Abstract
Mdm2 binding protein (MTBP) has been implicated in cell cycle arrest and the Mdm2-p53 tumor suppressor pathway through its interaction with Mdm2. To determine the function of MTBP in tumorigenesis and its potential role in the Mdm2-p53 pathway, we crossed Mtbp deficient mice to Eµ-myc transgenic mice, in which overexpression of the oncogene c-Myc induces B cell lymphomas primarily through inactivation of the Mdm2-p53 pathway. We report that Myc-induced B cell lymphoma development in Mtbp heterozygous mice was profoundly delayed. Surprisingly, reduced levels of Mtbp did not lead to an increase in B cell apoptosis or affect Mdm2. Instead, an Mtbp deficiency inhibited Myc-induced proliferation and the upregulation of Myc target genes necessary for cell growth. Consistent with a role in proliferation, Mtbp expression was induced by Myc and other factors that promote cell cycle progression and was elevated in lymphomas from humans and mice. Therefore, Mtbp functioned independent of Mdm2 and was a limiting factor for the proliferative and transforming functions of Myc. Thus, Mtbp is a previously unrecognized regulator of Myc-induced tumorigenesis.
Keywords: MTBP, Myc, Mdm2, p53, lymphoma
Introduction
MTBP, a 104 kDa protein with no known functional motifs, was identified through a yeast two-hybrid screen to bind to the E3 ubiquitin ligase Mdm2 (Boyd et al., 2000), a regulator of the p53 tumor suppressor (Marine & Lozano, 2009). MTBP overexpression inhibited cell cycle progression in a p53-independent, Mdm2-dependent manner following nocodazole treatment (Boyd et al., 2000). A subsequent study showed that MTBP regulates p53 through modulation of Mdm2 ubiquitin ligase activity (Brady et al., 2005). Specifically, overexpression of MTBP in tumor cell lines increased Mdm2 ubiquitin ligase activity increasing p53 degradation, and suppression of MTBP expression had the opposite effect. To determine the physiological role of MTBP, mice lacking Mtbp were generated (Iwakuma et al., 2008). Mtbp heterozygous mice were viable and did not have any obvious defects. However, loss of both alleles of Mtbp was embryonic lethal. In contrast to Mdm2 deletion, the lethality of Mtbp deletion could not be rescued by loss of p53 (Iwakuma et al., 2008), suggesting that Mtbp may not regulate Mdm2 and consequently p53 in vivo. Regardless of the Mtbp/Mdm2 relationship, decreased Mtbp expression has been linked to tumor metastasis (Iwakuma et al., 2008), and the chromosomal region where MTBP lies is frequently amplified in human colorectal cancer and multiple myeloma (Carrasco et al., 2006; Martin et al., 2007). Therefore, the function of MTBP in relationship to Mdm2 and p53 and in tumorigenesis is currently unclear.
Cell cycle and apoptosis are critical regulators of tumor development. Deletion of E2F1, a transcription factor essential for proliferation, inhibits c-Myc-induced B cell lymphomagenesis through upregulation of the cell cycle inhibitor p27 and suppression of cell cycle progression (Baudino et al., 2003). Moreover, reduced levels of ornithine decarboxylase (ODC), a transcriptional target gene of c-Myc required for polyamine synthesis, leads to decreased proliferation and inhibition of Myc-induced B cell lymphoma development (Nilsson et al., 2005). It is well established that Myc induces apoptosis in primary B cells, in part, by activating the ARF-Mdm2-p53 tumor suppressor pathway (Eischen et al., 1999). Myc activates ARF, which inhibits Mdm2 causing p53 activation and apoptosis. Inactivation of ARF or p53 or overexpression of Mdm2 are frequent events detected in B cell lymphomas that arise in mice overexpressing Myc (Eµ-myc transgenics) (Eischen et al., 1999). Moreover, mice that are deficient in ARF or p53 or overexpress Mdm2 have an acceleration of lymphoma development due to a reduction in B cell apoptosis (Alt et al., 2003; Eischen et al., 1999; Schmitt et al., 1999; Wang et al., 2008). In contrast, Mdm2 heterozygosity inhibits Myc-induced lymphomagenesis due to increased p53-dependent B cell apoptosis (Alt et al., 2003), which can be rescued with loss of one allele of ARF (Eischen et al., 2004). Therefore genes that influence Mdm2, as Mtbp is postulated to do, should have a significant effect on Myc-induced apoptosis and tumor development. However, our data show that loss of one allele of Mtbp did not impact apoptosis or function through Mdm2, yet lymphoma development in Mtbp+/−Eµ-myc transgenic mice was inhibited. Mtbp heterozygous cells had reduced rates of Myc-induced proliferation and decreased ability to upregulate Myc target genes necessary for cell growth. Our results indicate that Mtbp regulates Myc-induced lymphomagenesis not through Mdm2, but in cooperation with Myc.
