Abstract
Rapid, sensitive and accurate detection of analytes present in low concentrations in complex matrices is a critical challenge. One issue that affects many biosensor protocols is the number and nature of the interferents present in complex matrices such as plasma, urine, stool and environmental samples, resulting in loss of sensitivity and specificity. We have developed a method for rapid purification, concentration and detection of target analytes from complex matrices using antibody-coated superparamagnetic nanobeads (immunomagnetic beads, or IMBs). The SPR detection signal from Staphylococcal enterotoxin B (SEB) was dramatically increased when the IMBs were used as detection amplifiers. When SEB detection included a 10-fold concentration/purification IMB protocol, a substantial increase in detection sensitivity was observed. This procedure was used to successfully purify and concentrate SEB from serum and stool samples, then amplify the SPR detection signal. SEB at a concentration of 100 picograms/mL was easily detected in both buffer and stool samples using this procedure. The IMB protocol also served to verify the analyte detection by using two different anti-SEB antibodies, mouse monoclonal antibodies attached to the magnetic nanobeads and rabbit polyclonal antibodies on the SPR sensor surface. Multiple detections of SEB in stool were performed using the same sensor surface by regenerating the sensor surfaces with a pH 2.2 buffer wash.
INTRODUCTION
Surface plasmon resonance (SPR) biosensors, when derivatized with highly specific recognition elements, provide powerful tools for rapidly determining the presence and concentration of analytes in solution or suspension. Label-free detection and near real-time analysis, coupled with the recent development of small, easy to use portable instruments make SPR biosensor systems excellent candidates for point of care detection devices, environmental monitoring systems, and for general laboratory instruments.1
Two major challenges in developing detection systems for clinical and environmental testing are overcoming interference from complex sample matrices, and achieving the sensitivities required for detection or diagnosis. Samples from serum, saliva, and stool, as well as environmental samples such as lake or ocean water and soil contain many substances that can impede the binding of analyte to the SPR surface, or bind nonspecifically the SPR surface, interfering with the specific detection signal.
Several approaches have been used to enhance the sensitivity and reduce the background for analyte detection in complex solutions. They include selective enrichment of microorganisms prior to detection using PCR-based protocols (reviewed in Benoit and Donahue, 2003)2; immunomagnetic separation and concentration of analyte3 and use of tangential filtration membrane barriers to exclude interferent molecules larger than the analyte of interest from the sample stream.4 Each of these techniques has drawbacks such as the time required for selective enrichment, acidic elution steps (prior to detection) required for immunomagnetic separation, and the inability to remove interferents from large analytes of interest using filtration protocols.
Because SPR-based assays rely on a change in refractive index near the sensor surface in response to a binding event, one means of enhancing SPR signals is to introduce secondary amplifying antibodies following the initial binding of target analyte to the SPR surface. Earlier, we described the utility of the secondary antibody verification/amplification method with SPR biosensors.5 The use of an antibody specific for a different target epitope for the secondary amplification also provides verification of the analyte detection. The use of two complimentary antibodies for the detection protocol is similar to the enzyme-linked immunosorbant assay (ELISA) method used in many current antibody-based detection systems.6
Dense particles linked to secondary antibodies have also been used to amplify the detection signal. Both colloidal gold nanoparticles7,8 and magnetic nanoparticles9 have been shown to increase the SPR signal when added as amplifiers. Colloidal magnetic particles have desirable properties for SPR detection because they can function as both a concentration/purification agent as well as an amplifier for detection. This accomplishes the two goals of increasing the sensitivity of the SPR assay (concentration and amplification by colloidal beads) as well as reducing the background interference (purification) simultaneously.
Staphylococcal enterotoxin B (SEB) (molecular weight 28.4 KDa) is one of several toxins produced by the bacterium Staphylococcus aureus and is a common cause of food poisoning outbreaks. Bacterial toxins such as SEB, which have a resistance to heat and enzymatic digestion, can cause intestinal illness in the absence of their bacterial progenitor.10 SEB is also considered a risk for use in bioterrorism due to its heat stability and high toxicity when aerosolized and inhaled.11 A sensitive, rapid assay for analyzing SEB in complex matrices would be useful.
