Abstract
The suppressors of T cell receptor (TCR) signaling 1 and 2 (Sts-1 and -2, respectively) are multidomain proteins that negatively regulate the signaling of membrane-bound receptors, including TCR and the epidermal growth factor receptor (EGFR). Sts-1 was recently shown to be a new type of protein tyrosine phosphatase (PTP), with the phosphatase activity located within its C-terminal phosphoglycerate mutase (PGM) homology domain and key for the regulation of TCR signaling in T cells. The activity of the related Sts-2 enzyme is significantly less than that of Sts-1. Here we investigate the phosphatase activity of the PGM domain of Sts-2, Sts-2PGM. The crystal structure of Sts-2PGM is remarkably similar to Sts-1PGM, including conservation of all catalytic residues. Insight into mechanistic details is provided by the structures of the apo, tungstate-bound, and phosphate-bound enzyme. The active site shows stringent specificity, with the kcat optimum at pH 5.0 suggesting that Sts-2 might function as an acid-dependent phosphatase. Mutation of active site residues Gln372, Ala446, Glu481, Ser552, and Ser582 to their equivalents in Sts-1 increases the phosphatase activity of Sts-2PGM toward model substrates. Overall, our data demonstrate that Sts-2PGM adopts the conformation of an active phosphatase whose activity is fundamentally different from that of Sts-1 despite the strong structural homology. They also demonstrate that nonconserved active site residues are responsible for the difference in activity between the two isoforms. These differences reflect possible distinct physiological substrates.
Protein phosphorylation and dephosphorylation on tyrosine residues are key steps in regulating fundamental aspects of multicellular organisms (1–3). These steps are regulated by protein tyrosine kinases (PTKs) and PTPs,1 both of which are deregulated in human diseases, especially cancer (4, 5). The majority of known PTPs are receptor or nonreceptor proteins that harbor within their active site a nucleophilic Cys residue that is essential for catalysis (6–8). Asp-based PTPs, including members of the Eya gene products, have also been reported (9). Recently, we showed that Sts-1, a member of a newly characterized family of the suppressor of TCR signaling proteins has a His-based PTP activity (10). Sts-1 and Sts-2 belong to a gene family whose members can be found in an evolutionarily diverse group of organisms. Members of this family include Sts-1 and -2 and the insect ecdysteroid phosphate phosphatase, EPPase (11). Mouse Sts-1 was initially identified as a 70 kDa protein that binds to a phosphotyrosine-containing peptide derived from the Jak2 kinase (12), while mSts-2 was cloned on the basis of its sequence homology to mSts-1 (13). Independently, human Sts-2/Clip4 was also identified by a two-hybrid search for proteins interacting with the ubiquitin ligase c-Cbl (14) and by affinity chromatography as a c-Cbl associating protein (Sts-2/TULA) in T lymphoblastoid cells (15). In this report, Sts-1 and -2 will be used for the sake of consistency. Although Sts-1 and -2 are ~40% identical and ~75% similar, they have a distinct expression pattern. Sts-1 appears to be ubiquitously expressed, while Sts-2 is preferentially expressed in cells of the hematopoietic system (12, 15, 16).
Sts proteins are characterized by a unique tripartite structure, with an N-terminal UBA (ubiquitin association) domain, a central SH3 (Src-homology 3) domain, and a carboxyl region with similarity to the catalytic domain of enzymes of the PGM superfamily (17). In this regard, Sts proteins are the only described PGM proteins to contain the tandem UBA and SH3 homology domains (16). PGM enzymes, also termed 2H-phosphatases (18), encompass a group of structurally related enzymes that include the phosphoglycerate mutase (dPGM), fructose-2,6-bisphosphate phosphatase (Fru-2,6PPase), acid phosphatases (AcPs), and the bacterial phosphatase SixA. The substrates of these enzymes range from phosphorylated small molecules to large phosphoproteins. For example, lysophosphatidic acid is a substrate for the prostatic acid phosphatase (19), and the sensor kinase for the anaerobic respiratory control (Arc) response in Escherichia coli is a substrate for SixA (20). The sequence of these enzymes is characterized by a conserved “RHGE” signature motif that is essential for catalysis. In addition to the signature motif, one arginine and one histidine residue are strictly conserved while the rest of the primary sequence is weakly preserved among family members.
We have previously demonstrated that the 2H-phosphatase domain of Sts-1 has a phosphatase activity that targets phosphotyrosine-containing proteins, including the tyrosine kinase Zap-70 and EGFR, pTyr-containing peptides, and pTyr analogues like p-nitrophenyl phosphate (pNPP) (10). Site-directed mutagenesis confirmed that the mechanism of Sts-1PGM activity is similar to that of 2H-phosphatases and the crystal structure of the Sts-1PGM domain demonstrated its homology to members of this family of enzymes. These findings make Sts-1 the prototype of a new His-based PTP family of proteins that is different from the known family of PTPs that rely on a Cys or Asp residue for their phosphatase activity (6).
Interestingly, while the residues known to be important for the catalytic activity of Sts-1PGM are conserved in Sts-2PGM, the latter has a much weaker phosphatase activity toward pNPP (10). To shed light on the differences in catalytic activity of the Sts proteins, we used biochemical and structural approaches to characterize the activity of the Sts-2PGM domain.
EXPERIMENTAL PROCEDURES
Protein Preparation
The PGM domain of mouse Sts-2 (residues 354–624) was cloned as a His6-tagged protein in the pProEX-HTb vector (Invitrogen), expressed in the E. coli CodonPlus BL21(DE3) strain, and purified as described previously for Sts-1PGM (21). Mutants were generated with the QuikChange XL site-directed mutagenesis kit (Stratagene) and purified according to the same protocol described for the wild-type protein.
Phosphatase Assay
The phosphatase activity of wild-type and mutant Sts-2PGM was measured using pNPP (Sigma), a commonly used nonspecific phosphatase substrate. When pNPP was used, the reaction mixture contained 100 mM TAB (25 mM Tris, 50 mM acetic acid, and 25 mM bis-Tris), 150 mM NaCl, 0.1 mM EDTA, and 1 mM DTT. The reaction was initiated by adding the protein (10 µM) to the pNPP in the reaction mixture preincubated at 37 °C. The reaction was stopped by adding NaOH to a final concentration of 0.15 M and the mixture chilled on ice. The amount of the hydrolysis product pNP converted from pNPP was quantified by measuring the absorption at 405 nm using the following relationship: OD405 = εb[pNP], where ε = 1.78 × 104 M−1 cm−1, b is the light path in centimeters, and [pNP] is the pNP concentration. For the investigation of the phosphatase activity as a function of pH, TAB buffer was used over the pH interval from 4.5 to 8.5. Outside these pH values, the protein precipitated or no phosphatase activity was detected. Various steroid phosphates (Steraloids Inc.) were also tested as substrates for Sts-2PGM. Stock solutions (0.1 M) of prednisolone 21-phosphate disodium salt, dexamethasone 21-phosphate disodium salt, and β-methasone 21-phosphate disodium salt were prepared in water. The reaction mixture contained the same reaction buffer used for the pNPP, but phosphate release was assessed using the malachite green assay (22).
pTyr-Containing Proteins as Sts-2PGM Substrates
All assays were performed at 37 °C and either pH 5.0 or 7.2 as indicated. Tyrosine-phosphorylated proteins were obtained by anti-pTyr immunoprecipitations from TCR-stimulated murine T cells, and protein dephosphorylation assays were conducted as previously described (10).
kcat and Km Measurements
Initial velocities (υ) were measured at various pNPP concentrations as the slope of the linear part of the pNP concentration plotted as a function of time. To determine the kinetic parameters kcat and Km, υ was plotted as a function of pNPP concentration and the data points were fitted to a Michaelis–Menten equation
| (1) |
using SigmaPlot 5.05 (SPSS Science Inc., Chicago, IL).