Materials and Methods
Mice
Congenic C57Bl/6 Eµ-myc transgenic mice were from Drs. Alan Harris (Walter & Eliza Hall Institute, Melbourne, Australia) and Charles Sidman (University of Cincinnati, Cincinnati, OH), and ARF−/− mice were from Drs. Martine Roussel and Charles Sherr (St. Jude Children’s Research Hospital, Memphis, TN). Mtbp+/− (C57Bl/6X129/Sv backcrossed onto C57Bl/6 at least five generations) mice were crossed to male Eµ-myc transgenics to generate F1’s. F1’s were crossed to generate F2’s for analysis. F2’s were also crossed to ARF−/− or p53−/− mice to generate mice deficient in ARF or p53. Mice were carefully followed, and at signs of disease were sacrificed. Statistical significance was determined by log-rank tests. All experiments with mice were approved by the Vanderbilt Institutional Animal Care and Use Committee (IACUC) and followed all federal and state rules and regulations.
Culture and infection of primary pre-B cells and lymphocyte phenotype analysis
Primary pre-B cell (CD43−, B220+, CD19+, IgM−) cultures were generated from bone marrow of 5–8 week old mice. Pre-B cells were infected with MSCV-MycER-IRES-GFP and GFP+ cells were sorted by FACS. Phenotypic analyses of pre-B cells, whole spleens, bone marrow, and lymphomas were performed with fluorescently linked antibodies from Southern Biotechnology or BDPharMingen and flow cytometry. All procedures were previously described (Eischen et al., 1999).
Viability, apoptosis, proliferation, cell cycle, and chromosome analysis
Viability following explantation of bone marrow into IL-7 containing medium, after the addition of 1 µM 4-OHT (Sigma) to the culture medium of MycER expressing pre-B cells, or after IL-7 deprivation was determined at specific intervals by Trypan Blue Dye exclusion. Apoptosis and cell cycle was measured with propidium iodide (PI) staining and analysis on a FACScalibur. Quantification of fragmented (sub-G1) DNA was performed with CellQuest (BD Immunocytometry Systems). The percentage of cells in each phase of the cell cycle was determined by ModFIT (Verity House Software). Proliferation was determined by BrdU incorporation as previously described (Wang et al., 2008). MTS assays were performed at intervals as per manufacturer’s protocol (Promega) following transfection of four replicates of siRNAs (on-target SMART pools for Mtbp or non-targeting control, Dharmacon) with Lipofectamine2000 (Invitrogen). Metaphases of splenocytes were analyzed for breaks and aneuploidy as previously described (Wang et al., 2008).
Western and Southern blotting
Murine pre-B cells, lymphomas, and spleens, and normal human lymph node, spleen, peripheral blood lymphocytes, and lymphoma cell lines were lysed as previously described (Zindy et al., 1998). Antibodies specific for p19ARF (GeneTex), p53 (Ab-7, Calbiochem), Mdm2 (C-18, Santa Cruz), Myc (06–340, Upstate Biotechnology), murine Mtbp (Santa Cruz), human MTBP (PHL-1, Rockland), E2F1 (C20, Santa Cruz), p16 (M-156, Santa Cruz), p21 (SXM30, BD Biosciences), p27 (BD Biosciences), and β-actin (Sigma) were used to Western blot. HRP-linked secondary antibodies and ECL (GE Healthcare) or Supersignal (Pierce) to detect bound immunocomplexes were used. Southern blots for ARF, p53, and Mtbp were preformed as previously described (Eischen et al., 1999; Iwakuma et al., 2008).
Quantitative RT-PCR
Total RNA was isolated, cDNA was generated, and qRT-PCR with SybrGreen was performed as previously described (Wang et al., 2008). Primers for Mtbp and β-actin were previously described (Iwakuma et al., 2008; Wang et al., 2008).
Northern Blotting
Total RNA was prepared using RNAbee (tel-Test) according to manufacturer's instructions, separated by electrophoresis, transferred to hybond-N nylon membrane, and crosslinked by UV. Full-length human MTBP cDNA was labeled with 32P (random primed labeling kit; Boehringer Mannheim), and hybridizations were performed in Rapid-hyb (GE Healthcare) according to manufacturer's instructions. For the cycloheximide experiments, cycloheximide (10µg/ml) was added 30 min. prior to the addition of 4-OHT (Eischen et al., 2001).