We describe here assay protocols for immunomagnetic separation and concentration of SEB from analyte-spiked samples (buffer, stool, and serum) using small (50 nm) paramagnetic nanoparticles conjugated to monoclonal anti-SEB antibodies (IMBs). The nanoparticles also significantly amplify the SPR detection signal. Detection of picogram levels of SEB was achieved via this purification/concentration/amplification procedure. Verification of analyte presence was also achieved because each of the antibodies (bead conjugated and sensor surface immobilized) bound to different epitopes of the target analyte.
EXPERIMENTAL SECTION
The SPR Sensor System
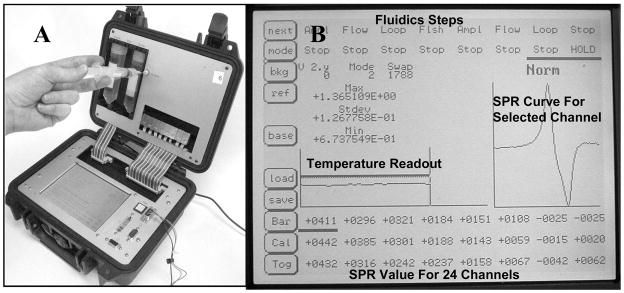
The portable 24 channel SPR biosensor system used for the experiments described here was constructed in our laboratory and was designed around the integrated three-channel Spreeta® chips developed by Texas Instruments.1 The Spreeta® SPR chips contain a light source and array detector integrated with the gold sensor surface (described in Naimushin et al. 2003, 2005; Stevens et al. 2007).12–14 The current system (Figure 1A) is an 8-chip, 24-channel system with an integrated digital signal processor (DSP). Readout and control functions are controlled with a touch screen interface (Figure 1B). The system uses a semi-automated sample handling system, in which a single sample solution is delivered via syringe through an injection port into a 2 mL sample loop (Figure 1A). The sample then flows through a sample temperature conditioning channel, then sequentially over the surfaces of the eight three-channel sensor chips. The sample flow is followed by an automated wash step to return the sensor flow stream back to starting buffer. Data from each sensor channel is reported as a refractive index value once per second. The resolution of the instrument is approximately 1×10−6 refractive index units (RIU). The numerical output from the system is a 106 multiple of the true value for ease of display and analysis, e.g. a 25 RIU value reported is actually 25×10−6 RIU. The sensor housing has an adjustable temperature controller that maintains the temperature to within ±0.01 °C, and was set to 25 °C for the experiments reported here. This portable SPR system is housed in a 10 in × 10 in × 5 in Pelican™ case, and weighs less than 6 pounds. The system operates from either line voltage (115V) or a 12V source such as a portable rechargeable battery.1
Figure 1.
(A) The 24-channel portable SPR sensor system. (B) A close up of the touch-screen display (bold labels added).
The Spreeta® sensor chip uses an 830 nm LED light source, providing a probing distance of approximately 400 nm into the sample medium. Thus, a 50 nm particle coupled to antibody (15 nm, 150 KDa) and SEB antigen (3 nm, 28.4 KDa) is well within the range of detection by the Spreeta® SPR sensor chip.15
Sensor Surface Preparation
The Spreeta® sensors were cleaned by bathing the gold sensor surfaces with a 10% nitric acid solution to remove organic materials. The surface was then rinsed with ddH2O (deionized, distilled H2O) and gently wiped with microscope lens paper wetted with 70% ethanol. Antibodies were attached to the cleaned gold sensor surface by direct physisorption. Twenty microliters of concentrated antibodies in Dulbecco’s Phosphate Buffered Saline (PBS) (Gibco #14200) were pipetted directly onto the horizontally oriented gold surface and allowed to incubate at room temperature for 2 hours. The gold surface was then rinsed with PBS to remove unbound antibodies and inserted into the flow cell housing. All antibodies used were supplied by the Department of Defense Critical Reagents Program (CRP), (Frederick, MD). For the experiments described here, a single 3-channel sensor was coated with polyclonal rabbit anti-SEB, while a second 3-channel sensor chip was coated with polyclonal rabbit anti-Bacillus anthracis and served as a reference sensor. The remaining six 3-channel sensors were not used, except when additional sensors were added for testing the regeneration of the sensor surface (Figure 7).
Figure 7.