Inhibition of Sts-2PGM
Tungstate and phosphate at various concentrations were incubated with the pNPP-containing buffer at 37 °C. Sts-2PGM (10 µM) was added to this buffer, and the phosphatase activity was measured as previously described. Inhibition by sodium vanadate was assessed by the malachite green assay read at 650 nm (22). Initial velocities (υ) were plotted as a function of the inhibitor concentration ([I]), and the data points were fitted with SigmaPlot to a competitive inhibition equation
| (2) |
where KI is the inhibition constant, [pNPP] = 1 mM, and Km = 2.0 mM. The reported IC50 values correspond to the inhibitor concentration at 50% inhibition.
Protein Crystallization
Initial crystals were grown at 20 °C by mixing 3 µL of 20 mg/mL Sts-2PGM [in 20 mM HEPES and 150 mM NaCl (pH 7.5)] and 3 µL of a reservoir solution. The reservoir consisted of 20% (w/v) polyethylene glycol 4000 (PEG4000), 0.2 M Li2SO4, 5–20 mM SrCl2, and 0.1 M Tris-HCl (pH 8.0). Sts-2PGM crystallized in space group P212121 with two dimers in the asymmetric unit corresponding to a Vm value (23) of 2.33 Å3/Da and an estimated solvent content of 47%.
Data Collection, Structure Determination, and Refinement
All diffraction intensities were collected at 100 K on beamline X6A at the National Synchrotron Laboratory Source (NSLS, Brookhaven National Laboratory), on a 2K × 2K CCD detector (ADSC), integrated and scaled with the HKL2000 package (24).
The structure of Sts-2PGM was determined using the single-wavelength anomalous dispersion (SAD) technique. A run consisting of 270° was collected at a wavelength of 1.0 Å to ensure a high anomalous signal on a single crystal soaked for 1 h in a solution containing 40 mM sodium tungstate. Bijvoet pairs were scaled and merged separately in HKL2000. The heavy-atom substructure was determined with SHELXD (25) with data truncated to 2.7 Å resolution. This resolution cut was on the basis of statistics reported by SHELXC. Four tungsten atoms were found using this approach. The coordinates of the heavy atoms were further refined and initial phases calculated with SOLVE (26). The electron density map calculated to 2.7 Å resolution after phasing and density modification with RESOLVE (27) proved to be of excellent quality. This electron density was used to build an initial model of four Sts-2PGM monomers with the automatic building procedure in RESOLVE (28). This model was further refined in REFMAC (29) to 1.95 Å resolution with a final round of TLS (30) to crystallographic residuals Rcryst and Rfree of 20.0 and 23.9%, respectively, and excellent stereochemistry. The final model has no density for residues A623, A624, B354–B357, B622–B624, C354–C357, C621–C624, D354–D357, and D622–D624. There was extra density N-terminal to A354 where the AMGS sequence was built in and refined. This extra sequence is a result of the way Sts-2PGM is cloned in the pProEX-HTb vector. Stereochemistry was checked with PROCHECK (31). Table 1 summarizes statistics on data collection and model refinement.
Table 1.
Statistics on X-ray Data Collection and Model Refinement
| native | tungstate | phosphate | H366A | |
|---|---|---|---|---|
| X-ray Data Collection | ||||
| unit cell dimensions (Å) | 77.9 × 116.1 × 121.4 | |||
| space group | P212121 | |||
| resolution (Å) | 50.0-1.95 | 50.0-2.25 | 50.0-2.8 | 50.0-2.05 |
| no. of unique reflections | 80016 | 101825h | 28141 | 68913 |
| completeness, overall (last shell)a (%) | 99.9 (100.0) | 99.4 (97.1)h | 98.6 (100.0) | 99.8 (100.0) |
| redundancy, overall (last shell)a | 7.6 (7.6) | 5.5 (4.7)h | 5.7 (6.0) | 7.8 (7.0) |
| 〈I〉/〈σ(I)〉, overall (last shell)a | 35.0 (4.5) | 18.5 (2.6)h | 21.1 (2.7) | 26.5 (3.0) |
| Rsymb (%) | 8.4 (50.0) | 9.8 (56.5)h | 10.1 (61.7) | 9.0 (65.4) |
| Model Refinement | ||||
| resolution (Å) | 50.0-1.95 | 50.0-2.25 | 50.0-2.8 | 50.0-2.05 |
| no. of reflections used | 79938 | 50569 | 26559 | 65215 |
| Rfreec (%), overall (last resolution shell) | 24.1 (26.3) | 24.2 (27.6) | 27.1 (34.4) | 26.8 (29.5) |
| Rworkd (%), overall (last resolution shell) | 20.3 (22.1) | 19.4 (22.6) | 23.5 (27.7) | 22.5 (22.7) |
| Rcryste (%) | 20.5 | 19.6 | 23.7 | 22.7 |
| rmsd for bond lengths (Å) | 0.013 | 0.012 | 0.014 | 0.011 |
| rmsd for bond angles (deg) | 1.427 | 1.345 | 1.523 | 1.290 |
| estimated standard uncertaintyf (Å) | 0.161 | 0.228 | 0.434 | 0.203 |
| average temperature factor (Å2) | ||||
| protein atoms | 28.1 | 40.3 | 61.5 | 33.7 |
| water molecules | 30.2 | 36.8 | 46.1 | 33.6 |
| ligand | - | 29.5 | 90.4 | - |
| ions | - | 44.3 | - | 28.3 |
| Ramachandran plotg (%) | 91.8/8.1 | 91.1/8.9 | 87.1/12.9 | 90.8/9.2 |
The last resolution shells are 2.02-1.95, 2.33-2.25, 2.90-2.80, and 2.12-2.05 Å for the native, tungstate-bound (SAD), phosphate-bound, and H366A data, respectively.
Rsym = Σi,hkl|〈I(hkl)〉 − Ii(hkl)|/Σi,hklIi(hkl).
Rfree = Σ(hkl)εT||Fobs| − |Fcalc||/Σ(hkl)εT|Fobs|, where T is the test set (37) obtained by randomly selecting 5% of the data. The last resolution shells are 2.0-1.95, 2.3-2.25, 2.85-2.77, and 2.12-2.05 Å for the native, tungstate-bound, phosphate-bound, and H366A structures, respectively.
Rwork = Σ(hkl)εW||Fobs| − |Fcalc||/Σ(hkl)εW|Fobs|, where W is the working set.
Rcryst = Σ(hkl)|||Fobs| − |Fcalc|||/Σ(hkl)|Fobs| calculated over the entire set of unique reflections.
Calculated from Rfree statistics.
Most favored regions/additional allowed regions.
These statistics were obtained after treating I+ and I− as independent reflections.
For the phosphate-bound structure, diffraction data (Table 1) were collected on one Sts-2PGM crystal soaked in 0.2 M Na/K phosphate. The H366A mutant crystallized using a reservoir solution of 20% (w/v) PEG4000, 0.2 M MgCl2, and 0.1 M Tris-HCl (pH 8.5). Both structures were determined by difference Fourier techniques and refined in REFMAC. Because of the low-resolution data of the phosphate-bound structure, tight noncrystallographic symmetry (NCS) restraints were applied in the refinement. A phosphate ion is missing from the active site of one monomer; instead, it is replaced by a water molecule located 2.8 Å from the NE2 atom of His366. In the H366A structure, an extra electron density was found at the site usually occupied by the His366 side chain. On the basis of the basic surrounding of this density and the refined temperature factor, a chloride ion was built in and refined. Coordinates of the various structures have been deposited in the Protein Data Bank (PDB) as follows: entry 3D4I for apo-Sts-2PGM, entry 3D6A for the tungstate-bound structure, and entry 3DB1 for the phosphate-bound structure.