Results
Mtbp expression is regulated by mitogens, oncogenes, and cell cycle
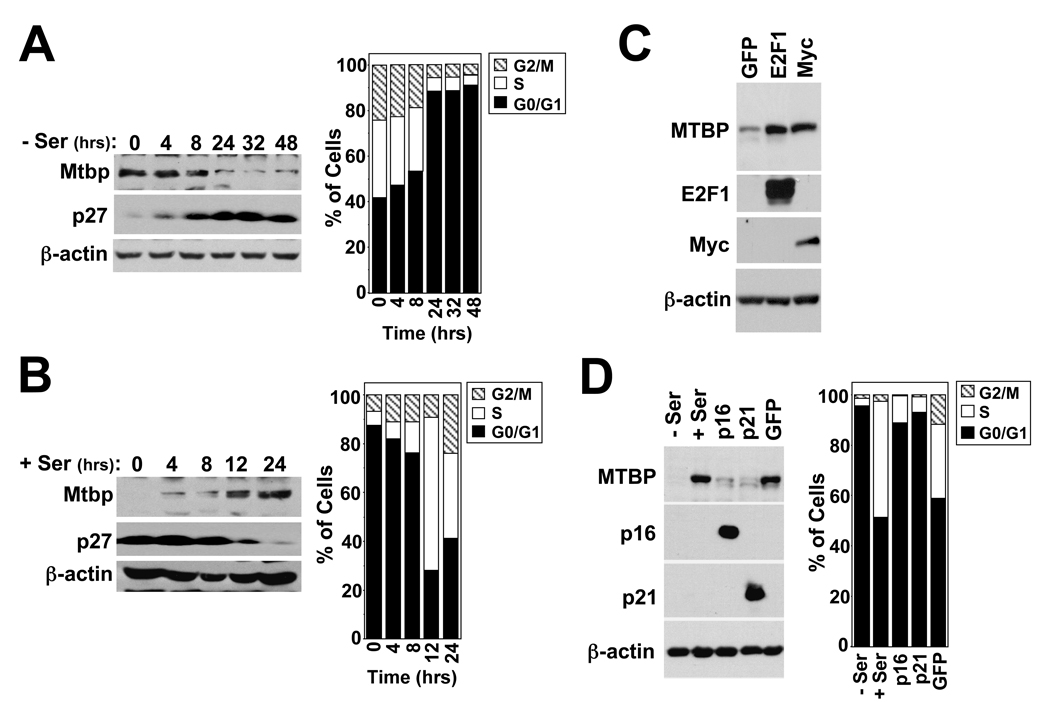
To obtain a better understanding of Mtbp, we explored how Mtbp expression was regulated. The G1 cell cycle arrest induced by depleting serum from cultured p53−/− mouse embryo fibroblasts (MEFs) resulted in a dramatic decrease in Mtbp protein expression with a concomitant increase in the cell cycle inhibitor p27, which is upregulated in G1 (Fig 1A). Re-addition of serum back to serum starved MEFs restored Mtbp expression as cells left G1 and moved into S phase (Fig. 1B). Similar results were observed in wild-type or ARF-null MEFs (data not shown). Overexpression of cyclin D or cyclin E or of the oncogenes c-Myc or E2F1, which drive cells into S phase, led to an increase in MTBP protein expression in human carcinoma cells and murine fibroblasts (Fig. 1C & data not shown). In contrast, expression of the cell cycle inhibitors, p21cip1 or p16Ink4a, which arrest cells in G1, resulted in decreased MTBP protein expression (Fig. 1D). Therefore, pro-proliferative signals upregulated and anti-proliferative signals downregulated Mtbp protein expression in human and murine cells, and this occurred independent of ARF and p53. These results suggest a positive role for Mtbp in cell growth.
Figure 1.
Mtbp expression is regulated by proliferative signals. (A) Western blot and cell cycle analysis of p53−/− MEFs deprived of serum (−Ser) for the indicated hours. (B) Following 24 hours of serum deprivation, serum was added back to the culture medium of p53−/− MEFs for the indicated number of hours and Western blot and cell cycle analyses were performed. (C) Western blots of H1299 cells infected with recombinant adenoviruses (30 plaque-forming units/cell) encoding GFP alone, E2F1 and GFP, or c-Myc and GFP. (D) Western blot and cell cycle analyses of H1299 cells grown in medium with (+Ser) or without (−Ser) serum or with serum and infected with adenoviruses expressing GFP alone, p16 and GFP, or p21 and GFP.
