Abstract
In many high-resolution structures of DNA there are ordered waters associated with the floor of the minor groove and extending outward in several layers. It is thought that this hydration structure, along with cations, reduces the Coulombic repulsion of the interstrand phosphates. In previous studies, the replacement of the 3-N atom of adenine with a C-H to afford 3-deazaadenine was shown to decrease the thermodynamic stability of DNA via a reduction in the enthalpic term. Using spectroscopic and calorimetric methods, we report herein a rigorous examination of the thermodynamics of DNA with 3-deazaadenine modifications, and report for the first time how the presence of a minor groove methyl group, i.e., 3-methyl-3-deazaadeine, affects DNA stability, hydration and cation binding. The methylation of adenine at the N3-position to yield N3-methyladenine represents an important reaction in the toxicity of many anticancer compounds. This minor groove lesion is unstable and cannot be readily studied in terms of its effect on DNA stability or structure. Our studies show that 3-methyl-3-deazaadenine, an isostere of N3-methyladenine, significantly destabilizes DNA (ΔΔG > 4 kcal•mol−1) due to a significant drop in the enthalpy (ΔH) term, which is associated with a lower hydration of the duplex relative to the unfolded state.
Introduction
In its simplest state, DNA is an ensemble of nucleic acids, water and salts, and the structure of the ensemble is dynamically dependent on all three components, e.g., sequence of nucleic acid bases, type and concentration of salts and the level of hydration.1 As a result, DNA can adopt multiple conformations upon environmental changes that differ subtly from each other, such as B-DNA vs. B′-DNA, or dramatically, as B-DNA vs. A- or Z-DNA.2 These alterations involve changes in the conformation of the bases and sugars, and in the concomitant reorganization of water molecules and cations.
To explore how alterations in hydration and cation binding affect structure and stability, studies have been performed on DNA that is site specifically modified with base analogs that are designed to affect the interactions of water and cations at the edges of the bases in the major and minor grooves. It turns out that even modest changes in DNA structure alter the thermodynamic stability, although associated structural changes are not always obvious from crystal structures or low temperature NMR studies. For example, the crystal3 and high-resolution NMR4 structure of 5’-CGCGAATTCZCG (Z = 7-deazaguanine; c7G) is indistinguishable from the unmodified sequence with one notable exception; a highly conserved cation-binding site is missing in the crystal structure.3 Despite the similarity in the crystal structure, the temperature-dependent imino proton NMR spectrum showed a specific increase in exchange of imino proton of the C•G pair that flanks the c7G modification toward the 3′-terminus.4 The exchange rate for the c7G•C base pair was the same as in the natural sequence. Thermodynamic studies revealed a slight drop in ΔΔG vs. the natural sequence, but with a significant change in the enthalpic term that was only partially compensated by entropic stabilization. Analysis of the relative change in hydration and cation binding between the duplex and random coil states for the natural and c7G modified sequence provided an important insight into the origin of the thermodynamic effects of the modification. The DNA with the c7G showed, after normalizing for the number of base pairs, a decrease of 10 water molecules per mole of duplex in the hydration level relative to the natural sequence. This calculates to a reduction of 5 water molecules per modification. The release of counterions from the c7G substituted duplex is also lower. A similar scenario has also been observed with the corresponding 7-deazaadenine (c7A) modification (manuscript in preparation). Of note, is that tethering a basic amine onto the 7-position of c7G (i.e., 7-NH2CH2-c7G) resulted in a duplex with thermodynamic properties similar to the unmodified DNA, while an isosteric hydroxyl group was destabilizing similar to the c7G DNA.5 In this case the cationic 7-NH2CH2-c7G substitution had a small effect on hydration and cation binding vs. the unmodified DNA.
Because of our observation that changes in the major groove cause an enthalpic destabilization that is associated with a reduction in duplex hydration and cation binding, we have performed thermodynamic measurements on DNA with minor groove alterations, i.e., a c3A residue. In addition, we included studies on 3-methyl-3-deazaA (3-Me-c3A) to determine the effect of a hydrophobic group in the minor groove. The 3-Me-c3A serves as a stable isostere of N3-methyladenine (3-MeA) that is a common methylation adduct formed by many carcinogens and anticancer drugs, e.g., temozolomide.6-10
The substitution of a single c3A into 5′-d(CG-c3A-TTGCG)•3′-d(GCTAACGC) at 120 mM Na+ has been shown to be destabilizing (~1.4 kcal/mol) by UV melting experiments.11 Multiple substitutions caused further destabilization and a significant drop in the van't Hoff ΔH term. It was suggested that the instability was due to the pKa of N-1 in c3A that would make it more prone to protonation, which would disrupt Watson-Crick H-bonding to its dT partner. In addition, disruption of the structure of minor groove hydration was also proposed as a potential destabilizing factor. A sequence-dependent effect of c3A on the flexibility of an intrinsically bent A-tract has also been noted.12 The reduction in bending was observed when the c3A was placed on the 3′-side of an A6 tract. It was suggested that the relative hydrophobicity of c3A might interfere with minor groove hydration and this could reduce bending at the A-tract.12 However, no studies have been performed that actually measure changes in hydration induced by the c3A or 3-Me-c3A residues. In studies presented herein, an adenine at positions -4 and -6, respectively, in the self-complementary duplexes 5′-d(GAGAGCGCTCTC)-3′ and 5′-d(CGCGAATTCGCG)-3′ was replaced by c3A or 3-Me-c3A. The results show that there are significant thermodynamic differences between the methylated and unmethylated deazaadenines. The origin of the difference is discussed in terms of the fundamental role of minor groove ions and water molecules in the stabilization of the DNA architecture.