RESULTS
Sts-2PGM Phosphatase Activity
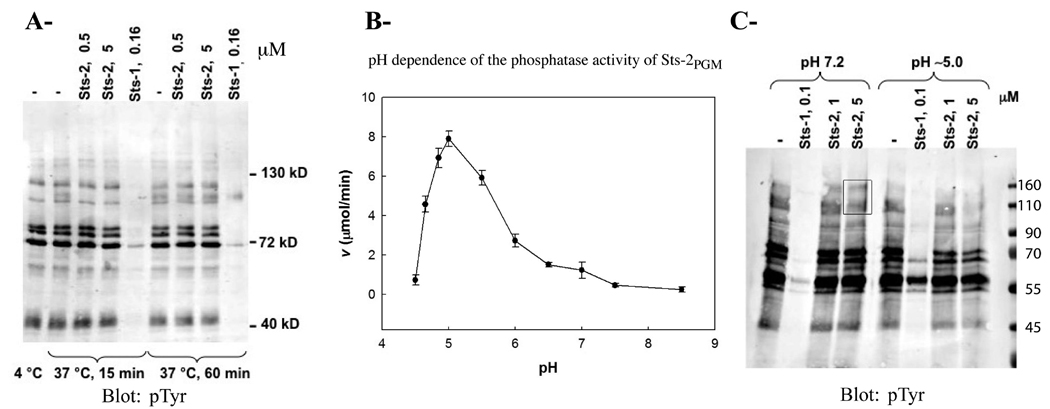
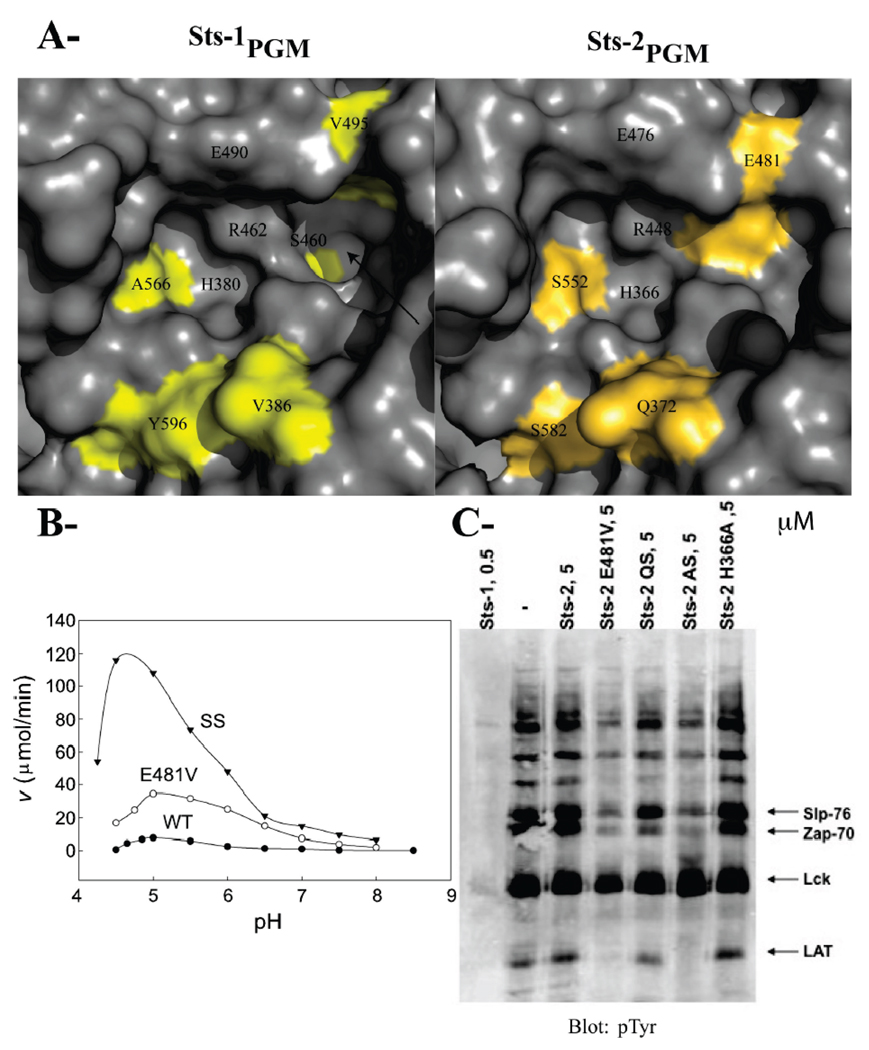
To evaluate the PTP activity of Sts-2PGM, we incubated it with a diverse array of tyrosine-phosphorylated proteins. As shown in Figure 1A, Sts-2PGM (5 µM) failed to dephosphorylate pTyr-containing proteins while Sts-1PGM (0.16 µM) efficiently dephosphorylated pTyr-containing proteins. This result is consistent with our previous finding that it has a weak phosphatase activity against the pTyr analogue pNPP (10). Because some of the 2H-phosphatases are acid phosphatases, we tested the idea that Sts-2PGM is an acid phosphatase and investigated its catalytic activity as a function of pH. We assessed the ability of recombinant Sts-2PGM to hydrolyze pNPP (1 mM) at different pH values. Because the natural physiological substrates of Sts-2PGM are unknown, pNPP was used as a model substrate. pNPP is a nonspecific phosphotyrosine analogue that is widely used in studying the reaction mechanism of various phosphatases, including PTPs and acid phosphatases. As shown in Figure 1B, the catalytic activity of Sts-2PGM is optimal at pH 5.0. At this acidic pH, the kcat (0.3 s−1) is 10 times higher than the value measured at pH 7.5; however, the Km value (1.5 mM) is similar (Table 2). In comparison, the catalytic activity of Sts-1PGM for pNPP hydrolysis is optimal at pH 7.0, and at pH 5.0, the Km value (0.02 mM) is 25 times lower than that measured at pH 7.5 (0.5 mM) while the kcat has not significantly changed [63.4 s−1 vs 70 s−1 (Table 2)]. Thus, the improvement in the specificity constant at pH 5.0 seen for both Sts domains is exhibited at different points in the catalytic trajectory and is due to distinct reasons, an increased kcat in the case of Sts-2PGM and a decreased Km in the case of Sts-1PGM. This result suggests important differences in function between the two phosphatases despite total conservation of the key catalytic residues.
FIGURE 1. Phosphatase activity of recombinant Sts-2PGM.
(A) Proteins from TCR-stimulated Jurkat cells were isolated by immunoprecipitation, eluted from the pTyr antibodies, and evaluated as Sts-2PGM substrates at 37 °C at the indicated concentration (micromolar) and time. Sts-1PGM was used as a positive control. Eluted proteins incubated at 4 and 37 °C in the absence of enzymes (two lanes to the left) were used as negative controls. (B) pH dependence of the phosphatase activity of Sts-2PGM. Initial velocities of pNPP (1 mM) hydrolysis by Sts-2PGM (10 µM) were measured and plotted at the indicated pH. All assays were conducted at 37 °C. (C) Tyrosine-phosphorylated proteins purified as described for panel A were evaluated as substrates for Sts-2PGM at two different concentrations (1 and 5 µM) and pH values (7.2 and 5.0) as indicated. Reaction products were evaluated by anti-phosphotyrosine Western analysis. The two high-molecular weight bands that are present at pH 7.2 but disappear at pH 5.0 are boxed. All assays were conducted at 37 °C with Sts-1PGM (0.1 µM) as a positive control. Molecular weight standards (kDa) are shown on the right. Each assay was repeated at least three times. Figures are representative of one experiment.
Table 2.