Mtbp transcription is induced by Myc
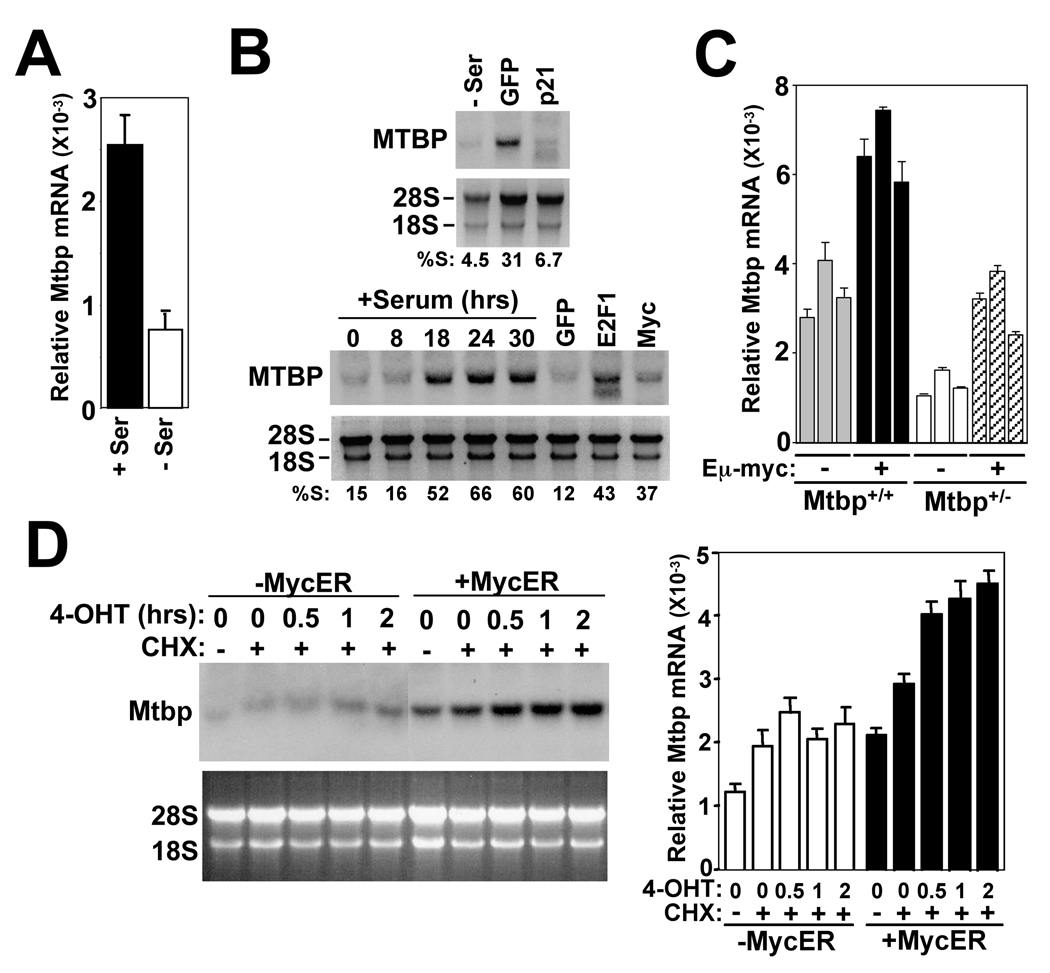
To address whether the increase in Mtbp protein levels from serum and oncogenes was due to changes in Mtbp transcripts, we evaluated RNA levels. Mtbp/MTBP mRNA, as measured by quantitative real-time PCR (qRT-PCR) or Northern blot, was significantly reduced by serum deprivation or following overexpression of p21 (Fig. 2A & 2B). Increased MTBP mRNA was detected with the re-addition of serum to serum starved cells or overexpression of Myc or E2F1 (Fig. 2B). Two bands are visible in the E2F1 lane, which could indicate an alternative spliced form of MTBP, but at this time it is unclear. Mtbp mRNA expression was also induced in vivo by c-Myc. qRT-PCR showed significantly elevated Mtbp mRNA in spleens of Mtbp+/+Eµ-myc transgenic mice, which overexpress c-Myc specifically in B cells, compared to levels in Mtbp+/+ non-transgenic littermate spleens (Fig. 2C). There were also increased levels of Mtbp mRNA in Mtbp+/−Eµ-myc spleens compared to levels in non-transgenic Mtbp+/− spleens. Interestingly, Mtbp levels in Mtbp+/+Eµ-myc spleens were higher than those in Mtbp+/−Eµ-myc spleens (Fig. 2C). To investigate whether Mtbp is directly induced by Myc, murine fibroblasts expressing a 4-hydroxytamoxifen (4-OHT) regulatable form of Myc (MycER)(Littlewood et al., 1995) were generated and treated with the protein synthesis inhibitor cycloheximide. Within 30 min. of MycER activation by 4-OHT, Mtbp mRNA expression increased (Fig. 2D), indicating that new protein synthesis was not required for Myc to induce Mtbp mRNA. There were modestly higher basal Mtbp levels in the MycER expressing cells in the absence of 4-OHT, likely due to the slight leakiness of the MycER construct. These results demonstrate that Myc may directly regulate Mtbp mRNA expression.
Figure 2.