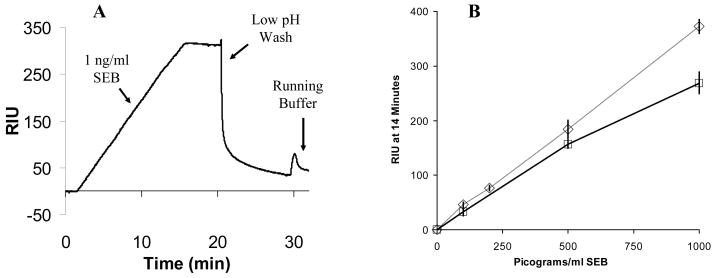
(A) SPR sensorgram showing a low pH wash to rapidly reset the sensor surface following a 1 ng/mL SEB detection in stool. (B) SEB detection in stool samples with zero (◇- ◇) and one (□- □) regeneration of the surface demonstrating the functional regeneration of the sensor surface following SEB detection from the complex (stool) sample. The data for “zero” regeneration are the same as the stool detection data shown in Figure 5 and are shown for comparative purposes. Results for one regeneration at 100 (33±7.4 RIU), 500 (156±7.1 RIU), and 1000 (269±19.8 RIU) pg/mL are shown. The control value (unspiked sample) was 15.5 RIU, and was subtracted from the data shown.
Preparation of Antibody-Coupled Nanoparticles (IMBs)
Murine monoclonal anti-SEB antibodies were provided by the CRP. Biotinylation of the antibodies was carried out using EZ-Link Sulfo-NHS-Biotin (Pierce #21338, Rockford, IL) following the standard protocols provided by the manufacturer for biotinylating immunoglobulins. A 40-molar excess EZ-link™ biotinylation solution was made by adding 19.2 μL of a 0.6 mg/mL stock solution in ddH2O to 2.5 mg monoclonal anti-SEB antibody in PBS buffer. The final reaction mixture was adjusted with PBS buffer to a final volume of 1 mL. The coupling solution was allowed to react for 1 hour, before dialysis overnight in a 3500 molecular weight cut off (MWCO) SlideA-Lyzer (Pierce #66330) against 2 liters of PBS. The extent of biotinylation was determined with the SPR system using a side-by-side comparison with commercially available biotinylated antibodies (Vector Labs #BA-2000). Three Spreeta® sensors were coated with the commercial biotinylated antibodies, the biotinylated monoclonal anti-SEB, or an unbiotinylated monoclonal anti-SEB. A 50 μg/mL avidin solution (Avidin D, Vector Labs #A2000) was then flowed through the SPR system. The unbiotinylated monoclonal anti-SEB antibodies served as a control and exhibited no avidin binding, while the commercially biotinylated antibodies from Vector Labs and the biotinylated monoclonal anti-SEB each bound similar amounts of avidin.
Biotinylated antibodies were then mixed with 50 nanometer streptavidin-coated nanobeads (Miltenyi μMACS, Gladbach, Germany #120-001-017) by incubating 100 #l of the bead solution with an excess (50 μg) of biotinylated antibodies. A 100 #L aliquot of μMACS Streptavidin nanobeads is reported by the manufacturer to bind approximately 15 μg of antibody. Use of excess antibody ensured a low degree of crosslinking of the antibody/bead complexes. The antibody/nanobead mixtures were stored at 4 °C. Prior to use, excess antibodies were removed from the mixture by magnetic separation using the magnetic wash column provided by the nanobead manufacturer (Miltenyi #130-042-701). The magnetically captured bead/antibody complexes were washed with 4 volumes of 250 μL PBS with 0.1% Tween 20 (Sigma #P1379) (PBS-T), and then eluted into 1 mL of PBS-T by adding the buffer and removing the magnet. Excess antibody was collected from the column flow-through for reuse. Two of the 1 mL samples were combined for use in each IMB experiment.
Sample Materials
The stool sample used for this study was obtained from the University of Washington Medical Center (UWMC), under a University of Washington IRB approved project for assay development. The sample obtained was mostly liquid, and stool solids were allowed to settle overnight at 4 °C before the sample was divided into 2 mL aliquots. This sample was considered a “blank” sample (containing no SEB), but was not analyzed for SEB content prior to the experiments described here. The unspiked control experiments in buffer and stool presented below indicate that little or no SEB was present in the stool sample prior to being spiked with purified SEB. Fetal bovine serum (FBS) was obtained from Gibco (#0437-028, Invitrogen Corporation, Carlsbad, CA). SEB (Sigma # S-4881) was diluted into 2 mL stool, serum, or buffer, and then added to an 18 mL nanobead mixture (2 mL nanobeads in 16 mL PBS-T) for a final volume of 20 mL.