Activity Constants of the Sts PGM Domain Mutants
| substrate/enzyme | Km (mM) | kcat (s−1) | kcat/Km (M−1 s−1) | relative activity (%) |
|---|---|---|---|---|
| pNPP | ||||
| Sts-1PGM | 0.5 ± 0.06 | 70 ± 2.0 | 141000 | 9400 |
| Sts-1PGM (pH 5.0) | 0.02 ± 0.009 | 63.4 ± 5.5 | 3150000 | 210000 |
| Sts-2PGM | 2.0 ± 0.13 | 0.03 ± 0.001 | 15 | 1 |
| Sts-2PGM (pH 5.0) | 1.5 ± 0.08 | 0.3 ± 0.005 | 200 | 13.0 |
| Sts-2PGM(H366A) | NDc | NDc | NDc | NDc |
| Sts-2PGM(E481V) | 1.0 ± 0.15 | 0.15 ± 0.01 | 150 | 10 |
| Sts-2PGM(E481V) (pH 5.0) | 0.2 ± 0.02 | 0.65 ± 0.01 | 3100 | 207 |
| Sts-2PGM(S552A/S582Y) | 1.9 ± 0.2 | 0.43 ± 0.02 | 226 | 15 |
| Sts-2PGM(S552A/S582Y) (pH 5.0) | 0.62 ± 0.1 | 2.9 ± 0.1 | 4700 | 312 |
| Sts-2PGM(Q372V/S582Y) | 0.7 ± 0.1 | 0.4 ± 0.01 | 571 | 38 |
| Sts-2PGM(A446S/S552A) | 0.64 ± 0.08 | 0.27 ± 0.009 | 420 | 28 |
| steroid phosphatea (pH 7.5 or 5.0) | NDc | |||
| pSer (pH 7.5 or 5.0) | NDc | |||
| pThr (pH 7.5 or 5.0) | NDc | |||
| pTyr (pH 7.5 or 5.0) | NDc | |||
| phosphorylated moleculesb | NDc |
Prednisolone 21-phosphate, β-methasone 21-phosphate, and dexamethasone 21-phosphate.
β-Glycerophosphate, phenyl phosphate, 1- or 2-naphthyl phosphate, and 4-methylumbelliferyl phosphate.
Not detectable.
The common phosphatase inhibitors tungstate, vanadate, and phosphate inhibited the activity of Sts-2PGM. Fitting the initial rates of pNPP hydrolysis plotted versus the inhibitor concentration to eq 2 demonstrates that the IC50 values of tungstate, vanadate, and phosphate are 0.8 ± 0.1, 5.3 ± 3.3, and 52 ± 6.2 mM, respectively (Table 3).
Table 3.
IC50 and KI Values of Various Sts-2PGM (10 µM) Inhibitors Measured with 1 mM pNPP
| inhibitor | IC50 (mM) | KI (mM) |
|---|---|---|
| tungstate | 0.82 ± 0.1 | 0.54 ± 0.07 |
| vanadate | 5.3 ± 3.3 | 3.55 ± 2.3 |
| phosphate | 52 ± 6.2 | 35.1 ± 4.1 |
Since Sts-2PGM is a better phosphatase at pH 5.0 than at pH 7.5, we compared the PTP activity of Sts-2PGM at these two pH values. Consistent with the pH dependence data, Sts-2PGM has a slightly better PTP activity at pH 5.0 than at pH 7.2 as shown for the disappearance of high-molecular weight bands in Figure 1C. Taken together, our in vitro data show that Sts-2PGM is an active phosphatase whose activity is optimal at acidic pH values.
Structure of the Sts-2PGM Domain
To further explore the mechanism underlying the reduced phosphatase activity of Sts-2PGM, we determined its crystal structure. Table 1 summarizes statistics on data collection and model refinement. The final model consists of two interacting dimers. The organization of Sts-2PGM into dimers is consistent with size exclusion data (not shown) and with the quaternary structures of homologous Sts-1PGM (10) and EPPasePGM (32), which also form dimers. The rmsds calculated after superposition of the Cα atoms of all four Sts-2PGM monomers on each other (33) range between 0.42 and 0.78 Å, indicating that all monomers are identical. The similarity between the monomers is remarkable in the active site. For the sake of simplicity, the chain with the lowest average temperature factor will be used to describe the structure.
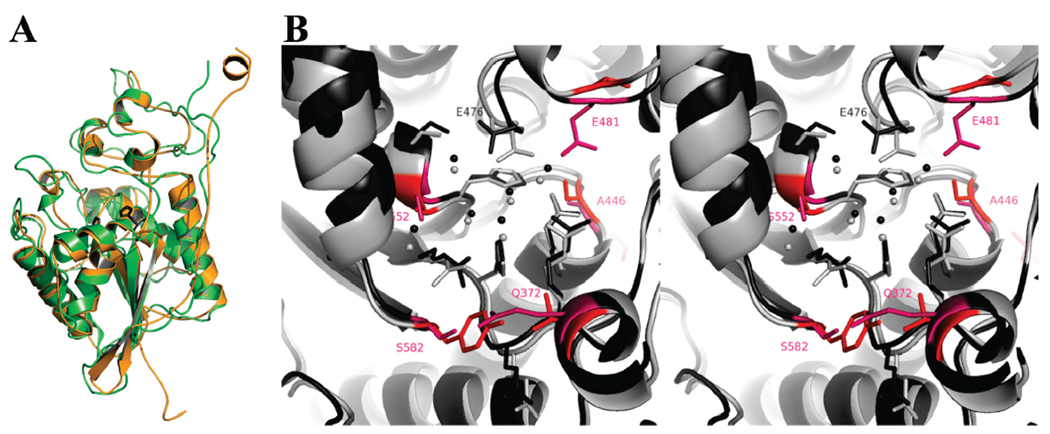
The Sts-2PGM monomer has the α/β topology characteristic of members of the 2H-phosphatase family of enzymes, including Sts-1PGM (17, 18) (see the Supporting Information). As expected from the sequence homology, Sts-2PGM is remarkably similar to Sts-1PGM [Figure 2A (10)] and EPPasePGM (32). The 365RHGE368 signature motif characteristic of all 2H-phosphatases is located in a cavity formed by the carboxyl end of the central β-sheet and by surrounding loops. This solvent-exposed cavity is the active site of all 2H-phosphatases. In Sts-2PGM, it contains the conserved Arg365, Arg448, and His551 residues in addition to the signature motif (Figure 2B). The α/β fold is stabilized by conserved interactions, which ensure that the catalytic cavity adopts a structure similar to those of many other active 2H-phosphatases (18). Thus, our structural data show that the active site of Sts-2PGM is solvent-exposed and well-suited for the binding of a phosphorylated substrate that can be hydrolyzed by the conserved basic arginine and histidine residues.
FIGURE 2. Comparison of mouse Sts-1PGM and Sts-2PGM.
(A) Superposition of Sts-1PGM (green, PDB entry 2H0Q) and Sts-2PGM (orange, PDB entry 3D4I). The two conserved His residues are shown in ball-and-stick format in dark gray to highlight the active site. The calculated rmsds between Sts-2PGM and Sts-1PGM and EPPasePGM are 1.10 and 1.45 Å, respectively. (B) Stereoview of the active sites of Sts-1PGM (light gray) and Sts-2PGM (dark gray) shown in ribbon and ball-and-stick format. The side chains of nonconserved residues are colored and labeled in red. Spheres represent water molecules. This figure was prepared with PyMOL (http://pymol.sourceforge.net/).