Mtbp is transcriptionally regulated by Myc. (A) qRT-PCR analysis of p53−/− MEFs growing in (+Ser) or deprived of (−Ser) serum for 24 hours. (B) Following 24 hours of serum deprivation (−Ser), serum was added to H1299 cells and cells harvested at intervals (hours). Northern blots for MTBP mRNA from these samples and samples in Figure 1C and 1D were performed. 28S and 18S are also shown. The percentage of cells in S-phase (%S) in each sample is indicated. (C) qRT-PCR analysis of splenocytes from littermate matched mice that were Mtbp+/+ or Mtbp+/− and either Eµ-myc negative (−) or Eµ-myc positive (+). (D) Murine fibroblasts infected with a bicistronic retrovirus encoding GFP alone (−MycER) or MycER and GFP (+MycER) were serum deprived for 24 hours and left untreated (−) or pre-treated (+) with cycloheximide (CHX) for 30 min; 4-OHT was then added for the indicated times. Northern blot and qRT-PCR for Mtbp expression were performed. 28S and 18S are also shown. (A, C, & D) qRT-PCR data were generated in triplicate and normalized to β-actin levels.
Mtbp regulates Myc-induced lymphomagenesis independent of Mdm2
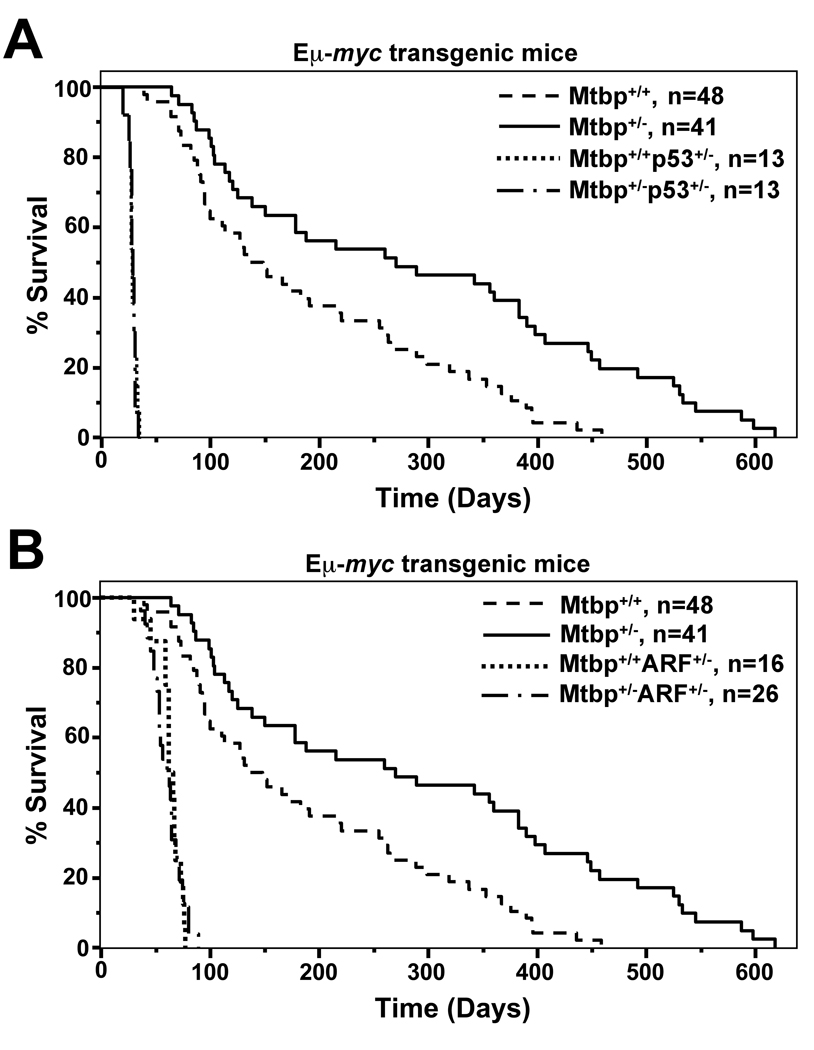
It was reported that MTBP levels impact Mdm2 expression and function (Brady et al., 2005). Since increased Mdm2 levels accelerate and decreased Mdm2 levels inhibit lymphoma development in Eµ-myc transgenic mice due to disregulation of p53 (Alt et al., 2003; Wang et al., 2008), we generated Mtbp+/−Eµ-myc mice to evaluate lymphoma development. Mtbp+/−Eµ-myc mice had a profound delay in lymphoma development compared to wild-type Eµ-myc littermates (Fig. 3; p=0.0004, log-rank test). At one year of age, 40% of the Mtbp+/−Eµ-myc transgenics were alive, whereas only 15% of the Mtbp+/+Eµ-myc mice were alive. The 270 days mean survival of Mtbp+/−Eµ-myc transgenics was twice that of the 135 days mean survival for Mtbp+/+Eµ-myc mice. The lymphomas that developed in Mtbp+/−Eµ-myc mice were typical pre-B/B cell lymphomas that arise in Eµ-myc mice (data not shown). Therefore, an Mtbp haploinsufficiency significantly hindered Myc-mediated B cell lymphomagenesis.