Sample Processing
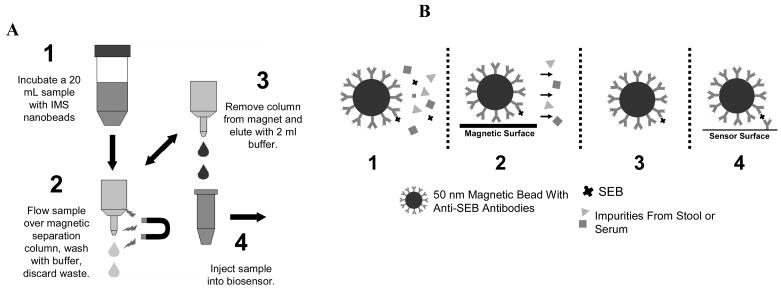
The control and SEB spiked and solutions were incubated with IMBs at room temperature for 1 hour. The 20 mL samples were processed as shown in Figure 3A, with four wash steps of 250 μL each. The bead/analyte complex was eluted with 2 mL PBS-T buffer for analysis by SPR.
Figure 3.
(A) Steps for processing samples with colloidal IMBs. (B) A cartoon of each of the processing steps.
Sensor System Operating Procedure
For each experiment, a 2 mL sample was injected into the holding loop and flowed through the system at 70 μL/min for 14 min, thus a total of 1 mL of sample was introduced to the sensor before the flush step removed any remaining sample from the injection coil. The flow was then returned to the PBS-T starting buffer, which was used as the running buffer for all experiments. For the regeneration step, low pH buffer [100mM Glycine (Fisher Scientific #BP381-1), adjusted to pH 2.2 with 1M HCl] was injected and flowed through the system for 10 minutes at a flow rate of 70 μL/min.
RESULTS AND DISCUSSION
Nanobeads Amplify the SPR Signal
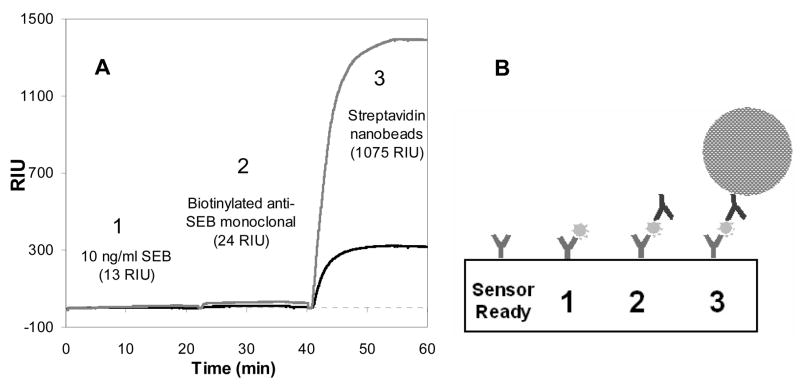
Step-wise reagent addition demonstrated the level of amplification provided by the nanobeads. Figure 2A shows the sequential three-step protocol for detection of SEB and amplification of the detection signal: 1) the direct detection of free SEB in buffer solution, 2) amplification with 50 μg/mL biotinylated anti-SEB antibodies and 3) a secondary amplification with streptavidin-nanobeads (200 μL stock μMACS beads in 2 mL total buffer) that bound to the captured biotinylated anti-SEB antibodies. Figure 2B shows a schematic of the three steps and the relative sizes of each particle. This multi-step introduction of amplifiers demonstrated the dramatic amplification that the colloidal IMBs provide to the SPR sensing protocol.
Figure 2.
(A) Stepwise detection in PBS-T buffer of (1) 10 ng/mL SEB followed by signal amplification/verification with (2) 50 μg/mL anti-SEB (monoclonal, biotinylated) and further amplification by (3) streptavidin/colloidal IMB nanoparticle conjugates (200 Nl bead stock solution in 2 mL PBS-T. This demonstrates the dramatic (80-fold) amplification of 2nd antibodies bound to nanoparticles compared with analyte alone. The reference channel (lower sensorgram, darker line) derivatized with rabbit anti-Bacillus anthracis antibody shows the background signal for each step. RIU values (in parentheses) are reference-subtracted. Channels were reset to zero between each step by the system software. (B) Illustration of the sensor surface showing the binding and amplification steps.