Experimental Methods
Materials
All oligodeoxynucleotides (ODNs) were synthesized by Invitrogen (Frederick, MD). The phosphoramidite derivative of 3-Me-c3A was prepared as previously described.13 The phosphoramidite of c3A was obtained commercially from Glen Research (Sterling, VA). The modified oligomers were purified using a 10 × 250 mm Phenomenex 5 μm phenyl-hexyl reverse phase HPLC column (Torrance, CA) equilibrated with 0.1 M diethylammonium acetate (pH 7.0). After desalting by gel-permeation chromatography using a Sephadex G-25 column, the sample was lyophilized to dryness. The samples were characterized by MALDI-TOF-MS. The dry oligomers were then dissolved in the appropriate buffer.
The concentrations of the oligomer solutions were determined at 80 °C using 260 nm and extinction coefficient ~ 1.11 × 105 M−1 cm−1 (ODN; 1-3, 4, 6 and 7), and 1.05 × 105 M−1 cm−1 (ODN-5; hairpin) at 260 nm and 25 °C assuming similar extinction coefficients for 3-Me-c3A and c3A. These values are obtained from the molar absorptivity at 25 °C, and derived from the tabulated values of the dimers and monomer bases,14,15 and extrapolated from high temperatures using the upper portions of the UV melting curves, following procedures described earlier.16 All measurements were performed in buffer solutions consisting of 10 mM sodium phosphate, adjusted to the appropriate salt and osmolyte concentrations with NaCl and ethylene glycol, respectively.
UV-Spectroscopy
Absorption vs. temperature profiles (UV melts) for each duplex were measured at either 260 nm and/or 275 nm using a thermoelectrically controlled Varian Cary 300 spectrophotometer (Palo Alto, CA), interfaced to a PC computer for data acquisition and analysis. The temperature was scanned at heating rates of 1.00 °C/min. Melting curves as a function of strand concentration (4–70 μM) were obtained to check for the molecularity of each molecule. Additional melting curves were obtained as a function of salt and osmolyte concentration to determine the differential binding of counterions and water molecules that accompanies their helix coil transitions.
UV melts were measured in the salt range of 10−200 mM NaCl at pH 7.0, and at a constant total strand concentration of 7 μM, to determine the differential binding of counterions, ΔnNa+, which accompanied their helix−coil melting. This linking number was measured experimentally with the assumption that counterion binding to the helical and coil states of each oligonucleotide took place with a similar type of binding using the relationship:17,18
| Eq. 1 |
The numerical factor corresponded to the conversion of ionic activities into concentrations. The first term in parentheses, (ΔHcal/RTM2), was a constant determined directly from DSC experiments, where R was the gas constant. The second term in parenthesis was determined from UV experiments from the dependencies of TM on salt concentration.
For the determination of Δnw, UV melts were measured in the ethylene glycol concentration range of 0.5- 3.0 m at pH 7.0 and 10 mM NaCl. and at a constant total strand concentration of 7 μM. The osmolalities of the solutions were obtained with a Wescor Vapro vapor pressure osmometer, Model 5520 (Logan, UT). These osmolalities were then converted into water activities, aw, using the following equation:19
| Eq. 2 |
where Osm is the solution osmolality and Mw is the molality of pure H2O, equal to 55.5 mol/kg H2O. Differential binding of water, Δnw, was calculated using the relationship:17,18
| Eq. 3 |
The [ΔHcal/R(TM)2] term used in the determination of Δnw was derived from the 100 mM salt concentration and the slope (∂TM/∂log aW) was calculated from the 10 mM salt data.
Differential Scanning Calorimetry
Heat capacities vs. temperature profiles were measured with a VP-DSC differential scanning calorimeter (Microcal Inc., Northampton, MA). The dry oligodeoxynucleotides were dissolved in 10 mM sodium phosphate buffer (pH 7.0) and adjusted to the desired ionic strength with NaCl for all unfolding experiments. The heat capacity profile for each DNA solution was measured against a buffer solution. In a typical experiment the reaction and the reference cells were each filled with 0.75 mL of solution. Temperature was scanned from 0 to 100 °C at a rate of 0.75 °C/min. The experimental curve was normalized by the heating rate, and a buffer vs. buffer scan was subtracted and normalized for the number of moles. The resulting curves were then analyzed with Origin version 7.0 (Microcal); their integration (∫ΔCp dT) yielded the molar unfolding enthalpy (ΔHcal), which was independent of the nature of the transition.16,20 The molar entropy (ΔScal) was obtained similarly, using ∫(ΔCp/T) dT. The free energy change at any temperature T was then obtained with the Gibbs equation:
Circular Dichroism
Circular dichroism (CD) measurements were conducted on a Jasco(model J-815) CD spectrometer (Easton, MD). The spectrum of each duplex was obtained using a strain-free 1 cm quartz cell at low temperatures to ensure 100% duplex formation. Typically, 1 OD of a duplex sample was dissolved in 1 mL of a buffer containing 10 mM sodium phosphate (pH 7.0). The reported spectra correspond to an average of three scans from 220 to 350 nm at a wavelength step of 1 nm.