Figure 3.
Myc-induced lymphomagenesis is inhibited by Mtbp heterozygosity. (A, B) Kaplan-Meier survival curves of Eµ-myc transgenic mice of the indicated genotypes (p=0.0004 log-rank test, Mtbp+/+Eµ-myc versus Mtbp+/−Eµ-myc). The number of mice in each group is denoted by “n” values. Lymphoma was documented in all mice.
To determine whether a deficiency in Mtbp would also inhibit the rapid lymphoma development in p53+/−Eµ-myc mice (Alt et al., 2003) and the contribution of p53 to Mtbp function, p53+/−Mtbp+/−Eµ-myc mice were generated. Deletion of one allele of p53 in Mtbp+/−Eµ-myc mice accelerated lymphomagenesis to rates analogous to those of p53+/−Mtbp+/+Eµ-myc transgenics (Fig. 3A); the mean survivals of both genotypes were 30 days. Thus, reduced levels of Mtbp suppressed Myc-induced lymphomagenesis, but the effect was abrogated by deletion of one p53 allele. These data imply that p53 functions downstream of Mtbp or that loss of p53 dominates B cell transformation and supersedes any effect Mtbp haploinsufficiency has in Myc-induced lymphomagenesis.
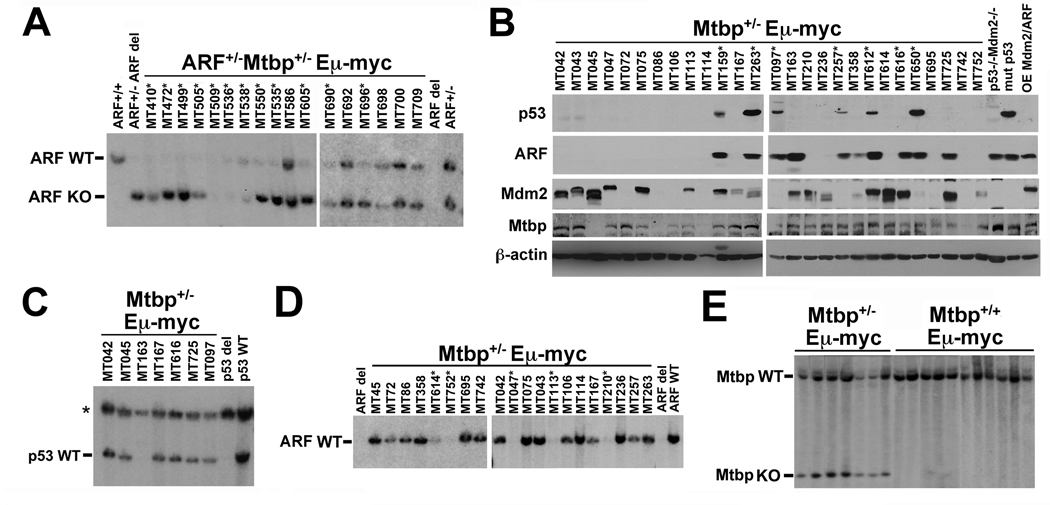
The levels of p53 accessible to Mdm2 are regulated by ARF levels in Eµ-myc mice (Eischen et al., 2004). If Mtbp regulates Mdm2, then altering the amount of ARF present should impact the ability of Mtbp to regulate Mdm2; we generated ARF+/−Mtbp+/−Eµ-myc transgenics to test this concept. In contrast to ARF+/−Mdm2+/−Eµ-myc mice (Eischen et al., 2004), loss of one allele of ARF did not restore lymphoma latency in Mtbp+/−Eµ-myc mice to wild-type Eµ-myc rates (Fig. 3B). In fact, ARF+/−Mtbp+/−Eµ-myc mice rapidly developed lymphoma and had a similar mean survival to ARF+/−Mtbp+/+Eµ-myc littermates (60 days versus 62 days, respectively). Therefore, Mtbp levels did not impact the ability of ARF to regulate Mdm2. To assess this concept further, we evaluated loss of heterozygosity (LOH) of the wild-type allele of ARF in ARF+/−Mtbp+/−Eµ-myc lymphomas. ARF LOH was observed in 12 of 17 (71%) ARF+/− Mtbp+/−Eµ-myc lymphomas analyzed, including three that had also deleted the knockout allele of ARF (Fig. 4A), a frequency analogous to that in ARF+/−Eµ-myc lymphomas (77%, (Eischen et al., 1999)). If Mtbp regulates Mdm2, the expected frequency of ARF LOH in the ARF+/−Mtbp+/−Eµ-myc lymphomas should have been similar to the 44% observed in lymphomas from ARF+/− Mdm2+/−Eµ-myc transgenics (Eischen et al., 2004). These genetic results indicate that Mtbp function is not influenced by ARF status, and that Mtbp does not appear to be functioning through Mdm2.
