogenous brown fat, however. In the case of Ucp1, several mouse models have suggested that even a small amount of ectopic Ucp1 expression can have profound effects on whole body fat metabolism similar to RIP140 depletion (Yamada et al. 2006, Almind et al. 2007).
At the molecular level, studies of the mechanism of RIP140 function have centred around analysis of its interaction with particular nuclear receptors. Among the nuclear receptors with established roles in adipogenesis and/or adipocyte metabolism are liver X receptor (Laffitte et al. 2001, Juvet et al. 2003, Seo et al. 2004), farnesoid X receptor (Cariou et al. 2006, Rizzo et al. 2006) and oestrogen-related receptor-α (ERR-α; Sladek et al. 1997, Vega & Kelly 1997, Nichol et al. 2006). ERR-α is of exceptional interest as it has been associated with regulation of genes associated with mitochondrial biogenesis, oxidative phosphorylation and fatty acid oxidation in several systems (Vega & Kelly 1997, Huss et al. 2004, Schreiber et al. 2004), thus it is reasonable that regulation of a similar cohort of genes by RIP140 is mediated in some measure by association with ERR-α. Indeed evidence that RIP140 may function in regulating several genes via ERR-α actions has been presented (Powelka et al. 2006, Debevec et al. 2007).
As a key regulator of adipogenesis and adipocyte metabolism, PPAR-γ looms as a strong candidate for acting as a mediator of RIP140 functions in adipocytes. Several lines of evidence for functional interaction of PPAR-γ and RIP140 have been presented: (1) PPAR-γ has been shown to directly interact with RIP140 in vitro and in the presence of the PPAR-γ ligand (L’Horset et al. 1996, Treuter et al. 1998, Tazawa et al. 2003); (2) Gene expression profiling of adipocytes depleted of PPAR-γ or RIP140 shows a substantial coincidence of genes upregulated by RIP140 depletion and downregulated by PPAR-γ depletion (M. Chouinard, J. Christianson, J.V. Virbasius and M.P. Czech, unpublished data), consistent with the notion of RIP140 as a negative regulator of PPAR-γ-mediated gene expression; (3) RIP140 depletion enhances activation of the majority of genes whose expression is stimulated by the PPAR-γ ligand rosiglitazone. This observation can be explained by the binding of RIP140 to PPAR-γ at rosiglitazone-sensitive promoters. Depletion of RIP140 from the cells should favour assembly of active transcription complexes including ligand-dependent activators in place of RIP140. (4) RIP140 has been detected by chromatin immunoprecipitation in an enhancer element, which includes a PPAR response element (PPRE) as well as an ERR-α binding site (Christian et al. 2005, Debevec et al. 2007). This PPRE also confers PPAR-γ ligand activation of the Ucp1 promoter only in RIP140 knockout cells. Evidence for direct in vivo association of RIP140 and PPAR-γ at regulated promoter sites is yet to be presented. However, the observations described above point to PPAR-γ as a primary target of RIP140 regulation.
Investigations into the regulation of RIP140 itself hint at important roles of RIP140 in homeostatic mechanisms in a number of systems. Transcription of RIP140 is in turn regulated by nuclear receptors including PPAR-γ, ERR-α and ER (Carroll et al. 2005, Augereau et al. 2006, Nichol et al. 2006), suggesting that RIP140 participates in feedback loops that modulate function of these nuclear receptor networks. Acute regulation of RIP140 activity by post-translational modifications including phosphorylation and acetylation has also been described (Gupta et al. 2005, Huq & Wei 2005, Mostaqul Huq et al. 2006), and may constitute an additional layer of regulation subject to metabolic or hormonal signals.
MAP4K4/NIK: a TNF-α -regulated protein kinase that functions as negative regulator of PPAR-γ and adipogenesis
MAP4K4/NIK was initially identified in an RNAi-based screen we developed to search for protein kinases that modulate insulin-sensitive deoxyglucose uptake in cultured adipocytes (Tang et al. 2006). Remarkably, silencing MAP4k4 caused an increase in the expression of PPAR-γ, along with a corresponding increase in the expression of GLUT4 and stimulation of insulin-mediated deoxyglucose uptake. Furthermore, the effect of MAP4K4/NIK silencing on deoxyglucose uptake is quite specific, as siRNA-mediated silencing of 22 other MAPK family members expressed in adipocytes did not enhance insulin signalling to deoxyglucose transport (Tang et al. 2006).
MAP4K4/NIK is a mammalian serine/threonine protein kinase that belongs to a large group of protein kinases related to Saccharomyces cerevisiae Sterile 20 (STE20) (Dan et al. 2001). There are 36 STE20-related protein kinases in the human genome, which can be divided into two groups: the germinal centre protein kinases and the p21-activated protein kinases. MAP4K4/NIK is a member of the germinal centre protein kinase IV group (Fig. 3a). This kinase was first identified a few years ago by Su et al. (1997) in the mouse through a screen for proteins that interact with the SH3 domain of Nck, using the two-hybrid system. Thus, MAP4K4/NIK appears to interact with the SH3 domain of Nck, and it was termed Nck Interacting Kinase (NIK) (see Fig. 3b). In this initial study, however, no link between MAP4K4 and adipocyte function was reported.
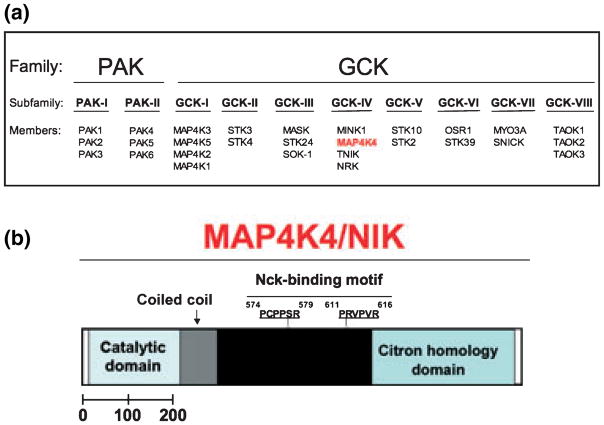
Figure 3.