Sts-2PGM Active Site
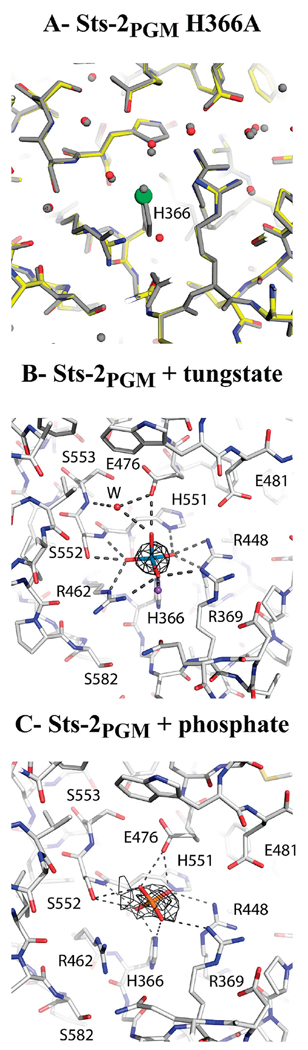
The structural similarity between Sts-2PGM and Sts-1PGM is very strong in the catalytic pocket. The conserved Arg365, His366, Arg448, and His551 residues adopt a configuration that is identical to that of their homologues in Sts-1PGM (Arg379, His380, Arg462, and His565) and in other 2H-phosphatases. For example, the imidazole group of His366 is stabilized on either side by the side chain of Arg365 and Arg448. In addition, the ND1 atom of His366 is within hydrogen bonding distance (2.8 Å) of the main chain carbonyl of Gly367, implying that this nitrogen is protonated and that NE2 is deprotonated and therefore capable of serving as a nucleophile in a hydrolysis reaction (Figure 2B). The four conserved residues are complemented in the active site of Sts-2PGM by Arg369 and Glu476, which are also conserved in Sts-1 as Arg383 and Glu490, respectively. Three water molecules sitting on top of His366 and coordinating the various catalytic residues are present in the active site. Remarkably, these water molecules are located similarly in Sts-1PGM (Figure 2B). The conservation of the arrangement of the key catalytic residues between Sts-2PGM and other 2H-phosphatases suggests not only that these residues are indeed the catalytic residues of Sts-2PGM but also that the reaction mechanism is conserved. In such a scenario, His366 serves as a nucleophile that targets the phosphorus atom of a phosphorylated substrate. The phosphate is transiently transferred to His366, which becomes temporarily phosphorylated. Arg365, Arg448, and His551 are believed to stabilize the negative charges that occur during the phosphate transfer and its subsequent hydrolysis, while Glu476 is believed to be the general acid (17, 18). Our results with the H366A mutant of Sts-2PGM confirm this hypothesis (see the Supporting Information). This mutant has no detectable activity, and the structure of its active site is preserved despite the mutation (Figure 3A) highlighting the role of the histidine of the RHGE motif as the attacking nucleophile during the phosphatase reaction in support of the accepted reaction mechanism (17). Thus, close inspection of the Sts-2PGM active site shows that the key catalytic residues for the phosphatase activity are present and adopt a conformation that is consistent with the stabilization of a phosphate moiety and therefore suitable for a phosphatase reaction.
FIGURE 3. Insight into catalysis.
(A) Superposition of the active sites of the H366A mutant (color) and wild-type Sts-2PGM (gray). Red and green spheres represent water molecules and the chloride ion, respectively. (B and C) Interactions made by the tungstate and phosphate ions, respectively, with Sts-2PGM. Active site residues within hydrogen bonding distance of the tungstate and phosphate ions are shown in ball-and-stick format. The difference electron density maps (Fo − Fc) calculated after omission of all nonprotein atoms are shown around the tungstate and phosphate moieties with cutoffs of 20σ, 4σ, and 3σ, respectively. A globular nonprotein density was also found within ionic distances (1.8–2.0 Å) of an oxygen atom of all four tungstates. On the basis of temperature factor values and residual electron density, a sodium ion (purple sphere in panel B) was built and refined into this density. The likely role of this solvent-exposed sodium ion is to cooperate with the protein basic residues in neutralizing the negative charges of the tungstate ion. The interactions made by the phosphate moiety with Sts-2PGM are similar to the ones reported for Sts-1PGM (10). Dashed lines represent hydrogen bond interactions.
Despite the strict conservation of catalytic residues, the three-dimensional structure, however, illustrates differences between Sts-1PGM and Sts-2PGM. Three active site residues, Gln372, Glu481, and Ser582 in Sts-2PGM, which surround the nucleophilic His366 from opposite sides of the active site are found to be Val386, Val495, and Tyr596, respectively, in Sts-1PGM (labeled red in Figure 2B). Glu481, which makes a strong hydrogen bond with the conserved Arg448 in Sts-2PGM, introduces a negative charge close to the conserved Glu476. The presence of this extra negative charge in the vicinity of Glu476 forces the side chain carboxylic group of this residue to adopt a conformation that is slightly different from the one it adopts in Sts-1 (Figure 2B). As a result, in Sts-2PGM Glu476 hydrogen bonds Ser553 while in Sts-1PGM Glu490 instead hydrogen bonds the water molecules that sit on top of His380. In addition, Glu481 reduces the active site volume by blocking the access to a small hydrophobic pocket adjacent to the active site. In contrast, the Glu481 equivalent in Sts-1, Val495, is a smaller hydrophobic residue that does not hydrogen bond the conserved arginine residue. These differences, which exchange small hydrophobic side chains into large acidic or hydrogen acceptor/donor ones, could underlie the relatively weaker phosphatase activity of Sts-2 (10) or affect substrate specificity (see below).
Structural Insight into the Phosphatase Reaction of Sts-2PGM
Since Sts-2PGM is an active phosphatase, it should stabilize a phosphate or a phosphate-like moiety in its active site. The presence of numerous basic residues in the active site contributes to its positive electrostatic potential in support of this hypothesis (Supporting Information). We thus determined the crystal structure of tungstate- and phosphate-bound Sts-2PGM. The structure of the tungstate complex was refined against the 2.25 Å resolution SAD data used to determine the Sts-2PGM structure (Table 1). The final electron density map unambiguously showed a four-coordinate, tetrahedral tungsten molecule bound in the active site of each Sts-2PGM chain, demonstrating that indeed the proposed active site of Sts-2PGM can stabilize a phosphate-like moiety. Sts-2PGM holds the tungstate ion tightly at the center of the active site where it replaces the three water molecules previously found in the apo form. As shown in Figure 3B, the anionic oxygen atoms of the tungstate make strong hydrogen bonds with His551, the three arginines (Arg365, Arg369, and Arg448), the main chain amide and hydroxyl groups of Ser552, and Glu476. In addition, the tungstate molecule interacts with the main chain amide of Ser553 and the side chain of Glu476 via a water molecule mediated-hydrogen bond. Except for Ser552, all these residues are conserved in Sts-1PGM. When compared to that of apo-Sts-2PGM, the binding of the tungstate to the active site resulted in subtle but important changes. For example, the guanidinium group of Arg365 reoriented such that it broke the interactions it made with Ser552 and Ser582 in apo-Sts-2PGM to interact tightly with the tungstate. Moreover, Glu481, which is close to Arg448 in apo-Sts-2PGM, has flipped to the solvent in two monomers such that it is not in the active site. In all four Sts-2PGM monomers, the NE2 atom of the active site His366 is within 2.3–2.5 Å of the tungsten atom. This geometry implies that the NE2 atom is deprotonated, consistent with the proposed mechanism of an in-line nucleophilic attack by His366 on the phosphorus atom of the phosphorylated substrate. The interactions made by the tungstate with the three arginines at positions 365, 369, and 448 are also consistent with the role put forward by the proposed mechanism for these residues in neutralizing the negative charge buildup on the transition state during phosphoryl transfer.
As with the tungstate-bound structure, the phosphate ion replaces the three water molecules in apo-Sts-2PGM close to His366 with no significant changes to the active site structure. The phosphate occupies two distinct positions in the active site of Sts-2PGM that differ by the distance between the phosphorus atom and the NE2 atom of His366. The two distinct positions occupied by the phosphate might reflect the difference in the involvement of the four monomers in the crystal packing. Interestingly and despite the conservation of the crystal packing, the tungstate has only one binding mode in the active site. The interactions of the phosphate ion with the active site depend on how close it is to His366. In the two Sts-2PGM monomers where the P–NE2 distance is 4.1 Å, the phosphate ion is within hydrogen bonding distance of Arg365 and Arg369. In the monomer where the P–NE2 distance is 3.2 Å, the phosphate makes additional hydrogen bonds with the side chains of His366, Arg448, Glu476, His551, and Ser552 (Figure 3C). In comparison to the tungstate, the phosphate is making weaker interactions with Sts-2PGM structures as shown by the fewer number of hydrogen bonds and their longer distances (Figure 3B,C). In addition, the W–NE2 distances are considerably shorter than the P–NE2 distances. Together, these observations suggest a higher affinity of the tungstate for Sts-2PGM and are consistent with its higher inhibition capability we observed in the inhibition assays.