Figure 4.
Normal frequencies of p53, ARF, and Mdm2 alterations in Mtbp+/−Eµ-myc lymphomas. (A) Southern blots for ARF in ARF+/−Mtbp+/−Eµ-myc transgenic lymphomas. Location of the ARF wild-type (WT) and knockout (KO) alleles are indicated. Asterisks denote lymphomas that have deleted ARF. (B) Levels of the indicated proteins in whole cell extracts of Mtbp+/−Eµ-myc lymphomas were assessed by Western blotting. Lysates from p53−/−Mdm2−/− MEFs, a lymphoma expressing mutant p53 (mut p53), and a lymphoma overexpressing Mdm2 and ARF (OE Mdm2/ARF) were controls. Asterisks indicate lymphomas with mutant p53. p53 in MT616 is not detectable due to a nonsense mutation. (C) Southern blot for p53 in Mtbp+/−Eµ-myc lymphomas. The asterisk denotes the location of the pseudogene. (D) Southern blot for ARF in Mtbp+/−Eµ-myc lymphomas. Asterisks indicate lymphomas that have deleted ARF. (E) Southern blot for Mtbp in Eµ-myc lymphomas. The location of Mtbp wild-type (WT) and knockout (KO) alleles are indicated.
Another means to evaluate whether Mtbp levels are regulating Mdm2 function in vivo is to analyze Mtbp+/−Eµ-myc lymphomas for p53 inactivation and Mdm2 levels. Of note, half of the Mdm2+/−Eµ-myc lymphomas have mutated p53 (Alt et al., 2003). As previously reported for Eµ-myc lymphomas (Eischen et al., 1999), p53 overexpression resulting from a mutation was present in a quarter of the lymphomas analyzed from Mtbp+/+Eµ-myc littermates (data not shown). Similarly, 26% (7 of 27) of the lymphomas analyzed from Mtbp+/−Eµ-myc transgenics contained mutant p53 (Fig. 4B). All mutations fell within the DNA-binding domain of p53, including one lymphoma (MT616) that had a nonsense mutation resulting in a truncated p53 that was not detected by Western blot. Only one of 27 Mtbp+/−Eµ-myc lymphomas (MT163) had deleted p53 (Fig. 4C), the same frequency as that observed in Mtbp+/+Eµ-myc lymphomas. Biallelic ARF deletions were detected in 19% (5 of 27) of Mtbp+/−Eµ-myc lymphomas (Fig. 4D), which is similar to the frequency of ARF deletions in wild-type Eµ-myc lymphomas (Eischen et al., 1999). At least one of the three isoforms of Mdm2 was overexpressed in 62% (17 of 27) of the Mtbp+/−Eµ-myc lymphomas (Fig. 4B), which is analogous to the frequency in Mtbp+/+Eµ-myc lymphomas. The levels of Mtbp protein in the lymphomas did not correlate to Mdm2 levels or p53 status. Moreover, Southern blots showed there was no LOH of the wild-type allele of Mtbp in Mtbp+/−Eµ-myc lymphomas, and Mtbp was not deleted in Mtbp+/+Eµ-myc lymphomas (Fig. 4E). Therefore, loss of one allele of Mtbp did not result in increased p53 activation, leading to an increased frequency of p53 mutations or deletions in Mtbp+/−Eµ-myc lymphomas, as would be expected if Mtbp regulated Mdm2. Moreover, Mdm2 levels were not influenced by Mtbp levels, and ARF deletions occurred at a normal frequency. Therefore, the data indicate Mtbp does not regulate Mdm2 or the ARF-Mdm2-p53 pathway during Myc-induced lymphomagenesis.