(a) Protein kinases related to Saccharomyces cerevisiae Sterile 20. Depicted are the two families, p21-activated protein kinases (PAK) and the germinal centre protein kinases (GCK), the subfamilies, PAK I and II and GCK-I to -VIII. Members from each one of the subfamilies are also shown. Depicted in red is MAP4K4, a member of the GCK-IV subfamily. (b) Schematic diagram depicting predicted structural motifs of mouse MAP4K4. Within the MAP4K4 sequence shown is the N-terminal catalytic domain in the hatched area, the coiled-coil region in grey and the Nck-binding motif in black, followed by the C-terminal CNH domain. The two proline-rich motifs that match consensus SH3 binding motifs and mediate the association of MAP4K4 with Nck are shown (amino acids 574–616).
More recent studies have shown that MAP4K4 appears to control cellular events ranging from cell motility, rearrangement of the cytoskeleton, cell proliferation and insulin resistance in muscle induced by tumour necrosis factor-α (TNF-α) (Nishigaki et al. 2003, Wright et al. 2003, Hu et al. 2004, Taira et al. 2004, Collins et al. 2006, Zohn et al. 2006, Bouzakri & Zierath 2007). This protein kinase also appears to specifically activate c-Jun N-terminal kinase (JNK) (Su et al. 1997, Yao et al. 1999). In fact, the majority of studies focusing on MAP4k4 propose that it acts as an upstream activator of the c-Jun-N-terminal kinases 1 and 2 (JNK1/2), extracellular signal-related kinase 1/2 (ERK1/2) and p38 SAP kinase (Wright et al. 2003, Collins et al. 2006, Zohn et al. 2006, Bouzakri & Zierath 2007). Consistent with this notion, the activation of JNK by MAP4K4 was found to be inhibited by the dominant-negative mutants of TAK1, MKK4 and MKK7, suggesting that this kinase may function through the TAK1-MKK4-MKK7-JNK kinase cascade, and mediate in part the TNF-α signalling pathway (Yao et al. 1999). Moreover, subsequent studies by Machida et al. (2004) also showed that MAP4K4 interacts with Rap2 (a Ras family small GTP-binding protein) through its C-terminal domain and that Rap2 enhances activation of JNK by MAP4K4. Together, these observations suggest a role for MAP4K4 on SAPK/JNK kinase cascade activation. However, loss of MAP4K4/NIK in 3T3-L1 adipocytes had no effect on the ability of TNF-α to induce phosphorylation of JNK1 and JNK2 or on the ability of TNF-α to induce inhibitor alpha of nuclear factor-kappa B (IκB) degradation (Tang et al. 2006). Thus, the enhancement of insulin-stimulated deoxyglucose transport observed in adipocytes depleted of MAP4K4 is not caused by disruption of TNF-α activation of JNK or nuclear factor-kappa B (NFκB) signalling cascades in cultured adipocytes. Moreover, there are almost certainly other routes of JNK activation, independent of MAP4K4, and in turn there are likely other cascades in which MAP4K4 plays key roles.
These observations, combined with the well-established role of TNF-α as a potent negative regulator of adipogenesis (Zhang et al. 1996) and the fact that large amounts of this cytokine are secreted by adipocytes and macrophages within adipose tissue of obese animals (Wellen & Hotamisligil 2003, 2005, Wellen et al. 2004), prompted us to investigate the effect of TNF-α on MAP4K4 expression in cultured adipocytes. We found that treatment of 3T3-L1 adipocytes with TNF-α for 24 h caused a three to fourfold increase in MAP4K4 mRNA levels (Tang et al. 2006, Tesz et al. 2007). This enhancement in MAP4k4 expression is mediated by TNF-α-receptor 1 (TNFR1), but not TNFR2. Remarkably, TNF action on MAP4k4 protein kinase expression is quite specific in cultured adipocytes, such that expression of other stress protein kinases such as JNK1/2 and p38 SAP kinase is unaffected by TNF. Moreover, the action of TNF MAP4k4 is not mimicked by other cytokines such as IL-1 and IL-6 (Tesz et al. 2007). We also demonstrated that this specificity of TNF responsiveness appears due to a unique robust and prolonged phosphorylation of JNK1/2 and p38 SAP kinase that leads to activation of the transcription factors c-Jun and ATF2. These latter factors are in turn required for regulated MAP4k4 expression (Tesz et al. 2007). Taken together, these results indicate that the increase in MAP4k4 expression in response to TNF-α is mediated through a signalling pathway elicited selectively by TNFR1 activation leading to c-Jun and ATF2 activation in cultured adipocytes (see Fig. 4a). Our findings also suggest that MAP4K4 acts as a negative regulator of adipogenesis and insulin-stimulated glucose transport and that it may play a role in signalling by TNF-α, a cytokine known to downregulate GLUT4, PPAR-γ and adipogenesis (Fig. 4a).
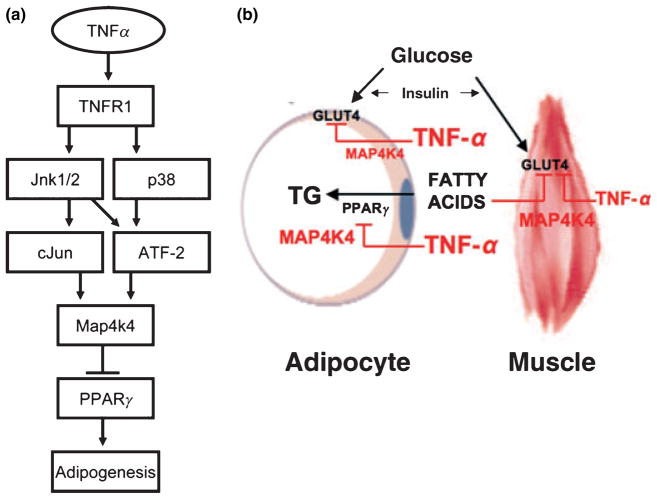
Figure 4.