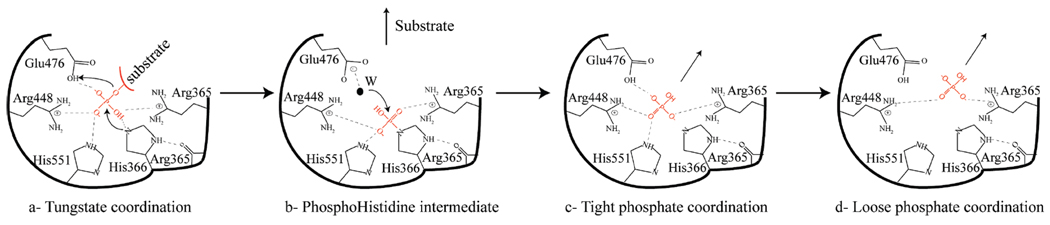
The comparison of the phosphate- and tungstate-bound structures has important implications for our understanding of the mechanism of the phosphatase reaction carried out by Sts-2PGM and more generally by other 2H-phosphatases. Because of the short distances (2.3–2.5 Å) between the tungsten and His366, we propose that the tungstate-bound structure (Figure 3B) mimics the structure of an early step during catalysis after the active site stabilized the phosphorylated substrate but before the nucleophilic attack by His366 has taken place. Our data thus support a model in which the active site residues Arg365, His366, Arg369, Arg448, Glu476, Ser552, and His551 stabilize and orient a high-affinity phosphorylated substrate close to His366 such that the P–NE2 distance is shorter than 2.3 Å. This distance allows for a nucleophilic attack by His366 on the phosphorus atom. As the reaction progresses, the P–NE2 distance becomes shorter, leading to the transfer of the phosphate to His366, the formation of pHis366, and the release of the dephosphorylated substrate into the solvent. The hydrolysis of pHis366 by a nucleophilic water attack regenerates the apoenzyme and releases an inorganic phosphate molecule that initially makes several hydrogen bonds with the active site as shown in Figure 3C. We propose that the water molecule labeled W in Figure 3B, which bridges the tungstate to Glu476 and Ser553, is well-suited for the nucleophilic attack. This water molecule is 4.5 Å from the tungsten atom and can be activated by Glu476. As the reaction proceeds, the distance from the phosphate to His366 increases and the number of hydrogen bonds it makes with the protein decreases before it is released to the solvent (Scheme 1). Together, the tungstate and phosphate structures are consistent with a model in which the cluster of basic residues located at the carboxyl end of the central β sheet structure forms the active site Sts-2PGM.
Scheme 1.
Model of the Phosphatase Reaction As Suggested by the Tungstate- and Phosphate-Bound Structuresa
a Along the reaction path, the phosphorus of the phosphorylated substrate moves within an attacking distance of the nucleophilic His (His366) such that the phosphate’s interactions with the active site are mimicked by the tungstate (a). The phosphate is then transferred to His366 to generate the phosphorylated intermediate (b). pHis366 is next hydrolyzed by the nucleophilic water molecule (W), and the released phosphate is initially tightly bound by the active site as mimicked by the interactions made by the phosphate 3.2 Å from His366 (c). These interactions weaken (phosphate 4.2 Å from His366) before the phosphate is released into the solvent (d).
Combined, our structural data support a model in which Sts-2PGM is a catalytically active phosphatase that can stabilize a phosphate moiety in its active site.
Specific Determinants of the Phosphatase Activity of Sts-2PGM
If the active site structure and the key catalytic residues are strictly conserved between Sts-2PGM and Sts-1PGM, why is Sts-2PGM a poorer phosphatase at least for the model substrate pNPP? One plausible explanation for this weak activity is that noncatalytic nonconserved residues interfere with the binding of phosphorylated substrates to the active site of Sts-2. Comparison of the two PGM domains indicates that such residues, which are colored yellow in Figure 4A, include Gln372, Ala446, Glu481, Ser552, and Ser582 (see also Figure 2B). To test this hypothesis, we generated and characterized mutants in which these residues were changed to their counterparts in Sts-1. We focused on those residues because they are located in the active site or surrounding it.
FIGURE 4. Residues that differentiate Sts-1PGM from Sts-2PGM.
(A) Surface representation of the active site of Sts-1PGM and Sts-2PGM. Nonconserved residues are colored yellow and orange, respectively. The hydrophobic cavity that is solvent-exposed in Sts-1PGM but blocked by Glu481 in Sts-2PGM is indicated with an arrow. (B) pH dependence of the phosphatase activity of Sts-2PGM. Initial velocities of pNPP (1 mM) hydrolysis by wild-type (●), E481V (○), and SS (S552A/S582Y) (▼) mutants of Sts-2PGM (1 µM) were measured and plotted at the indicated pH and 37 °C. (C) Proteins from TCR-stimulated Jurkat cells were isolated by immunoprecipitation, eluted from the pTyr antibodies, and evaluated as substrates for Sts-2PGM mutants at the indicated concentration in micromolar. QS is the Q372V/S582Y double mutant and AS the A446S/S552A double mutant. Arrows indicate the bands for the Slp-76, Zap-70, Lck, and LAT proteins. The assay was repeated at least three times, and the gel is representative of one experiment.
We purified the E481V single mutant and the A446S/S552A, Q372V/S582Y, and S552A/S582Y double mutants of Sts-2PGM. We checked their in vitro phosphatase activity by assessing their ability to hydrolyze pNPP and pTyr-containing proteins (Figure 4B,C). As mentioned earlier, Glu481 introduces a negative charge in the vicinity of Glu476 and makes a hydrogen bond interaction with Arg448, which might impinge on the action of these conserved residues during substrate dephosphorylation. In Sts-1, the equivalent residue is the small apolar Val495. Mutating Glu481 to Val should thus relatively increase the phosphatase activity of Sts-2PGM. Indeed, the E481V mutant has a 10-fold better specificity constant (150 M−1 s−1) when compared to wild-type Sts-2PGM (15 M−1 s−1). This improvement is due to a 2-fold reduction in Km (1 mM) and a 5-fold increase in kcat [0.15 s−1 (Table 2)]. In addition, the E481V mutant exhibited a better ability to hydrolyze pTyr-containing proteins than wild-type Sts-2PGM (Figure 4C). Together, these results demonstrate that Glu481 is among the residues responsible for the weaker phosphatase activity of Sts-2.
If the conclusion that mutating Glu481 makes Sts-2PGM more Sts-1PGM-like were true, then the E481V mutant would behave more like Sts-1PGM and show an intermediate improvement in its Km and kcat values at pH 5.0. We therefore measured the catalytic constants of the E481V mutant of Sts-2PGM at pH 5.0. As shown in Table 2, when compared to that at pH 7.5, the Km value for this mutant improves by a factor of 5 (0.2 mM vs 1.0 mM), which is more in line with the changes experienced by Sts-1PGM than Sts-2PGM. Similarly, the kcat value for this mutant improves by a factor of 4 (0.65 s−1 vs 0.15 s−1), which is less than the 10-fold improvement experienced by wild-type Sts-2PGM. This result confirms that the E481V mutation makes Sts-2 behave more like Sts-1, supporting the conclusion of the pNPP hydrolysis results that Glu481 is among the residues responsible for the difference in activity between the two phosphatases. Given that Glu481 is the only acidic active site residue that is not conserved between the two Sts domains, we tested the hypothesis that it is responsible for the higher Sts-2PGM activity at acidic pHs and measured the catalytic rate of the E481V mutant as a function of pH. The active site of this mutant in principle has the same overall charge as Sts-1PGM. As reported in Figure 4B, the activity of this mutant is maximal at pH 5.0, similar to wild-type Sts-2PGM, demonstrating that Glu481 is not responsible for its acid-dependent phosphatase activity and suggesting that other factors are the cause for this behavior.