Detection of SEB in Buffer and Stool Using the IMB
Separation/Concentration Protocol
In subsequent experiments, monoclonal anti-SEB antibody coated nanobeads were mixed with solutions containing the target antigen (SEB) prior to injection into the SPR system. Figure 3A illustrates the procedure used to concentrate and purify the samples. In Figure 3B a cartoon diagram of the interactions are shown. In addition to amplifying the signal, the magnetic properties of the beads were used to separate the analyte from the sample matrix (analyte affinity purification) and concentrate the sample prior to introduction to the sensor system.
Due to their small size, simply holding a magnet up to the solution is not adequate for immobilizing the nanoparticles. Magnetic nanoparticles require a column of larger packed magnetic beads (illustrated in step 2 and 3 of figure 3A) that, when placed in a magnetic field, capture the smaller nanoparticles. Removal of the magnet from the column allowed the beads to be eluted into a smaller volume of buffer prior to injection into the sensor system.
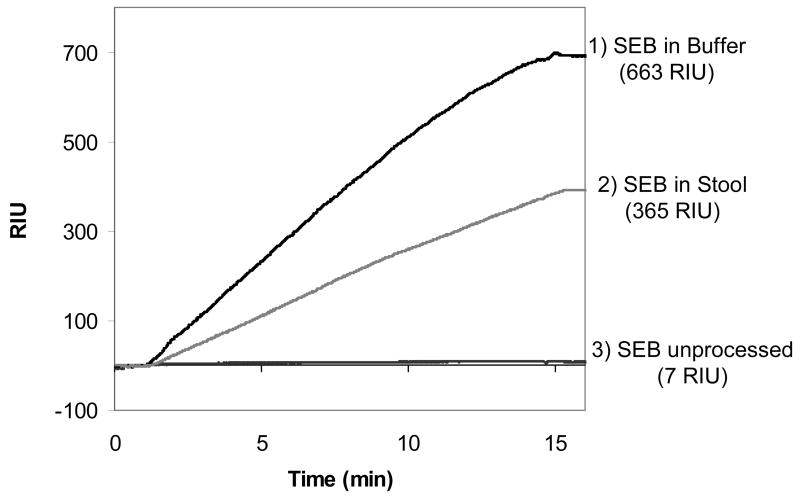
Figure 4 shows the results of three separate SEB detection experiments, two of which use the bead processing steps described in Figure 3. The largest signal (663 RIU, 58.2 RIU/min for the initial 5 min) was from a 1 ng/mL sample of SEB in buffer, processed according to the procedure in Figure 3. The intermediate signal (365 RIU, 30.6 RIU/min for the initial 5 min) was from a 1 ng/mL sample of SEB in stool processed according to the procedure in Figure 3, and the lowest signal (7 RIU, 0.5 RIU/min for the initial 5 min) was from a direct detection (no IMB processing or amplification) of a sample of 10 ng/mL SEB in buffer that was run for comparison. Unpurified stool and serum samples were not run through the instrument.
Figure 4.
The SPR response curve for a processed and unprocessed sample: 1) The result of step 4 from Figure 3, detection of 1 ng/mL SEB with the nanobead protocol in 20 mL buffer (PBS-T) starting solution, 2) The result of step 4 from Figure 3, detection of 1 ng/mL SEB with the nanobead protocol in a 20 mL stool sample (2mL stool in 18 mL PBS-T), and 3) detection of 10 ng/mL SEB in PBS-T by direct detection (no bead processing/amplification) for comparison. The reference channel signals (Rabbit anti-Bacillus anthracis antibody surface, not shown) were subtracted from the numerical value shown in parentheses, and the results are the average of triplicate samples.
Figure 4 demonstrates the dramatic increase in sensitivity provided by the IMB protocol vs. the detection of SEB alone. The 7 RIU signal from 10ng/mL SEB without IMB processing (compressed in this figure) was significantly above background and was easily detected using the current system (with a background of approximately ±1 RIU). The 1 ng/mL samples processed with the IMB procedure provided a much larger signal upon binding of the analyte/bead complex. The concentration/amplification protocol rapidly purified analyte free from complex matrices and amplified the detection signal compared to the signal from the direct detection assay.