(a) Model for the increase in MAP4k4 expression via TNF-α signalling. TNF-α activates the catalytic activity of MAP4k4, but in addition elevates its expression. Our data are consistent with the following hypothesis. Treatment of 3T3-L1 adipocytes with TNF-α stimulates TNFR1 and causes enhanced activation of JNK1/2 and p38 SAP kinase. In turn, activated JNK1/2 and p38 SAP kinase cause increased phosphorylation and activation of c-Jun and ATF2. Increased activation of c-Jun and ATF2 leads to increased MAP4k4 transcription, thus increasing MAP4k4 expression. This increase in MAP4k4 expression then negatively regulates PPAR-γ expression and adipogenesis in 3T3-L1 adipocytes (adapted from Tesz et al. 2007). (b) MAP4k4 is required for TNF-α signalling in adipocytes and skeletal muscle. Model for MAP4k4 downregulation of GLUT4-mediated glucose transport in muscle and adipose cells and the effect of this kinase in insulin receptor signalling in skeletal muscle. Increased levels of circulating fatty acids lead to lipid overload and impairment of insulin-stimulated glucose transport through GLUT4 in muscle. PPAR-γ activity promotes fatty acid storage as triglycerides (TG) in adipocytes. TNF-α signals through MAP4K4 and downregulates PPAR-γ expression and TG biosynthesis in adipocytes. MAP4K4 may also mediate in part the actions of TNF-α on insulin-stimulated glucose transport through GLUT4 in adipocytes as well as in muscle cells.
Consistent with this hypothesis, siRNA-based silencing of MAP4k4 partially protected against the TNF-induced depletion of both PPAR-γ and GLUT4 (Tang et al. 2006). Furthermore, we showed in a subsequent study (unpublished data) that depletion of MAP4K4 in 3T3-L1 adipocytes enhances PPAR-γ protein levels and function, suggesting that MAP4K4 is required for TNF-α signalling in adipocytes. However, we have found that MAP4k4 is not required for many of the TNF-α effects in adipocytes, such as the decrease in PPAR-γ mRNA. Interestingly, TNF-α appears to require MAP4K4 to cause insulin resistance in muscle, as siRNA silencing of MAP4k4 completely restores insulin sensitivity in muscle tissue from diabetic humans, in part by downregulating TNF-α activation of JNK1/2 and ERK1/2 (Bouzakri & Zierath 2007). Thus, based on the recent results published by our group (Tang et al. 2006, Tesz et al. 2007) and others (Bouzakri & Zierath 2007), MAP4k4 seems to function in the signalling pathways that mediate at least some of the inhibitory effects of TNF-α on adipose and muscle tissue processes, as depicted in Figure 4b.
Based on the results of TNF-α action on MAP4K4 expression in 3T3-L1 adipocytes described above, the increased levels of TNF-α within adipose tissue of both obese humans and animals may also upregulate the expression of MAP4K4 in adipose tissues. Thus, a key question raised by the above studies is whether MAP4K4 mediates some of the insulin resistance induced by obesity. Few years ago, mice deficient in MAP4K4 were generated by homologous recombination at the MAP4K4 gene (Xue et al. 2001). However, MAP4K4 (−/−) mice die between embryonic day 9.5 and 10.5, making it difficult to test a potential role of MAP4K4 in insulin resistance associated with increased levels of TNF-α in obesity. Nevertheless, this question should soon be addressed with conditional knockout models, where mice deficient in MAP4K4 in skeletal muscle or adipose tissue may be protected from insulin resistance caused by high fat feeding.
Another important question that is currently under investigation in our laboratory is what are the molecular mechanisms by which MAP4K4 depletion upregulates PPAR-γ function? Remarkably, RNAi-mediated silencing of MAP4K4 elevated the levels of both PPAR-γ1 and PPAR-γ2 proteins two to threefold in 3T3-L1 adipocytes without affecting PPAR-γ mRNA levels, suggesting that MAP4K4 regulates PPAR-γ at a post-transcriptional step (K.V.P. Guntur, A. Guilherme, G.J. Tesz & M.P. Czech, unpublished data). Moreover, PPAR-γ degradation rates are remarkably rapid as measured in the presence of cylcohexamide (t1/2 = 2 h), and silencing MAP4K4 had no effect on PPAR-γ degradation (Guntur et al. 2007, unpublished data). Thus, based on our preliminary results, it appears that MAP4K4 regulates PPAR-γ protein expression by inhibiting protein translation in cultured adipocytes. Further investigations will be required to confirm these results, as well as to identify the elements downstream of MAP4K4 that mediates its effects on protein translation in cultured adipocytes.