Results
UV melting studies
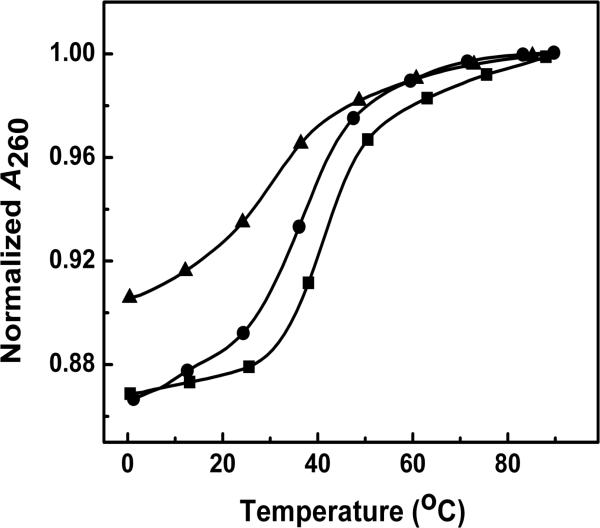
The unfolding of duplexes was studied by temperature-dependent UV spectroscopy. Absorption spectra at low and high temperatures revealed a higher hyperchromic effect at 260 nm than other wavelengths for all the oligomers (data not shown). This was chosen as the optimum wavelength for all UV melting studies. Typical UV melting curves are shown in Figure 1. All the curves follow the characteristic sigmoidal behavior for the unfolding of a nucleic acid helix structure. The helix-coil transition of ODNs 1-3 exhibit monophasic transition under all conditions studied. TM values were determined from the first derivative of the melting curves and shape analysis. The TM values follow the order: ODN-1 (41.0 °C) > ODN-2 (34.9 °C) > ODN-3 (30.3 °C) in 10 mM salt and ODN-1 (59.5 °C) > ODN-2 (51.2 °C) > ODN-3 (47.9 °C) in 100 mM salt at a DNA concentration of 10 μM. The presence of c3A residues in DNA reduces the TM confirming earlier reports.11 The data also indicates the lower stability of duplexes with 3-Me-c3A·T base pairs. ODN-3 also shows lower hyperchromicity which is indicative of lower stacking contributions. To confirm the molecularity of each complex, melting curves were obtained as a function of strand concentration. Strand concentration-dependent experiments done at both 10 mM and 100 mM salt concentrations showed an increasing TM with an increase in the total strand concentration in the range of ~ 4-120 μM (Figure 2). This is evidence of duplex to random coil denaturation.
Figure 1.
UV melting curves in 10 mM sodium phosphate buffer (pH 7.0) at ~ 10 μM total strand concentration at 260 nm for ODN-1 (■), ODN-2 (●) and ODN-3 (▲).
Figure 2.
TM dependence on strand concentration for duplexes in 10 mM sodium phosphate buffer (pH 7.0), 4-120 μM strand concentration for ODN-1 (■), ODN-2 (●) and ODN-3 (▲).
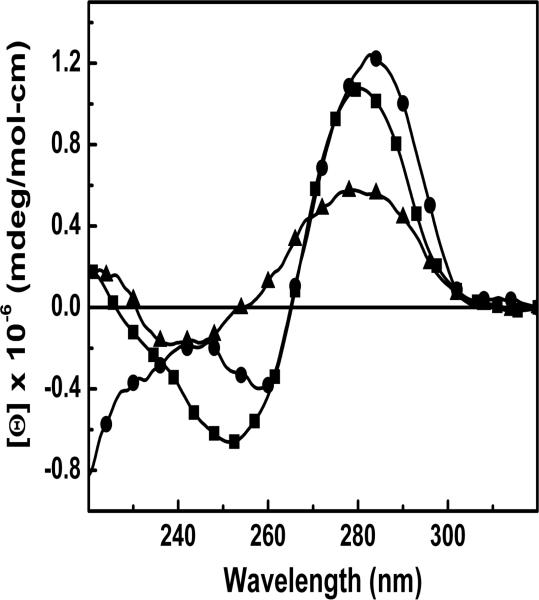
Circular dichroism
The average solution conformation of the oligomers was obtained using CD spectroscopy, which was performed in low salt conditions (10 mM salt). All spectra show a positive Cotton effect around 290 nm and a negative Cotton effect around 250 nm, which are characteristics of a right-handed helix in the B-conformation. The CD spectra of ODNs 1-3 are shown in Figure 3. The intensities of the negative band near 250 nm reflect base stacking contributions. The intensity of the differential CD band at 250 nm for ODN-3 is 75% lower relative to corresponding band for ODN-1 and 45% lower relative to ODN-2. These changes suggest a partial disruption of base stacking due to the presence of methyl group in 3-Me-c3A, and consistent with its lower hyperchromicity.