Detection Limits in Buffer and Stool Samples Using the IMB Protocol
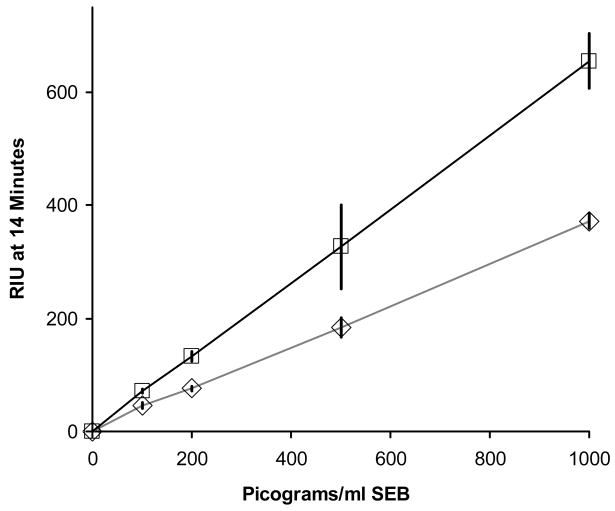
Figure 5 illustrates the analysis of both buffer and stool samples containing four different concentrations of SEB (100, 200, 500, and 1000 pg/mL) using the procedure outlined in Figure 3. Each data point represents the average of three separate sensor channels within the same experiment, with standard deviation error bars displayed. Verification of analyte identity was achieved simultaneously, since antibodies to one analyte epitope were used on the nanobead (murine monoclonal anti SEB) and antibodies to multiple analyte epitopes were immobilized on the sensor surface (rabbit polyclonal anti-SEB). Reference channels derivatized with rabbit polyclonal anti-Bacillus anthracis antibody were run simultaneously for each experiment and showed negligible nonspecific binding. Control experiments (samples run with no SEB present) were also performed and this value was subtracted from the SEB spiked result. The control value for unspiked buffer was 70 RIU, and the unspiked stool sample control was 17 RIU. The results shown had the reference channel and control value subtracted. Detection in stool involved a very complex matrix and did result in some loss of sensitivity. A comparison of direct detection of SEB in stool to SEB in buffer was not possible due to the potential for fouling of the fluidics system by particulates in the stool sample.
Figure 5.
Detection of SEB in buffer (□- □) and human stool sample (◇- ◇) solutions using colloidal IMBs for concentration of analyte and amplification of signal. Twenty mL starting volumes were processed according to the procedure outlined in Figure 3, providing 2 mL samples that were used for the SPR analyses. Results are shown for SEB in buffer at 100 (71±3.7 RIU), 200 (132±8.8 RIU), 500 (326±74.2 RIU), and 1000(655±48.8 RIU) pg/mL. Results are shown for SEB in stool samples at 100 (46±4.6), 200 (76±4.3), 500 (183±17.0 RIU), and 1000 (372±12.9 RIU) pg/mL. Standard deviation values were calculated from three simultaneous replicates run for each experiment. Values were recorded at 14 minutes following sample injection, after a brief buffer wash.
SEB Detection in Fetal Bovine Serum (FBS)
Detection of SEB with the IMB procedure in a second complex matrix (FBS) was also demonstrated. A single concentration of SEB (1 ng/mL) diluted 1:10 in FBS solution was processed as shown in Figure 3. The results are shown in Figure 6. The lack of pre-existing anti SEB antibodies in FBS eliminated potential complications from pre-existing anti SEB antibodies often seen in adult sera.12 Sensitivity of detection of SEB in serum resulted in a signal that was comparable to that achieved in buffer.
Figure 6.
Detection of 1 ng/mL SEB in FBS by the procedure outlined in figure 3A. A 2 mL sample of FBS was spiked with SEB and diluted to 20 mL with PBS-T buffer, with a final concentration of 1 ng/mL SEB. The control experiment was a repeat of the same procedure in FBS without SEB.
Regeneration of Sensor Surfaces
Removal of bound SEB-nanoparticle complexes from the sensor surface was achieved with a low pH buffer (100 mM glycine, pH 2.2) wash. Figure 7A shows the regeneration of the sensor following detection of a sample of 1 ng/mL SEB in stool. Figure 7B shows the detection of SEB at different concentrations with a low pH regeneration wash step between each detection event. Regeneration appears to lower the signal slightly, but the signal remains robust for the concentrations tested. This figure demonstrates the potential for multiple uses of the same sensor surface for repeated assays, which can greatly reduce the cost and effort of repeated assays.