FSP27: a lipid droplet protein that controls fat storage
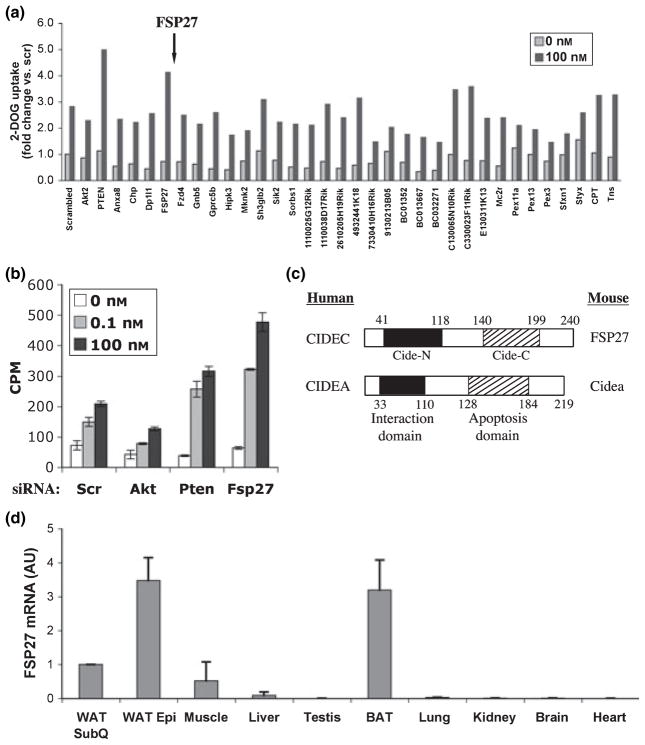
Another siRNA-based gene silencing screen we performed revealed the highly expressed protein FSP27 as a novel negative regulator of insulin-stimulated deoxyglucose uptake (Fig. 5a). Subsequent assays performed under standard conditions confirmed that depletion of FSP27 from cultured adipocytes enhanced insulin signalling to hexose transport, probably by increasing GLUT4 expression (Fig. 5b and data not shown). These latter results also show that depletion of the protein kinase Akt, a known positive regulator of insulin signalling (Jiang et al. 2003), inhibited insulin stimulation of deoxyglucose uptake, as expected, while depletion of a known negative regulator of insulin signalling, the PtdIns(3,4,5)P3 phosphatase PTEN, enhanced this process. In many experiments, depletion of FSP27 mRNA levels by siRNA was observed to be in the range of 60–90% in 3T3-L1 adipocytes (Fig. 5c).
Figure 5.
siRNA-based screen identifies FSP27 as a negative regulator of 2-deoxyglucose (2-DOG) uptake in 3T3-L1 adipocytes. (a) Four days after the induction of differentiation, 3T3-L1 adipocytes were transfected with pools of siRNA against the panel of genes shown (GenBank accession numbers will be provided upon request). The effect of each knockdown on 2-DOG uptake was determined by using a 2-DOG uptake assay. Shown is the average of two independent experiments. siRNA was purchased from Dharmacon (Chicago, IL, USA). Pools of siRNA consisting of a mix of four individual oligonucleotides against each gene were used for the screen. (b) Dose-dependent insulin-stimulated 2-DOG uptake in 3T3-L1 adipocytes. Cells on their fourth day of differentiation were transfected with scrambled siRNA or siRNA against, Akt, Pten or FSP27 and assayed for 2-DOG uptake after 72 h. Results are representative of at least six independent experiments each performed in triplicate. (c) Graphic representation of FSP27 and Cidea showing N-terminal CIDE-N domain and COOH-terminal CIDE-C domain. (d) Real-time PCR analysis of mRNA levels of FSP27 in mouse inguinal and axillary subcutaneous white adipose tissue, epididymal white adipose tissue, interscapular brown adipose tissue, liver, muscle, kidney, heart, lung, brain and testis. These tissues or organs were excised from 6-week-old C57Bl/6J male mice that were housed on a 12-h light/dark schedule and had free excess to water and food. After procurement, the samples were immediately stored flash frozen and stored at −80 °C. All procedures were carried according to the guidelines of the University of Massachusetts Medical School Institutional Animal Care and Use Committee (UMMS-IACUC). 36B4 gene was used as a reference gene for quantitative analysis.
FSP27 is a member of the CIDE family of proteins that share a conserved N-terminal CIDE-N domain and COOH-terminal CIDE-C domain (Fig. 5c). Three CIDEs have been reported in mice (Cidea, Cideb and FSP27) and humans (CIDEA, CIDEB and CIDEC) (Liang et al. 2003). The CIDE-N domain of CIDEs has significant homology to the regulatory domains of the apoptotic DNA fragmentation factors DFF40 (caspase activated-nuclease) and DFF45 (inhibitor) (Danesch et al. 1992, Inohara et al. 1998). FSP27 shares significant sequence similarity to Cidea (Inohara et al. 1998), a BAT protein in mouse that has been reported to be mitochondrial (Zhou et al. 2003), and Cideanull mice have a lean phenotype with resistance to diet-induced obesity and diabetes (Tansey et al. 2001, Zhou et al. 2003). As shown in Figure 5d, we could confirm high expression of FSP27 in WAT and BAT by RT-PCR, although detectable expression was also observed in mouse skeletal muscle. The Novartis GNF gene atlas shows that the human homologue of FSP27, CIDEC, is highly expressed in human adipocytes (Su et al. 2002), with little or no expression detectable in other tissues.
FSP27 itself was originally identified as an FSP of unknown function (Danesch et al. 1992, Inohara et al. 1998). In attempting to determine the biological mechanisms of FSP27 function in adipocytes, our preliminary studies unexpectedly revealed that FSP27 is associated with lipid droplets in 3T3-L1 adipocytes (V. Puri et al., 2007). Interestingly, a previous report based on the proteomic analysis on lipid droplets isolated from adipocytes listed FSP27 as one of the proteins that was present in isolated lipid droplet preparations (Brasaemle et al. 2004). In another set of studies, we found that siRNA-mediated knockdown of FSP27 in 3T3-L1 adipocytes leads to increased basal lipolysis as measured by glycerol release into the medium (V. Puri et al., 2007). Zhou et al. (2003) reported enhanced lipolysis in brown adipose tissue of Cideanull mice and Nordstrom et al. (2005) reported increased basal lipolysis in Cidea-depleted human pre-adipocytes in vitro. Thus, our new data on FSP27 provide an explanation for these previously published results – FSP27 and related CIDE proteins such as Cidea are lipid droplet proteins that may shield and protect the lipid droplets from lipolysis. It will be interesting in future studies to determine the structural basis for interaction of FSP27 with lipid droplets and regulation of lipolysis. Nonetheless, our findings already show that this ‘hit’ in the RNAi-based screening plays a role in a major function of adipocytes – storage of triglyceride in lipid droplets.