Figure 3.
Differential CD spectra in 10 mM sodium phosphate buffer for ODN-1 (■), ODN-2 (●) and ODN-3 (▲).
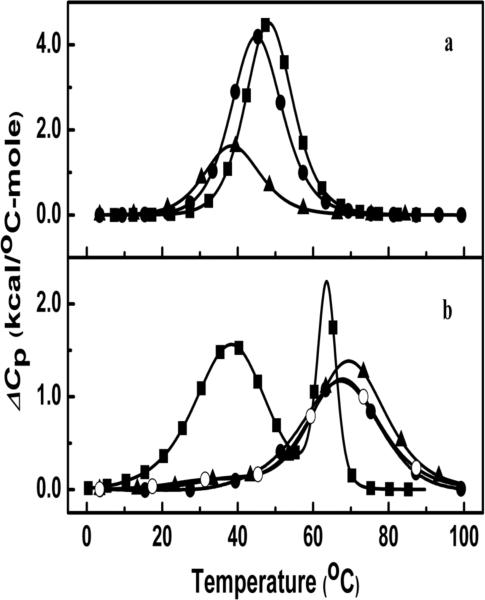
DSC unfolding studies of duplexes
The DSC melting curves for ODNs 1-3 are shown in Figure 4a. Additional DSC curves for ODNs 4-6 are shown in Figure 4b and the corresponding thermodynamic parameters are provided in Table 2. The reported thermodynamic values are an average of the analysis of three consecutive scans. ODNs 1-3 exhibit monophasic melting profiles at both 10 mM and 100 mM salt concentrations confirming their unfolding through a duplex to random coil transition as seen in the UV studies. The TM values at the concentrations used in the DSC experiments follow the order: ODN-1 (48.7 °C) > ODN-2 (45.2 °C) > ODN-3 (38.8 °C) in 10 mM salt and ODN-1 (66.1 °C) > ODN-2 (61.3 °C) > ODN-3 (58.8 °C) in 100 mM salt. ODNs 4-7 unfold via biphasic transitions at 10 mM salt. The DSC thermogram of ODN-4, as reported earlier, has a broad first transition and a sharp second transition that is typical of the helix → hairpin → random coil transitions of this Dickerson-Drew dodecamer duplex.21 DSC thermograms for ODN-6 and -7 reveals biphasic curves with a small shoulder at low temperature for the first transition and a broad second transition at higher temperature with second transition TM similar to that of the TM of hairpin ODN-5 (Table 2) reported in the literature.4 Enthalpies reported in Table 2 were determined by deconvolution of the DSC graphs.
Figure 4.
(a) DSC curves in 10 mM sodium phosphate buffer (pH 7.0) at ~120 μM strand concentration for ODN-1 (■), ODN-2 (●) and ODN-3 (▲). (b) DSC curves in 10 mM sodium phosphate buffer (pH 7.0) at ~200 μM strand concentration for ODN-4 (■), ODN-5 (●), ODN-6 (▲) and ODN-7 (○) .
Table 2.
Thermodynamic profiles for the formation of duplexes at 20 °Ca
| Oligomer | NaCl mM | TMb °C | ΔG°c kcal/mol | ΔH kcal/mol | TΔS kcal/mol | ΔnNa+ per mol DNA | Δnw per mol DNA |
|---|---|---|---|---|---|---|---|
| ODN-1 | 10 | 48.7 | -6.9 | -78.2 | -71.3 | -3.4 ± 0.2 | -41 ± 3 |
| 100 | 66.1 | -12.5 | -92.0 | -79.5 | -3.6 ± 0.2 | -43 ± 4 | |
| ODN-2 | 10 | 45.2 | -6.0 | -75.8 | -69.8 | -3.00 ± 0.1 | -35 ± 3 |
| 100 | 61.3 | -11.3 | -91.4 | -80.1 | -3.2 ± 0.2 | -38 ± 3 | |
| ODN-3 | 10 | 38.8 | -2.4 | -39.0 | -36.6 | -1.4 ± 0.1 | -24 ± 2 |
| 100 | 58.8 | -4.6 | -39.6 | -35.0 | -1.2 ± 0.1 | -21 ± 2 | |
| ODN-4 | 10 | 63.6 | -6.9 | -116.0 | -109.0 | - | - |
| ODN-5 | 10 | 68.4 | -4.4 | -31.0 | -26.6 | - | - |
| ODN-6 | 10 | 70.0 | -5.3 | -36.7 | -31.4 | - | - |
| ODN-7 | 10 | 68.9 | -4.7 | -32.9 | -28.2 | - | - |
Parameters are measured from UV (TM) and DSC melting curves in 10 mM sodium phosphate buffer (pH 7.0). The observed standard deviations are: TM (± 0.5 °C), ΔHcal (± 3%), ΔG°20 (± 5%), TΔScal (± 3%).