CONCLUSIONS
Toxins such as SEB are ideal candidates for detection by SPR because competing techniques such as PCR and microbial enrichment techniques require the presence of the toxin-producing organism for detection. This technique for detecting SEB with both increased sensitivity and lower background will enhance efforts aimed at improving food safety and rapid diagnosis of intoxication, as well as detecting the presence of SEB in the environment in the event of a deliberate release.
The assay sensitivities reported in this study should be adequate for detection of toxin in stool samples of affected individuals. Detection of staphylococcal enterotoxins in patient stool samples with a commercial ELISA with a reported sensitivity of 1 ng/mL (TECRA #BP-211, Frenchs Forest, Australia, developed for use with food samples) and reverse passive latex agglutination (RPLA), (Oxoid #TD0940, Cambridge, UK, a kit for the detection of staphylococcal toxic shock syndrome toxin in culture filtrates) with a sensitivity of 2 ng/mL have been reported. These samples were correlated with isolates of methicillin-resistant Staphylococcus aureus (MRSA) producing the specific enterotoxin detected by the ELISA or RPLA.16
This IMB-SPR system should be adaptable for dramatically decreasing the time and expense of diagnosing the presence of many other enteric pathogens and/or their related toxins, for example invasive fungal infections such as Aspergillus and Cryptococcus that are difficult to detect with current culture based assays17, and for detection of microbial-produced toxic proteins such as Clostridium difficile toxins (a common cause of nosocomial diarrhea with an increasing incidence of infection in hospitals worldwide).18 In the in the event of a Shiga toxin outbreak (the toxin produced by enterohemorrhagic Escherichia coli), rapid, sensitive detection of the toxin in stool is important for directing treatment because the bacterial progenitor requires significant time for isolation/demonstration.19 Environmental analyses, such as detection of domoic acid in clams or seawater, also require low level detection14 as would detection of toxic proteins in the environment due to deliberate releases.
Advantages of Nanoscale Paramagnetic Particles
Larger (micron-sized) particles can also amplify the SPR signal, however these particles rapidly sediment, and are not easily mixed or directed to the sensor surface. Thus, analyte must either be eluted from the beads prior to analysis with the SPR biosensor, or complex magnetic systems must be used to direct the bead-analyte complex to the SPR surface (magnetic transport, reviewed in Gijs, 2004).20 Colloidal paramagnetic particles disperse evenly into solution with minimal mixing, do not settle out of solution like larger micron-sized paramagnetic particles and diffuse rapidly to the sensor surface. These features of paramagnetic nanoparticles are important for automated applications and in-line processing.
The two-antibody assay described here is ideal for analytes with multiple unique epitopes, such as the protein detection described here. To use this procedure on small molecules the procedure must be modified. A small molecule without multiple determinants requires a competition assay (such as that described in Stevens et al. 2007) 14, which could be modified for use with nanobeads. This would add sensitivity and dynamic range to the assay, and the ability to concentrate large volumes. Larger analytes (such as microorganisms and viruses) could also benefit from nanobead purification, increasing the sensitivity of detection.
The SPR platform used for the experiments reported here provides several important features, including portability, flexibility to analyze a broad range of analytes, high throughput capability, ease of use, and high sensitivity and rapid confirmation of positive results that make it attractive for use in a broad range of applications in the medical laboratory and beyond. The portability of the SPR system, together with its multi-analyte capabilities also make it an ideal system for rapidly identifying biological agents that may be intentionally released, and for systems to monitor and detect environmental toxins. We have previously reported adaptations of this system for detection of small molecules such as domoic acid in clam extracts14 and cortisol in saliva via a flow filtration system.4
A significant advantage of the IMB protocol is the ease of concentrating analytes prior to introduction to SPR analysis. The experiments described here made use of a 10-fold concentration of SEB from stool and serum samples; however much greater concentration ratios should be achievable for more dilute samples. With an added pre-filtering step, 100- or even 1000-fold concentration should be achievable for dilute samples such as fresh or seawater.
The nanobead processing procedure is also amenable for automation. Control of the magnet and flow through the column can be integrated into the existing SPR fluidics system. The automated protocols can also include regeneration of the sensor surface and recycling of the assay materials, dramatically reducing the per-assay cost.