We have also further analysed the expression of FSP27 during adipogenesis in culture (Fig. 6a) and in response to PPAR-γ depletion in mature 3T3-L1 adipocytes using Affymetrix GeneChip microarrays (Fig. 6b). These data indicate a 59-fold increase in FSP27 mRNA expression during adipogenesis, consistent with previous findings, and a corresponding decrease in FSP27 expression upon depletion of PPAR-γ in fully differentiated 3T3-L1 adipocytes. Real-time quantitative PCR analysis shows PPAR-γ itself as well as adipose-specific genes GLUT4, AP2 and perilipin are upregulated upon FSP27 silencing (Fig. 6c). It is intriguing that FSP27, discovered by us to be a lipid droplet protein as described above, was revealed as a hit in the RNAi screen for regulators of insulin-sensitive deoxyglucose uptake (Fig. 5). One explanation is that in response to FSP27 depletion, increased intracellular fatty acids or other lipids released from lipid droplets mediate secondary responses that lead to increased GLUT4 expression (Fig. 6d). Indeed fatty acid derivatives such as eicosanoids have been shown to be sequestered in lipid droplets (Finstad et al. 1998). PPAR-γ serves as a key regulator of adipocyte differentiation and lipid storage (Tontonoz et al. 1995, Hamm et al. 1999, Rosen et al. 2000, Lazar 2002) and it is involved in regulation of GLUT4 in adipocytes (MacDougald & Lane 1995). Perhaps PPAR-γ or other nuclear receptors respond to ligands liberated from lipid droplets in response to FSP27 depletion to mediate this effect on GLUT4 transcription (Fig. 6d). In our laboratory, further studies in progress are designed to reveal functional roles of FSP27 as a lipid droplet protein and the related proteins Cidea and Cideb in adipocytes as well as in whole body metabolism in mice and humans.
Figure 6.
The expression of FSP27 is greatly upregulated during differentiation of 3T3-L1 adipocytes. (a) mRNA levels of FSP27 in 3T3-L1 adipocytes at different days of differentiation. Data were generated using Affymetrix GeneChip microarrays. (b) Fold changes, indicating the difference in expression of PPAR-γ and FSP27, from day 0 to day 6 during adipogenesis and after transfecting adipocytes (day 4) with scrambled siRNA or siRNA against PPAR-γ (P < 0.0001). (c) Real-time PCR analysis of mRNA levels of various adipogenic markers in 3T3-L1 adipocytes transfected with scrambled siRNA or siRNA against FSP27 on day 4 of their differentiation. Results are an average of three or more independent experiments. (d) Working model and hypothesis. We hypothesize that FSP27 is a lipid droplet protein that localizes to lipid droplets in adipocytes. Its presence on lipid droplets inhibits basal lipolysis, thereby promoting net triglyceride storage. Hypothetically, FSP27 depletion in our RNAi screen may have increased expression of GLUT4 through the release of PPAR-γ ligand(s) from lipid droplets, with the resultant increase in adipogenesis and adipocyte-specific genes (see text for details).
Summary and conclusions
Although much has been learned about adipocyte functions over the past 10 years, it is likely there remain multiple additional layers of unknown pathways and their components yet to be discovered. Biological screens of relatively high throughput have proved to be powerful approaches to discovery, in part because little or no bias is instilled into experimental designs. The RNAi-based screens for novel regulators of adipocyte function that we were able to develop, described in this review, confirm this attribute. The ‘hits’ described here, RIP140, MAP4k4 and FSP27, were all novel proteins with respect to regulation of adipocyte biology when they appeared in our screens. They are remarkably diverse and belong to three different classes of proteins: transcriptional corepressor, protein kinase and lipid droplet protein. Each regulates adipocyte biology through a different initiating mechanism; but they were revealed as an ultimate consequence of effects of their perturbations on expression of GLUT4, which is highly responsive to PPAR-γ and adipogenesis. It is likely that one of the targets of each of these novel adipocyte regulators is PPAR-γ itself, as described in detail in this review. RIP140 directly interacts with PPAR-γ and negatively influences its transcriptional activity; MAP4k4, likely through a protein kinase cascade, regulates the expression level of PPAR-γ; and FSP27 acts as a lipid droplet protein to restrain lipolysis, which in turn may regulate PPAR-γ through controlling the release of ligands. Thus, these novel adipocyte regulators have revealed new modes for modulating PPAR-γ, a crucial player in determining the adipocyte phenotype. In addition, each of these new regulators operates within a complex set of pathways to play a major role in regulating processes of central importance to adipocyte biology – adipogenesis, triglyceride storage, fatty acid release, fatty acid oxidation and mitochondrial oxidative phosphorylation. Determining further details of their mechanisms of action will be of high interest in the search for potential therapeutic approaches to metabolic diseases.
Acknowledgments
We thank our laboratory colleagues for their contributions and discussions about this work. The studies from our laboratory described here were supported by National Institutes of Health grants DK30898, DK60564, Diabetes Genome Project Consortium Grant DK60837-03 and University of Massachusetts Medical School Diabetes and Endocrinology Center (DK325220) Core Facilities (Genomics Core, Biomedical Imaging Core and Bioinformatics Core).
Footnotes
Conflict of interest
M.P.C. and A.G. own, or may be provided, equity in CytRx, Inc., based on intellectual property licensed from the University of Massachusetts Medical School, which shares royalties with inventors.