120-200 μM strand concentration.
measured at 20 °C.
Analysis of thermograms reveal that ODN-2, which has two c3A substitutions in the molecule, has an endothermic enthalpy of 75.8 kcal/mol that is similar to the unmodified oligomer, ODN-1, in 10 mM NaCl (Table 2). This pattern remains unchanged at the higher salt concentration of 100 mM NaCl. Introduction of two 3-Me-c3A residues in ODN-3 results in a reduction of enthalpy by 39.2 kcal/mol and 52.4 kcal/mol relative to unmodified ODN-1 (Table 2) in 10 and 100 mM buffer, respectively. The data suggest that incorporation of this methyl group destabilizes duplex structure at both low and high salt concentrations. This contrasts with the placement of methyl groups in the major groove where dT•dA base pairs are more stable than dU•dA.22
On the other hand, incorporation of c3A and 3-Me-c3A residues in the AATT core of ODN-4 stabilizes the hairpin conformation. Analysis of the data does not show any significant change in enthalpy when compared with the unmodified hairpin molecule ODN-5, indicating a similar extent of duplex structure.
The DSC melting curves at the two salt concentrations provides an indirect measurement of the heat capacity (ΔCp) in the unfolding of each duplex. The ΔCp (cal/°K•mol) values measured at 10 and 100 mM NaCl for ODN-1, ODN-2 and ODN-3 are 793, 969 and 30, respectively. This heat capacity effect indicates that ODN-3 behaves more like a random coil. Assuming that hydration of the random coils at high temperature will be similar for all of the sequences, by definition the more hydrophilic DNA will release more waters upon denaturation. This is consistent with the measured heat capacity effect. Therefore, the greater release of water from ODN-1 and ODN-2 duplexes means that they are more hydrophilic than ODN-3.
Thermodynamic profiles for the formation of duplex
The complete thermodynamic profiles for the folding of each duplex at 20 °C are shown in Table 2. Analysis of the thermodynamic data indicates that the stable formation of each duplex is accompanied by favorable Gibbs free energy, which results from the characteristic compensation of a favorable enthalpy term with an unfavorable entropy term. The favorable enthalpies arise from the formation of base-pairs and base pair stacks, uptake of electrostricted water and release of structural water. The unfavorable entropy contribution is mainly due to the ordering of a random coil into a duplex structure, condensation of counterions and immobilization of water molecules.
Relative to the unmodified oligomer ODN-1, both c3A and 3-Me-c3A modified oligomers were destabilized at low and high salt concentrations. The inclusion of two c3A modifications inODN-2 yielded a mild decrease in ΔG of 0.9 and 1.2 kcal/mol in 10 mM and 100 mM NaCl, respectively, a value that is similar to the destabilization observed in a different sequence.11 In contrast, substitution of two 3-Me-c3A residues results in a marked reduction of ΔG by 4.5 and 7.9 kcal/mol in low and high salt, respectively.
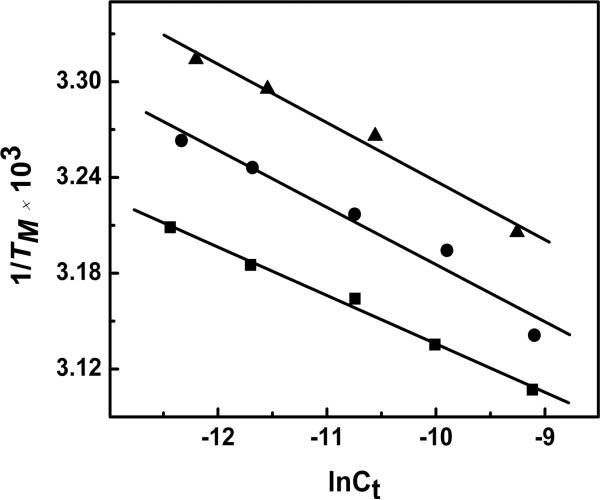
Differential association of counterions
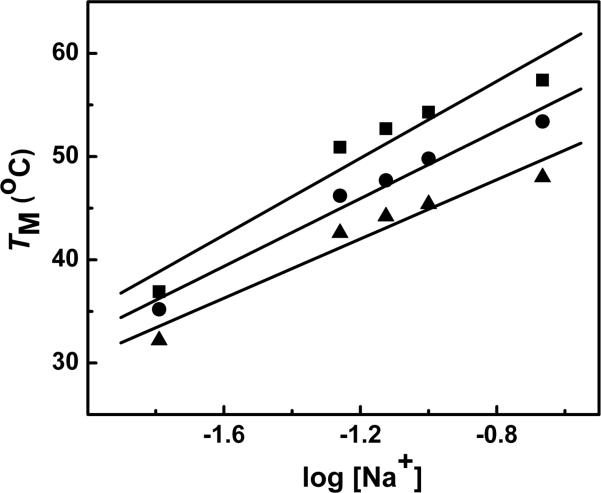
UV melting curves as a function of Na+ concentration were performed in the 10 to 200 mM NaCl range to examine the thermodynamic association of counterions with the DNA duplexes. The TM of ODNs 1-3 linearly increased with salt concentration (data not shown), consistent with the fact that the duplex states have higher charge density parameters. The TM dependence on salt concentration is shown in Figure 5. Equation 1 was used to calculate ΔnNa+ values shown in Table 2, using the slopes ∂TM/∂log [Na+] from Figure 5 in conjunction with the ΔH/R(TM)2 terms. Data indicates that in low salt, the Na+ uptake (mol Na+ per mol duplex) were in the order ODN-1 (3.4) > ODN-2 (3.0) > ODN-3 (1.4) and a similar pattern ODN-1 (3.6) > ODN-2 (3.2) > ODN-3 (1.2) was observed in the higher salt concentration of 100 mM (Table 2).