Future studies should include a more detailed examination of the reproducibility of the assay, including studies with spiked stool samples from multiple patients to determine if a generic concentration curve for stool samples can be generated, and studies from individuals with diarrhea from which enterotoxin producing Staphylococcus have been isolated. While chip-to-chip variation was not examined systematically in the experiments described here, evidence that the variability is low is provided by data such as the regeneration graph in figure 7B, where different sensors were used for two separate assays at each concentration of analyte.
Acknowledgments
This work was supported in part by grants from NIEHS/NSF #OCE-0434087, the US Army #A054-022-0121 and by Grant #66-0618 from the University of Washington Center for Process Analytical Chemistry,.
Footnotes
Disclosures
Clement Furlong, Scott Soelberg and Richard Stevens are shareholders in Seattle Sensor Systems, a company that has licensed the technology for the 24-channel SPR system used for these experiments.
References
- 1.Chinowsky TM, Soelberg SD, Baker P, Swanson NR, Kauffman P, Mactutis A, Grow MS, Atmar R, Yee SS, Furlong CE. Biosen Bioelectron. 2007;22:2268–2275. doi: 10.1016/j.bios.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Benoit PW, Donahue DW. J Food Prot. 2003;66:1935–1948. doi: 10.4315/0362-028x-66.10.1935. [DOI] [PubMed] [Google Scholar]
- 3.Straub TM, Dockendorff BP, Quinonez-Diaz MD, Valdez CO, Shutthanandan JI, Tarasevich BJ, Grate JW, Bruckner-Lea CJ. J Microbiol Methods. 2005;62:303–316. doi: 10.1016/j.mimet.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Stevens RC, Soelberg SD, Near S, Furlong CE. Anal Chem. 2008;80:6747–6751. doi: 10.1021/ac800892h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naimushin AN, Soelberg SD, Nguyen DK, Dunlap L, Bartholomew D, Elkind J, Melendez J, Furlong CE. Biosen Bioelectron. 2002;17:573–584. doi: 10.1016/s0956-5663(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 6.Engvall E, Perlmann P. Immunochem. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 7.Lyon LA, Musick MD, Natan MJ. Anal Chem. 1998;70:5177–5183. doi: 10.1021/ac9809940. [DOI] [PubMed] [Google Scholar]
- 8.Yao X, Li X, Toledo F, Zurita-Lopez C, Gutova M, Momand J, Zhou FM. Anal Biochem. 2006;354:220–228. doi: 10.1016/j.ab.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Teramura Y, Arima Y, Iwata H. Anal Biochem. 2006;357:208–215. doi: 10.1016/j.ab.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 10.Dinges MM, Orwin PM, Schlievert PM. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulrich RG, Sidell S, Taylor TJ, Wilhelmsen CL, Franz DR. Medical aspects of chemical and biological warfare. TMM Publications; Washington, DC: 1997. pp. 621–630. [Google Scholar]
- 12.Naimushin AN, Soelberg SD, Bartholomew DU, Elkind JL, Furlong CE. Sens Actuators, B. 2003;96:253–260. [Google Scholar]
- 13.Naimushin AN, Spinelli CB, Soelberg SD, Mann T, Chinowsky T, Kauffman P, Yee S, Furlong CE. Sens Actuators, B. 2005;104:237–248. [Google Scholar]
- 14.Stevens RC, Soelberg SD, Eberhart BTL, Spencer S, Wekell JC, Chinowsky TM, Trainer VL, Furlong CE. Harmful Algae. 2007;6:166–174. [Google Scholar]
- 15.Homola J, Yee SS, Gauglitz G. Sens Actuators, B. 1999;54:3–15. [Google Scholar]
- 16.Boyce JM, Havill NL. Am J Gastroenterol. 2005;100:1828–1834. doi: 10.1111/j.1572-0241.2005.41510.x. [DOI] [PubMed] [Google Scholar]
- 17.Playford EG, Kong F, Sun Y, Wang H, Halliday C, Sorrell TC. J Clin Microbiol. 2006;44:876–880. doi: 10.1128/JCM.44.3.876-880.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyne L, Hamel MB, Polavaram R, Kelly CNP. Clin Infectious Diseases. 2002;34:346–353. doi: 10.1086/338260. [DOI] [PubMed] [Google Scholar]
- 19.Park CH, Kim HJ, Hixon DL. J Clin Microbiol. 2002;40:3542–3543. doi: 10.1128/JCM.40.9.3542-3543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gijs MAM. Microfluid Nanofluid. 2004;1:22–40. [Google Scholar]