References
- Ahren B. Type 2 diabetes, insulin secretion and beta-cell mass. Curr Mol Med. 2005;5:275–286. doi: 10.2174/1566524053766004. [DOI] [PubMed] [Google Scholar]
- Almind K, Manieri M, Sivitz WI, Cinti S, Kahn CR. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proc Natl Acad Sci USA. 2007;104:2366–2371. doi: 10.1073/pnas.0610416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augereau P, Badia E, Fuentes M, Rabenoelina F, Corniou M, Derocq D, Balaguer P, Cavailles V. Transcriptional regulation of the human NRIP1/RIP140 gene by estrogen is modulated by dioxin signalling. Mol Pharmacol. 2006;69:1338–1346. doi: 10.1124/mol.105.017376. [DOI] [PubMed] [Google Scholar]
- Bouzakri K, Zierath JR. MAP4K4 gene silencing in human skeletal muscle prevents tumor necrosis factor-alpha-induced insulin resistance. J Biol Chem. 2007;282:7783–7789. doi: 10.1074/jbc.M608602200. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, Abdelkarim M, Caron S, Torpier G, Fruchart JC, Gonzalez FJ, Kuipers F, Staels B. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J Biol Chem. 2006;281:11039–11049. doi: 10.1074/jbc.M510258200. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. Embo J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Kiskinis E, Debevec D, Leonardsson G, White R, Parker MG. RIP140-targeted repression of gene expression in adipocytes. Mol Cell Biol. 2005;25:9383–9391. doi: 10.1128/MCB.25.21.9383-9391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Tullet JM, Parker MG. Characterization of four autonomous repression domains in the core-pressor receptor interacting protein 140. J Biol Chem. 2004;279:15645–15651. doi: 10.1074/jbc.M313906200. [DOI] [PubMed] [Google Scholar]
- Civitarese AE, Ukropcova B, Carling S, Hulver M, DeFronzo RA, Mandarino L, Ravussin E, Smith SR. Role of adiponectin in human skeletal muscle bioenergetics. Cell Metab. 2006;4:75–87. doi: 10.1016/j.cmet.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CS, Hong J, Sapinoso L, Zhou Y, Liu Z, Micklash K, Schultz PG, Hampton GM. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci USA. 2006;103:3775–3780. doi: 10.1073/pnas.0600040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Danesch U, Hoeck W, Ringold GM. Cloning and transcriptional regulation of a novel adipocyte-specific gene, FSP27. CAAT-enhancer-binding protein (C/EBP) and C/EBP-like proteins interact with sequences required for differentiation-dependent expression. J Biol Chem. 1992;267:7185–7193. [PubMed] [Google Scholar]
- Debevec D, Christian M, Morganstein D, Seth A, Parker M, White R. RIP140 regulates expression of Uncoupling Protein 1 in adipocytes through specific PPAR isoforms and ERR{alpha} Mol Endocrinol. 2007;21:1581–1592. doi: 10.1210/me.2007-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finstad HS, Drevon CA, Kulseth MA, Synstad AV, Knudsen E, Kolset SO. Cell proliferation, apoptosis and accumulation of lipid droplets in U937-1 cells incubated with eicosapentaenoic acid. Biochem J. 1998;336:451–459. doi: 10.1042/bj3360451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova O, Marcus-Samuels B, Graham D, Kim JK, Shulman GI, Castle AL, Vinson C, Eckhaus M, Reitman ML. Surgical implantation of adipose tissue reverses diabetes in lipoatrophic mice. J Clin Invest. 2000;105:271–278. doi: 10.1172/JCI7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Huq MD, Khan SA, Tsai NP, Wei LN. Regulation of co-repressive activity of and HDAC recruitment to RIP140 by site-specific phosphorylation. Mol Cell Proteomics. 2005;4:1776–1784. doi: 10.1074/mcp.M500236-MCP200. [DOI] [PubMed] [Google Scholar]
- Gurevich I, Flores AM, Aneskievich BJ. Corepressors of agonist-bound nuclear receptors. Toxicol Appl Pharmacol. 2007;223:288–298. doi: 10.1016/j.taap.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haluzik M, Parizkova J, Haluzik MM. Adiponectin and its role in the obesity-induced insulin resistance and related complications. Physiol Res. 2004;53:123–129. [PubMed] [Google Scholar]
- Hamm JK, el Jack AK, Pilch PF, Farmer SR. Role of PPAR gamma in regulating adipocyte differentiation and insulin-responsive glucose uptake. Ann N Y Acad Sci. 1999;892:134–145. doi: 10.1111/j.1749-6632.1999.tb07792.x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Leo C, Yu S, Wang H, Shen M, Luo Y, Daniel-Issakani S, Payan DG, Xu X. Identification and functional characterization of a novel human misshapen/Nck interacting kinase-related kinase, hMINK beta. J Biol Chem. 2004;279:54387–54397. doi: 10.1074/jbc.M404497200. [DOI] [PubMed] [Google Scholar]
- Huang S, Czech MP. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Huq MD, Wei LN. Post-translational modification of nuclear co-repressor receptor-interacting protein 140 by acetylation. Mol Cell Proteomics. 2005;4:975–983. doi: 10.1074/mcp.M500015-MCP200. [DOI] [PubMed] [Google Scholar]
- Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Koseki T, Chen S, Wu X, Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZY, Zhou QL, Coleman KA, Chouinard M, Boese Q, Czech MP. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc Natl Acad Sci USA. 2003;100:7569–7574. doi: 10.1073/pnas.1332633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JR, Barrick C, Kim KA, Lindner J, Blondeau B, Fujimoto Y, Shiota M, Kesterson RA, Kahn BB, Magnuson MA. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc Natl Acad Sci USA. 2005;102:6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvet LK, Andresen SM, Schuster GU, Dalen KT, Tobin KA, Hollung K, Haugen F, Jacinto S, Ulven SM, Bamberg K, Gustafsson JA, Nebb HI. On the role of liver X receptors in lipid accumulation in adipocytes. Mol Endocrinol. 2003;17:172–182. doi: 10.1210/me.2001-0210. [DOI] [PubMed] [Google Scholar]
- Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Yamauchi T, Kubota T, Moroi M, Matsui J, Eto K, Yamashita T, Kamon J, Satoh H, et al. Disruption of adiponectin causes insulin resistance and neointimal formation. J Biol Chem. 2002;277:25863–25866. doi: 10.1074/jbc.C200251200. [DOI] [PubMed] [Google Scholar]
- L’Horset F, Dauvois S, Heery DM, Cavailles V, Parker MG. RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol Cell Biol. 1996;16:6029–6036. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte BA, Repa JJ, Joseph SB, Wilpitz DC, Kast HR, Mangelsdorf DJ, Tontonoz P. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc Natl Acad Sci USA. 2001;98:507–512. doi: 10.1073/pnas.021488798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar MA. Becoming fat. Genes Dev. 2002;16:1–5. doi: 10.1101/gad.964002. [DOI] [PubMed] [Google Scholar]
- Lehrke M, Lazar MA. The many faces of PPAR-gamma. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A, White R, Parker MG. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci USA. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Houseknecht KL, Stenbit AE, Katz EB, Charron MJ. Reduced glucose uptake precedes insulin signaling defects in adipocytes from heterozygous GLUT4 knockout mice. FASEB J. 2000;14:1117–1125. doi: 10.1096/fasebj.14.9.1117. [DOI] [PubMed] [Google Scholar]
- Li J, Larocca JN, Rodriguez-Gabin AG, Charron MJ. Expression and signal transduction of the glucagon receptor in betaTC3 cells. Biochim Biophys Acta. 1997;1356:229–236. doi: 10.1016/s0167-4889(96)00170-x. [DOI] [PubMed] [Google Scholar]
- Liang L, Zhao M, Xu Z, Yokoyama KK, Li T. Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. Biochem J. 2003;370:195–203. doi: 10.1042/BJ20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Cabrero A, Saha PK, Kojima H, Li L, Chang BH, Paul A, Chan L. Increased beta -oxidation but no insulin resistance or glucose intolerance in mice lacking adiponectin. J Biol Chem. 2002;277:34658–34661. doi: 10.1074/jbc.C200362200. [DOI] [PubMed] [Google Scholar]
- MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- Machida N, Umikawa M, Takei K, Sakima N, Myagmar BE, Taira K, Uezato H, Ogawa Y, Kariya K. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J Biol Chem. 2004;279:15711–15714. doi: 10.1074/jbc.C300542200. [DOI] [PubMed] [Google Scholar]
- Marchetti P, Del Prato S, Lupi R, Del Guerra S. The pancreatic beta-cell in human Type 2 diabetes. Nutr Metab Cardiovasc Dis. 2006;16(Suppl 1):S3–S6. doi: 10.1016/j.numecd.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Mitra P, Zheng X, Czech MP. RNAi-based analysis of CAP, Cbl, and CrkII function in the regulation of GLUT4 by insulin. J Biol Chem. 2004;279:37431–37435. doi: 10.1074/jbc.C400180200. [DOI] [PubMed] [Google Scholar]
- Mostaqul Huq MD, Gupta P, Tsai NP, White R, Parker MG, Wei LN. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25:5094–5104. doi: 10.1038/sj.emboj.7601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol D, Christian M, Steel JH, White R, Parker MG. RIP140 expression is stimulated by estrogen-related receptor alpha during adipogenesis. J Biol Chem. 2006;281:32140–32147. doi: 10.1074/jbc.M604803200. [DOI] [PubMed] [Google Scholar]
- Nishigaki K, Thompson D, Yugawa T, Rulli K, Hanson C, Cmarik J, Gutkind JS, Teramoto H, Ruscetti S. Identification and characterization of a novel Ste20/germinal center kinase-related kinase, polyploidy-associated protein kinase. J Biol Chem. 2003;278:13520–13530. doi: 10.1074/jbc.M208601200. [DOI] [PubMed] [Google Scholar]
- Nordstrom EA, Ryden M, Backlund EC, Dahlman I, Kaaman M, Blomqvist L, Cannon B, Nedergaard J, Arner P. A human-specific role of cell death-inducing DFFA (DNA fragmentation factor-alpha)-like effector A (CIDEA) in adipocyte lipolysis and obesity. Diabetes. 2005;54:1726–1734. doi: 10.2337/diabetes.54.6.1726. [DOI] [PubMed] [Google Scholar]
- Pajvani UB, Scherer PE. Adiponectin: systemic contributor to insulin sensitivity. Curr Diab Rep. 2003;3:207–213. doi: 10.1007/s11892-003-0065-2. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Shulman GI. Pathogenesis of skeletal muscle insulin resistance in type 2 diabetes mellitus. Am J Cardiol. 2002;90:11G–18G. doi: 10.1016/s0002-9149(02)02554-7. [DOI] [PubMed] [Google Scholar]
- Powelka AM, Seth A, Virbasius JV, Kiskinis E, Nicoloro SM, Guilherme A, Tang X, Straubhaar J, Cherniack AD, Parker MG, Czech MP. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Invest. 2006;116:125–136. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri V, Chakladar A, Virbasius JV, Konda S, Powelka AM, Chouinard M, Hagan GN, Perugini R, Czech MP. RNAi-based gene silencing in primary mouse and human adipose tissues. J Lipid Res. 2007;48:465–471. doi: 10.1194/jlr.D600033-JLR200. [DOI] [PubMed] [Google Scholar]
- Puri V, Konda S, Ranjit S, Aouadi M, Chawla A, Chouinard M, Chakladar A, Czech MP. Fat specific protein 27: A novel lipid droplet protein that enhances triglyceride storage. J Biol Chem. 2007;282:34213–34218. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- Rizzo G, Disante M, Mencarelli A, Renga B, Gioiello A, Pellicciari R, Fiorucci S. The farnesoid X receptor promotes adipocyte differentiation and regulates adipose cell function in vivo. Mol Pharmacol. 2006;70:1164–1173. doi: 10.1124/mol.106.023820. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- Rossetti L, Stenbit AE, Chen W, Hu M, Barzilai N, Katz EB, Charron MJ. Peripheral but not hepatic insulin resistance in mice with one disrupted allele of the glucose transporter type 4 (GLUT4) gene. J Clin Invest. 1997;100:1831–1839. doi: 10.1172/JCI119711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsali A, Nathan M. A review of types 1 and 2 diabetes mellitus and their treatment with insulin. Am J Ther. 2006;13:349–361. doi: 10.1097/00045391-200607000-00012. [DOI] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JB, Moon HM, Kim WS, Lee YS, Jeong HW, Yoo EJ, Ham J, Kang H, Park MG, Steffensen KR, Stulnig TM, Gustafsson JA, Park SD, Kim JB. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2004;24:3430–3444. doi: 10.1128/MCB.24.8.3430-3444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268:22243–22246. [PubMed] [Google Scholar]
- Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem. 2001;276:34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- Sovik O, Vestergaard H, Trygstad O, Pedersen O. Studies of insulin resistance in congenital generalized lipodystrophy. Acta Paediatr Suppl. 1996;413:29–37. doi: 10.1111/j.1651-2227.1996.tb14263.x. [DOI] [PubMed] [Google Scholar]
- Stenbit AE, Tsao TS, Li J, Burcelin R, Geenen DL, Factor SM, Houseknecht K, Katz EB, Charron MJ. GLUT4 heterozygous knockout mice develop muscle insulin resistance and diabetes. Nat Med. 1997;3:1096–1101. doi: 10.1038/nm1097-1096. [DOI] [PubMed] [Google Scholar]
- Su YC, Han J, Xu S, Cobb M, Skolnik EY. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira K, Umikawa M, Takei K, Myagmar BE, Shinzato M, Machida N, Uezato H, Nonaka S, Kariya K. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem. 2004;279:49488–49496. doi: 10.1074/jbc.M406370200. [DOI] [PubMed] [Google Scholar]
- Tang X, Guilherme A, Chakladar A, Powelka AM, Konda S, Virbasius JV, Nicoloro SM, Straubhaar J, Czech MP. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPAR-gamma, adipogenesis, and insulin-responsive hexose transport. Proc Natl Acad Sci USA. 2006;103:2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, Reitman ML, Deng CX, Li C, Kimmel AR, Londos C. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci USA. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H, Osman W, Shoji Y, Treuter E, Gustafsson JA, Zilliacus J. Regulation of subnuclear localization is associated with a mechanism for nuclear receptor corepression by RIP140. Mol Cell Biol. 2003;23:4187–4198. doi: 10.1128/MCB.23.12.4187-4198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesz GJ, Guilherme A, Guntur KV, Hubbard AC, Tang X, Chawla A, Czech MP. Tumor necrosis factor alpha (TNFalpha) stimulates Map4k4 expression through TNFalpha receptor 1 signaling to c-Jun and activating transcription factor 2. J Biol Chem. 2007;282:19302–19312. doi: 10.1074/jbc.M700665200. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Regulation of adipocyte gene expression and differentiation by peroxisome proliferator activated receptor gamma. Curr Opin Genet Dev. 1995;5:571–576. doi: 10.1016/0959-437x(95)80025-5. [DOI] [PubMed] [Google Scholar]
- Tozzo E, Shepherd PR, Gnudi L, Kahn BB. Transgenic GLUT-4 overexpression in fat enhances glucose metabolism: preferential effect on fatty acid synthesis. Am J Physiol. 1995;268:E956–E964. doi: 10.1152/ajpendo.1995.268.5.E956. [DOI] [PubMed] [Google Scholar]
- Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson JA. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12:864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- Vega RB, Kelly DP. A role for estrogen-related receptor alpha in the control of mitochondrial fatty acid beta-oxidation during brown adipocyte differentiation. J Biol Chem. 1997;272:31693–31699. doi: 10.1074/jbc.272.50.31693. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest. 2003;112:1785–1788. doi: 10.1172/JCI20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Uysal KT, Wiesbrock S, Yang Q, Chen H, Hotamisligil GS. Interaction of tumor necrosis factor-alpha- and thiazolidinedione-regulated pathways in obesity. Endocrinology. 2004;145:2214–20. doi: 10.1210/en.2003-1580. [DOI] [PubMed] [Google Scholar]
- Wright JH, Wang X, Manning G, LaMere BJ, Le P, Zhu S, Khatry D, Flanagan PM, Buckley SD, Whyte DB, Howlett AR, Bischoff JR, Lipson KE, Jallal B. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol Cell Biol. 2003;23:2068–2082. doi: 10.1128/MCB.23.6.2068-2082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wang X, Li Z, Gotoh N, Chapman D, Skolnik EY. Mesodermal patterning defect in mice lacking the Ste20 NCK interacting kinase (NIK) Development. 2001;128:1559–1572. doi: 10.1242/dev.128.9.1559. [DOI] [PubMed] [Google Scholar]
- Yamada T, Katagiri H, Ishigaki Y, Ogihara T, Imai J, Uno K, Hasegawa Y, Gao J, Ishihara H, Niijima A, Mano H, Aburatani H, Asano T, Oka Y. Signals from intra-abdominal fat modulate insulin and leptin sensitivity through different mechanisms: neuronal involvement in food-intake regulation. Cell Metab. 2006;3:223–229. doi: 10.1016/j.cmet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Yao Z, Zhou G, Wang XS, Brown A, Diener K, Gan H, Tan TH. A novel human STE20-related protein kinase, HGK, that specifically activates the c-Jun N-terminal kinase signaling pathway. J Biol Chem. 1999;274:2118–2125. doi: 10.1074/jbc.274.4.2118. [DOI] [PubMed] [Google Scholar]
- Zhang B, Berger J, Hu E, Szalkowski D, White-Carrington S, Spiegelman BM, Moller DE. Negative regulation of peroxisome proliferator-activated receptor-gamma gene expression contributes to the antiadipogenic effects of tumor necrosis factor-alpha. Mol Endocrinol. 1996;10:1457–1466. doi: 10.1210/mend.10.11.8923470. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Yon Toh S, Chen Z, Guo K, Ng CP, Ponniah S, Lin SC, Hong W, Li P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat Genet. 2003;35:49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
- Zhou QL, Park JG, Jiang ZY, Holik JJ, Mitra P, Semiz S, Guilherme A, Powelka AM, Tang X, Virbasius J, Czech MP. Analysis of insulin signalling by RNAi-based gene silencing. Biochem Soc Trans. 2004;32:817–21. doi: 10.1042/BST0320817. [DOI] [PubMed] [Google Scholar]
- Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125:957–969. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]