Figure 5.
TM dependence on salt concentration for duplexes in 10 mM sodium phosphate buffer (pH 7.0), ~ 8.0 μM strand concentration for ODN-1 (■), ODN-2 (●) and ODN-3 (▲).
Assuming that the random coil states of the single strand oligomers are thermodynamically equivalent at higher temperature, it can be concluded that the introduction of c3A or 3-Me-c3A into the duplex DNA causes a decreased association of counterions. For instance, there is a ΔΔnNa+ of ~0.4 between ODN-1 and ODN-2 at 10 and 100 mM NaCl, respectively. Introduction of the methyl group causes a larger ΔΔnNa+ of 1.6 and 2.0 between the pair ODN-2 and ODN-3 at low and high salt, respectively.
Differential association of water molecules
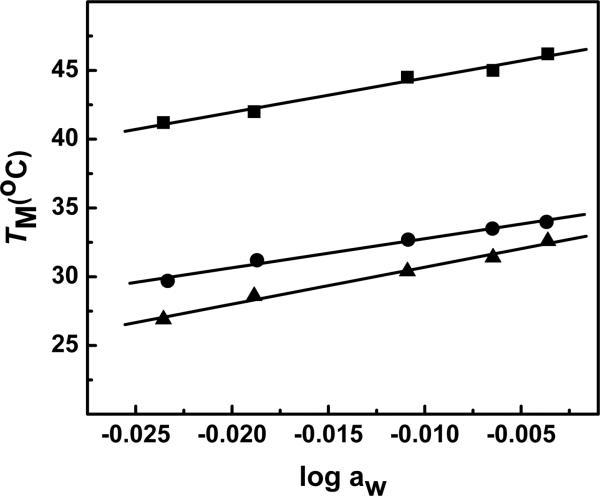
UV melting curves as a function of osmolyte (ethylene glycol) concentration were performed in the 0.5-3.0 m range at 10 mM salt concentration in order to determine the thermodynamic association of water molecules to DNA duplexes. Increasing the concentration of ethylene glycol (decreasing water activity) had a marginal effect on the TM. UV melting curves show that the TMs of the dodecamers ODNs 1-3 linearly decreased with increasing osmolyte concentrations i.e., decreasing activity of water (data not shown). The TM dependence on water activity of dodecamers is shown in Figure 6. The values for the formation of each duplex in 10 mM NaCl are shown in Table 2. Equation 3 was used to calculate Δnw values shown in Table 2, using the slopes ∂TM/∂log aw from Figure 6 in conjunction with the ΔH/R(TM)2 terms. Water uptake, expressed as mol H2O per mol duplex, measured in low salt, were in the order: 41 for ODN-1 > 35 for ODN-2 > 24 for ODN-3. A similar pattern was observed in the higher salt concentration of 100 mM: 43 for ODN-1 > 28 for ODN-2 > 21 for ODN-3.
Figure 6.
TM dependence on osmolyte concentration (as a function of ethylene glycol) in 10 mM sodium phosphate buffer (pH 7.0), ~ 8.0 μM strand concentration for ODN-1 (■), ODN-2 (●) and ODN-3 (▲).
The overall effect, again assuming that the random coil state of all the duplexes behave similarly at higher temperature, is that the substitution of c3A and 3-Me-c3A into the duplex DNA causes a decreased association of water molecules. Specifically, there is a ΔΔnw of 6 and 11 between ODN-1 and ODN-2 at 10 mM and 100 mM NaCl, respectively, and ΔΔnw of 11 and 17 between the pair ODN-2 and ODN-3 at low and high salt, respectively.
Discussion
The minor groove of DNA in the relatively few crystal structures that have been solved with high resolution show a well defined hydration scheme where waters are layered from the floor of the groove on outward toward the bulk solvent. This is especially true in sequences that have narrow minor grooves, such as those with an A2T2 or A3T3 central core.23-25 There is some debate whether the A/T rich sequences, without amino groups that project into the minor groove, drives the structure which in turn allows a well-defined hydration spine or whether the structural water in the narrow groove allows the groove to further narrow.