Next, we focused on Ala446 and Ser552. In Sts-2PGM, Ala446 is close to His551 and Ser552 hydrogen bonds Arg365 in the apo and phosphate-bound but not in the tungstate-bound structures of Sts-2PGM. Ser552 also interacts with the tungstate and phosphate moieties (Figure 3). The equivalent residues in Sts-1 are Ser460 and Ala566, respectively (Figure 2B), which do not make hydrogen bonds with His565, Arg379, or the phosphate (10). Introducing the A446S/S552A double mutation into Sts-2PGM results in a 28-fold better specificity constant (Table 2). Consistent with the pNPP result, the phosphatase activity of this double mutant against pTyr-containing proteins has also increased (Figure 4C).
Gln372 and Ser582 are located at the edge of the active site (Figure 2B). While Gln372 is solvent-exposed and does not interact with any Sts-2PGM residue, Ser582 makes a hydrogen bond with Arg365 of the RHGE motif in apo and phosphate-bound Sts-2PGM but not in the tungstate-bound structure (Figure 2 and Figure 3). The equivalent residues in Sts-1, Val386 and Tyr596, occupy similar positions but do not interact with any catalytic residue. As reported in Table 2, the Q372V/S582Y double mutation increases by a factor of 38 the specificity constant of Sts-2PGM. This significant increase is due to the reduction in Km by a factor of 3 (0.7 mM) and the increase in kcat by a factor of 10 (0.4 s−1). Likewise, this double mutant exhibited increased phosphatase activity against pTyr-containing proteins but not to the extent of the E481V mutant or the A446S/S552A double mutant of Sts-2PGM (Figure 4C).
As mentioned earlier, Ser552 and Ser582 interact with the tungstate and with Arg365. Mutating those two serines should therefore improve the rate of phosphate release during catalysis. As reported in Table 2, the S552A/S582Y double mutation increases by a factor of 13 the specificity constant of Sts-2PGM for pNPP hydrolysis. When compared to wild-type Sts-2PGM, this increase is mainly due to an improvement in kcat by a factor of 15, while the Km value of this mutant did not change, consistent with the hypothesis that the two serines are involved in the phosphate release steps. The plot of the initial velocities as a function of pH shows that it has a bell-shaped curve with an optimum at pH ~4.6 (Figure 4B). The reason for the shift in the catalytic rate maximum to a more acidic pH when compared to that of wild-type or E481V Sts-2PGM is currently unclear. At pH 5.0, the Km and kcat values are 0.6 mM and 2.9 s−1, respectively (Table 2).
The site-directed mutagenesis data clearly show that making the Sts-2PGM active site more Sts-1PGM-like improves its ability to hydrolyze pNPP and pTyr-containing proteins. The single and double mutants of Sts-2PGM significantly improved on the kcat and moderately on the Km values of pNPP hydrolysis (Table 2). However, they failed individually to significantly increase the kcat value to the Sts-1PGM level. The Sts-2PGM mutants also appeared less efficient than Sts-1PGM, as judged by the need to utilize 10-fold more Sts-2PGM to achieve any level of dephosphorylation of pTyr-containing proteins. We therefore propose that residues Gln372, Ala446, Glu481, Ser552, and Ser582 are among the residues responsible for the difference in activity between the Sts PGM domains. We predict that the Sts-2PGM quintuple mutant should have in theory an activity that approaches that of Sts-1PGM. Unfortunately, we could not test this prediction since any combination of these mutations resulted in an insoluble Sts-2PGM mutant protein. We note that the residues we mutated make hydrogen bond interactions with the conserved catalytic residues in Sts-2 but not in Sts-1. We thus propose that these Sts-2 specific hydrogen bonds between nonconserved and catalytic residues might be responsible for the slower phosphatase reaction since removing them increases the specific activity. The hydrogen bonds seem to influence the kcat since removing them partially restores the phosphatase activity in Sts-2. However, other factors might also be involved.
DISCUSSION
Despite the strong sequence homology between the two Sts isoforms, three major differences distinguish Sts-2 from Sts-1. First, Sts-1 is ubiquitously expressed in human tissues while Sts-2 expression is restricted to the hematopoietic system. Second, the observation that both genes need to be deleted for efficient TCR hyper-reactivity (13) suggests that Sts proteins have nonoverlapping functions. Third, while Sts-1 has a robust phosphatase activity located on its 2H-phosphatase homology domain that appears to be essential for its cellular function, Sts-2 is by far a weaker phosphatase despite strict conservation of all essential catalytic residues. In this work, we focused on investigating the phosphatase activity of Sts-2 using biochemical and structural techniques.
Our data show that recombinant Sts-2PGM has a substrate specificity that differs from that of its closest homologues, Sts-1PGM and EPPasePGM (32, 34). Sts-2PGM shows no steroid phosphate phosphatase activity (see Table 2 and the Supporting Information) and hydrolyzes the pTyr analogue pNPP significantly more poorly than Sts-1PGM: the measured kcat and Km values are 2330 and 4 times lower, respectively. The pH dependence of the catalytic activity also shows a discrepancy between the two Sts domains: the kcat value is optimal at pH 7.0 for Sts-1PGM but optimal at pH 5.0 for Sts-2PGM, suggesting that the latter might function in cells as an acid-dependent phosphatase. The 25-fold decrease in the Km value for pNPP hydrolysis for Sts-1PGM between pH 7.5 and 5.0 also does not compare to the modest decrease for Sts-2PGM (Table 2), suggesting fundamental differences between the two isoforms that might reflect differences in the physiological substrates. In addition, Sts-2PGM is unable to dephosphorylate pTyr-containing proteins. Taken together, these observations suggest that either Sts-2PGM is not a PTP or it is highly specific for a subset of pTyr-containing proteins that we have not yet identified.
Five nonconserved residues are among the residues responsible for the difference in activity between the two PGM domains. Since these nonconserved residues are involved in hydrogen bond interactions with key conserved catalytic residues and with the tungstate and phosphate moieties, it is likely that they are responsible for the weak Sts-2PGM phosphatase activity by impinging on the movement of the catalytic residues, modifying the charge distribution on the transition state structures during catalysis, or both. Arguments in favor of the former reason include the observation that the interactions between Arg365 and Ser552 and Ser582 are broken in the tungstate-bound Sts-2PGM structure. Arguments in favor of the latter reason include the short distance between the hydroxyl group of Ser552 and the tungstate. Interestingly, our mutagenesis results are consistent with mutagenesis data obtained with Cys-based PTPs, which concluded that structural determinants necessary for specific substrate binding reside outside the essential catalytic residues (7, 35, 36).
In light of the weak phosphatase activity of Sts-2PGM, it is interesting to note that several Cys-based phosphatases are expressed as inactive enzymes. Examples include the second phosphatase domain of CD45, the myotubularin-related proteins MTMR5 and MTMR13, the STYX family of proteins, and the PTEN-related tensin (reviewed in ref 6). In these proteins, key residues, including the nucleophilic Cys, are mutated rendering them inactive. Curiously, these inactive phosphatases have been shown to be essential for downregulating key cellular signaling pathways. The Sts-2PGM case seems unique since all the catalytic residues are conserved, an observation that suggests that its catalytic activity although relatively weak is important in vivo. However, the phosphatase activity of Sts-2PGM should be discussed in the context of the true substrates, which remain to be identified. The advantages of such a weak activity and how it relates to its cellular function are areas of ongoing research in our laboratories.
Supplementary Material
ACKNOWLEDGMENT
We thank Ben Sondgeroth for expert technical assistance, Todd Miller and Miguel Garcia-Diaz for helpful discussions and comments on the manuscript, and Vivian Stojanoff for help with data collection at X6A.