Our thermodynamic studies cannot directly answer this question. However, the lack of the 3-nitrogen atom in c3A has a potent impact on stability of the 5′-CGCGA-(c3A)-TTCGCG sequence (ODN-6) as it preferentially hairpins (Figure 4b). The same preference for the hairpin structure for the 5′-CGCGA-(3-Me-c3A)-TTCGCG sequence (ODN-7) is observed in the DSC experirments (Figure 4b). The unmodified dodecamer (ODN-4), with it's A/T rich central core, exists as a mixture of hairpin and duplex (Figure 4b). The ΔG, ΔH and TΔS values for the true hairpin (ODN-5) are very similar to those for ODN-6 and -7. In all cases, the decrease in the number of base pairs from 12 to 4, and resulting decrease in base stacking, causes a significant reduction in the ΔH term. The small differences that exist between ODNs 5-7 can be attributed to differences from the modifications in the loop regions. To avoid the formation of hairpin structures, which complicate the analysis, we chose a self-complementary sequence (5’-GAGAGCGCTCTC) with a G/C rich center that tends not to hairpin. In addition, we introduced the c3A or 3-Me-c3A lesions at position-4 that will not promote the hairpin structure. Therefore, the thermodynamic parameters presented for ODNs 1-3 are for the unfolding of the duplex structure. The observed increase in TM with increasing strand concentration is an example of proof of duplex formation. In the 5′-GAG-(c3A)-GCGCTCTC and 5′-GAG-(3-Me-c3A)-GCGCTCTC sequences the impact of c3A and 3-Me-c3A are still apparent, with the latter being highly destabilizing relative to the natural sequence by 4.5 and 7.9 kcal/mol at low and high salt, respectively. If disruption of the hydration structure near the floor of the groove by the hydrophobic methyl group is responsible for the change in ΔG then the hydration of ODN-3 would be expected to be lower than for ODN-2 and ODN-1. This is certainly the case with a ΔΔnW of 17 water/mol DNA for ODN-3 vs. ODN-1. This calculates to ~9 H2O per mol DNA per lesion. Interestingly, neither the enthalpy term nor the hydration parameter for ODN-3 is particularly sensitive to the salt concentration. The inclusion of the minor groove methyl group also causes a major reduction in cation binding (~2.2 Na+/mol) or 1.1 Na+/mol per 3-Me-c3A residue, assuming a localized effect. Clearly, the hydration effect is significant and might be the major driving force for the observed reduced enthalpy (ΔΔH° ~ 40 kcal/mol). The change in hydration may also (directly or indirectly) afford reduced base-pair stacking contributions. The band intensities at 250 nm in CD data are consistent with some reduction in base stacking in ODN-3 even at low temperature.
The differential thermodynamic profiles obtained indicate that the A → c3A substitution yielded marginal changes in ΔG, ΔH and ΔS, lower uptake of ions (0.2 per lesion corresponding to the loss of one ion site) and a marginal release of water (3/lesion). In contrast, the dA → 3-Me-c3A substitution yielded substantial changes in all thermodynamic parameters. Comparison of the signs of ΔΔG (i.e., ΔΔH-ΔTΔS) and ΔΔV yields insight about the type of water released.26 Similar signs for ΔΔG and ΔΔV indicate the participation of electrostricted water, while opposite signs indicate structural (i.e., hydrophobic) water. This is based upon the release of heat in the immobilization of electrostricted water where the water dipoles are compressed, while the energetic contribution for the release of structural water is close to zero, or even slightly positive due to improved packing around hydrophobic groups that eliminates void spaces. The data (Tabale 2) indicate that the c3A modification results in the participation of electrostricted water. In contrast, the placement of a hydrophobic methyl group in 3-Me-c3A results in a decrease in electrostricted water.
N3-methyladenine (3-MeA) is a cytotoxic but weakly mutagenic lesion that is formed by the reaction of DNA with a number of environmental and endogenous methylating agents, as well as by methylating anticancer drugs.6-10 Because the adduct is hydrolytically unstable, it has been difficult to study its effects in biochemical and biological systems. Since 3-Me-c3A is a stable isostere of 3-MeA it has been used as a surrogate in recent studies that show it to be a potent block of DNA polymerization.27 Although 3-Me-c3A, and presumably 3-MeA, can form a normal Watson-Crick base pair with T, the studies presented herein indicate that this may not be the case. We propose that the thermodynamic instability induced by the minor groove methyl group will allow DNA repair glycosylase enzymes to rapidly find and remove the 3-MeA adduct. The instability induced by the methyl group may also be, in part, responsible for their inhibition of DNA polymerases when 3-MeA (Settles and Gold, unpublished) or 3-Me-c3A 27 are in the template strand. We have preliminary evidence that 3-Me-c3A can be by-passed within A-tracts due to it becoming extrahelical during polymerization (Settles and Gold, unpublished).
Conclusion
It has been demonstrated that relatively subtle changes in the structure of a DNA base, in this case a C-H or C-CH3 group in the minor groove in place of the aromatic nitrogen at the N3-position of adenine, can have a dramatic effect on the enthalpic and entropic contributions to thermodynamic stability. The dynamic change in DNA appears to be associated with DNA hydration and cation association. This appears to be a common scenario; changes to the natural Watson-Crick base pairs reduces stability due to a loss of enthalpic stabilization that is associated with reduced hydration and cation binding.4,5
Table 1.
Structures and oligomer sequences
| ODN | sequence |
|---|---|
| 1 | 5′-GAGAGCGCTCTC-3′ |
| 2 | 5'-GAGZGCGCTCTC-3′ |
| 3 | 5′-GAGXGCGCTCTC-3′ |
| 4 | 5′-CGCGAATTCGCG-3′ |
| 5 | 5′-CGCGTTTTCGCG-3′ |
| 6 | 5′-CGCGAZTTCGCG-3′ |
| 7 | 5′-CGCGAXTTCGGC-3′ |
Acknowledgement
This work was supported by NIH grant CA29088 (BG).
References
- 1.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag Inc.; New York: 1984. [Google Scholar]
- 2.Leslie AG, Arnott S, Chandrasekaran R, Ratliff RL. Journal of Molecular Biology. 1980;143:49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- 3.Wang F, Li F, Ganguly M, Marky LA, Gold B, Egli M, Stone MP. Biochemistry. 2008;47:7147–7157. doi: 10.1021/bi800375m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganguly M, Wang F, Kaushik M, Stone MP, Marky LA, Gold B. Nucleic Acids Res. 2007;35:6181–6195. doi: 10.1093/nar/gkm670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganguly M, Wang R-W, Marky LA, Gold B. J. Am. Chem. Soc. 2009;131:12068–12069. doi: 10.1021/ja904930p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartsch H, Ohshima H, Shuker DE, Pignatelli B, Calmels S. Mutat. Res. 1990;238:255–267. doi: 10.1016/0165-1110(90)90017-6. [DOI] [PubMed] [Google Scholar]
- 7.Hecht SS. Mutat. Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 8.Bennett RA, Pegg AE. Cancer Res. 1981;41:2786–2790. [PubMed] [Google Scholar]
- 9.Tentori L, Graziani G. Curr. Med. Chem. 2002;13:1285–1301. doi: 10.2174/0929867023369916. [DOI] [PubMed] [Google Scholar]
- 10.Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Cancer Treat Rev. 1997;23:35–61. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- 11.Lever C, Li X, Cosstick R, Ebel S, Brown T. Nucleic Acids Res. 1993;21:1743–1746. doi: 10.1093/nar/21.8.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seela F, Grein T. Nucleic Acids Res. 1992;20:2297–2306. doi: 10.1093/nar/20.9.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irani RJ, SantaLucia J. Nucleosides, Nucleotides and Nucleic Acids. 2002;21:737–751. doi: 10.1081/NCN-120016477. [DOI] [PubMed] [Google Scholar]
- 14.Cantor CR, Warshow MM, Shapiro H. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 15.Marky LA, Blumenfeld KS, Kozlowski S, Breslauer KJ. Biopolymers. 1983;22:1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- 16.Marky LA, Breslauer KJ. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 17.Cantor CR, Schimmel PR. Biophysical Chemistry. W.H. Freeman and Company; New York: 1980. [Google Scholar]
- 18.Kaushik M, Suehl N, Marky LA. Biophysical Chemistry. 2007;126:154–164. doi: 10.1016/j.bpc.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Courtenay ES, Capp MW, Anderson CF, Record MTJ. Biochemistry. 2000;39:4455–4471. doi: 10.1021/bi992887l. [DOI] [PubMed] [Google Scholar]
- 20.Rentzeperis D, Marky LA, Dwyer TJ, Geierstanger BH, Pelton JG, Wemmer DE. Biochemistry. 1995;34:2937–2945. doi: 10.1021/bi00009a025. [DOI] [PubMed] [Google Scholar]
- 21.Shikiya R, Li JS, Gold B, Marky LA. Biochemistry. 2005;44:12582–12588. doi: 10.1021/bi050897i. [DOI] [PubMed] [Google Scholar]
- 22.Soto AM, Gmeiner WH, Marky LA. Biochemistry. 2002;41:6842–6849. doi: 10.1021/bi025715o. [DOI] [PubMed] [Google Scholar]
- 23.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 24.Drew HR, Dickerson RE. J. Mol. Biol. 1981;151:535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- 25.Woods KK, Maehigashi T, Howerton SB, Sines CC, Tannenbaum S, Williams LD. J. Am. Chem. Soc. 2004;126:15330–15331. doi: 10.1021/ja045207x. [DOI] [PubMed] [Google Scholar]
- 26.Marky LA, Kupke DW. Methods Enzymol. 2000;323:419–441. doi: 10.1016/s0076-6879(00)23376-4. [DOI] [PubMed] [Google Scholar]
- 27.Plosky BS, Frank EG, Berry DA, Vennall GP, McDonald JP, Woodgate R. Nucleic Acids Res. 2008;36:2152–2162. doi: 10.1093/nar/gkn058. [DOI] [PMC free article] [PubMed] [Google Scholar]