Footnotes
Research in N.N.’s laboratory is supported in part by grants from the National Institutes of Health (CA-115611) and the U.S. Department of Defense (NF060060). Research in N.C.’s laboratory is supported by grants from The Arthritis Foundation (LI07), the National Institute of Allergy and Infectious Diseases (R21AI075176), and The National Multiple Sclerosis Society through a Collaborative MS Research Center Award (CA1044A1). Research carried out at beamline X6A, National Synchrotron Light Source, Brookhaven National Laboratory, is supported by the U.S. Department of Energy under Contract DE-AC02-98CH10886. X6A is funded by the National Institute of General Medical Sciences under Grant Y1 GM-0080-03.
Abbreviations: DTT, dithiothreitol; EPPase, ecdysteroid phosphate phosphatase; PGM, phosphoglycerate mutase; pNP, p-nitrophenol; pNPP, p-nitrophenylphosphate; PTP, protein tyrosine phosphatase; rmsd, root-mean-square deviation; Sts, suppressor of T cell signaling; TCR, T cell receptor; pTyr, phosphotyrosine.
SUPPORTING INFORMATION AVAILABLE
Structure of the Sts-2PGM domain, dimerization of the Sts-2PGM domain, structure of the H366A mutant of Sts-2PGM, specificity of the Sts-2PGM phosphatase activity, ribbon diagram representation of Sts-2PGM, and electrostatic potential representation of Sts-2PGM. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Sibley DR, Strasser RH, Benovic JL, Daniel K, Lefkowitz RJ. Phosphorylation/dephosphorylation of the β-adrenergic receptor regulates its functional coupling to adenylate cyclase and subcellular distribution. Proc. Natl. Acad. Sci. U.S.A. 1986;83:9408–9412. doi: 10.1073/pnas.83.24.9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter T. A thousand and one protein kinases. Cell. 1987;50:823–829. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- 3.Mustelin T, Abraham RT, Rudd CE, Alonso A, Merlo JJ. Protein tyrosine phosphorylation in T cell signaling. Front. Biosci. 2002;7:d918–d969. doi: 10.2741/A821. [DOI] [PubMed] [Google Scholar]
- 4.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 5.Tonks NK. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 6.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 2001;21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denu JM, Stuckey JA, Saper MA, Dixon JE. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 9.Jemc J, Rebay I. The eyes absent family of phosphotyrosine phosphatases: Properties and roles in developmental regulation of transcription. Annu. Rev. Biochem. 2007;76:513–538. doi: 10.1146/annurev.biochem.76.052705.164916. [DOI] [PubMed] [Google Scholar]
- 10.Mikhailik A, Ford B, Keller J, Chen Y, Nassar N, Carpino N. A phosphatase activity of Sts-1 contributes to the suppression of TCR signaling. Mol. Cell. 2007;27:486–497. doi: 10.1016/j.molcel.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada R, Sonobe H. Purification, kinetic characterization, and molecular cloning of a novel enzyme ecdysteroid-phosphate phosphatase. J. Biol. Chem. 2003;278:26365–26373. doi: 10.1074/jbc.M304158200. [DOI] [PubMed] [Google Scholar]
- 12.Carpino N, Kobayashi R, Zang H, Takahashi Y, Jou ST, Feng J, Nakajima H, Ihle JN. Identification, cDNA cloning, and targeted deletion of p70, a novel, ubiquitously expressed SH3 domain-containing protein. Mol. Cell. Biol. 2002;22:7491–7500. doi: 10.1128/MCB.22.21.7491-7500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpino N, Turner S, Mekala D, Takahashi Y, Zang H, Geiger TL, Doherty P, Ihle JN. Regulation of ZAP-70 activation and TCR signaling by two related proteins, Sts-1 and Sts-2. Immunity. 2004;20:37–46. doi: 10.1016/s1074-7613(03)00351-0. [DOI] [PubMed] [Google Scholar]
- 14.Kowanetz K, Crosetto N, Haglund K, Schmidt MH, Heldin CH, Dikic I. Suppressors of T-cell receptor signaling Sts-1 and Sts-2 bind to Cbl and inhibit endocytosis of receptor tyrosine kinases. J. Biol. Chem. 2004;279:32786–32795. doi: 10.1074/jbc.M403759200. [DOI] [PubMed] [Google Scholar]
- 15.Feshchenko EA, Smirnova EV, Swaminathan G, Teckchandani AM, Agrawal R, Band H, Zhang X, Annan RS, Carr SA, Tsygankov AY. TULA: An SH3- and UBA-containing protein that binds to c-Cbl and ubiquitin. Oncogene. 2004;23:4690–4706. doi: 10.1038/sj.onc.1207627. [DOI] [PubMed] [Google Scholar]
- 16.Wattenhofer M, Shibuya K, Kudoh J, Lyle R, Michaud J, Rossier C, Kawasaki K, Asakawa S, Minoshima S, Berry A, Bonne-Tamir B, Shimizu N, Antonarakis SE, Scott HS. Isolation and characterization of the UBASH3A gene on 21q22.3 encoding a potential nuclear protein with a novel combination of domains. Hum. Genet. 2001;108:140–147. doi: 10.1007/s004390000453. [DOI] [PubMed] [Google Scholar]
- 17.Jedrzejas MJ. Structure, function, and evolution of phosphoglycerate mutases: Comparison with fructose-2,6-bisphosphatase, acid phosphatase, and alkaline phosphatase. Prog. Biophys. Mol. Biol. 2000;73:263–287. doi: 10.1016/s0079-6107(00)00007-9. [DOI] [PubMed] [Google Scholar]
- 18.Rigden DJ. The histidine phosphatase superfamily: Structure and function. Biochem. J. 2008;409:333–348. doi: 10.1042/BJ20071097. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka M, Kishi Y, Takanezawa Y, Kakehi Y, Aoki J, Arai H. Prostatic acid phosphatase degrades lysophosphatidic acid in seminal plasma. FEBS Lett. 2004;571:197–204. doi: 10.1016/j.febslet.2004.06.083. [DOI] [PubMed] [Google Scholar]
- 20.Ogino T, Matsubara M, Kato N, Nakamura Y, Mizuno T. An Escherichia coli protein that exhibits phosphohistidine phosphatase activity towards the HPt domain of the ArcB sensor involved in the multistep His-Asp phosphorelay. Mol. Microbiol. 1998;27:573–585. doi: 10.1046/j.1365-2958.1998.00703.x. [DOI] [PubMed] [Google Scholar]
- 21.Kleinman H, Ford B, Keller J, Carpino N, Nassar N. Crystallization and initial crystal characterization of the C-terminal phosphoglycerate mutase homology domain of Sts-1. Acta Crystallogr. 2006;F62:218–220. doi: 10.1107/S1744309106003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Arthur G. Improved procedures for the determination of lipid phosphorus by malachite green. J. Lipid Res. 1992;33:1233–1236. [PubMed] [Google Scholar]
- 23.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. 2003;A59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 25.Sheldrick GM. A short history of SHELX. Acta Crystallogr. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. 2000;D56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. 2003;D59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Jakoncic J, Wang J, Zheng X, Nassar N. Structural and Functional Characterization of the C-terminal Domain of the Ecdysteroid Phosphate Phosphatase from Bombyx mori Reveals a New Enzymatic Activity. Biochemistry. 2008;47:12135–12145. doi: 10.1021/bi801318w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47(Part 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Davies L, Anderson IP, Turner PC, Shirras AD, Rees HH, Rigden DJ. An unsuspected ecdysteroid/steroid phosphatase activity in the key T-cell regulator, Sts-1: Surprising relationship to insect ecdysteroid phosphate phosphatase. Proteins. 2007;67:720–731. doi: 10.1002/prot.21357. [DOI] [PubMed] [Google Scholar]
- 35.Denu JM, Dixon JE. Protein tyrosine phosphatases: Mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol. 1998;2:633–641. doi: 10.1016/s1367-5931(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 36.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 37.Brünger AT. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.