Loss of one allele of Mtbp does not alter Myc-induced apoptosis
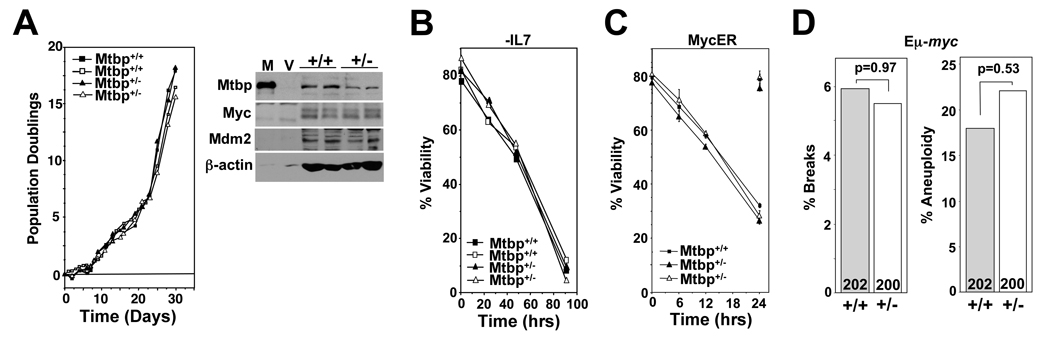
Since our genetic results indicate that Mtbp does not function through Mdm2, but decreased levels of Mtbp did inhibit Myc-induced lymphomagenesis, we investigated the role of Mtbp in Myc functions. We first evaluated the effects of decreased Mtbp levels on the growth and survival of pre-B cells. Pre-B cells from Mtbp+/− and Mtbp+/+ littermates emerged from bone marrow cultures at a similar rate and had analogous doubling times and percentages of spontaneous apoptotic cells (Fig. 5A & data not shown). These results are in contrast to those obtained with Mdm2+/− bone marrow, which underwent apoptosis in culture (Alt et al., 2003). Western blots showed lower levels of Mtbp protein in Mtbp+/− pre-B cells in comparison to Mtbp+/+ pre-B cells, whereas Myc and Mdm2 levels were equivalent in both genotypes (Fig. 5A). Since Mtbp expression is regulated by factors that induce growth (Fig. 1), and IL-7 is essential for pre-B cell growth and survival, we evaluated the susceptibility of Mtbp+/− pre-B cells to cytokine deprivation-induced apoptosis. Mtbp+/− pre-B cells underwent apoptosis from IL-7 deprivation at the same rate as Mtbp+/+ pre-B cells (Fig. 5B). Therefore, pre-B cells with decreased levels of Mtbp grew and underwent spontaneous apoptosis at normal rates, and were equally as sensitive to apoptosis from growth factor deprivation as wild-type pre-B cells.
Figure 5.
Normal rates of Myc-induced apoptosis and chromosome instability in Mtbp heterozygous B cells. (A) Bone marrow from two Mtbp+/+ (squares, +/+) and two Mtbp+/− (triangles, +/−) littermates was placed into IL-7 containing medium on day 0. Viable cells were counted at intervals. Data is representative of three independent experiments. Whole cell lysates from pre-B cells that emerged from the cultures were Western blotted. Protein lysates from 293T cells transfected with a vector encoding Mtbp (M) or empty vector (V) were controls (1/6 of the amount of protein of the other samples). (B) Littermate matched pre-B cells of the indicated genotypes were deprived of IL-7, and viability determined at intervals with Trypan Blue Dye exclusion. (C) 4-OHT was added at time 0 to MycER expressing pre-B cells (littermate matched) to activate MycER, and their viability was determined at intervals thereafter by Trypan Blue Dye exclusion. Apoptosis was confirmed by analysis of subdiploid DNA content after staining with PI. Symbols in upper right corner are 24 hour vehicle (ethanol) control treated MycER expressing pre-B cells. (D) Metaphases from splenocytes from Mtbp+/− Eµ-myc transgenic (+/−) and Mtbp+/+Eµ-myc littermates (+/+) prior to any detectable lymphoma. The number of total metaphases analyzed is indicated at the bottom of each bar. Breaks were chromosome or chromatid. Aneuploidy is defined as any metaphase having more or less than 40 chromosomes. Chi-squared test was used to determine significance.
To determine whether reduced Mtbp levels impact Myc functions, we evaluated Myc-induced apoptosis. pre-B cells were infected with a bicistronic retrovirus encoding MycER and green fluorescent protein (GFP). Both wild-type and Mtbp+/− GFP+ pre-B cells underwent apoptosis at a similar rate upon MycER activation with 4-OHT (Fig. 5C). We also assessed Myc-induced apoptosis in cultures of pre-B cells from Eµ-myc mice. Bone marrow cultures from Mtbp+/−Eµ-myc and Mtbp+/+Eµ-myc littermates prior to any detectable disease consistently had similar numbers of dead or dying cells (data not shown). Additionally, overexpression of Myc results in DNA breaks, which is thought to contribute to apoptosis and chromosomal instability (Ray et al., 2006; Vafa et al., 2002; Wang et al., 2008). We evaluated metaphases from splenocytes from Mtbp+/−Eµ-myc and Mtbp+/+Eµ-myc littermates prior to detectable lymphoma. There were similar percentages of metaphases with DNA breaks (chromosome and chromatid) and aneuploidy (greater or less than 40 chromosomes) in both genotypes (Fig. 5D). Therefore, decreased Mtbp levels did not appear to influence Myc-induced apoptosis, DNA breakage, or chromosomal instability.
Loss of one allele of Mtbp inhibits Myc-driven proliferation
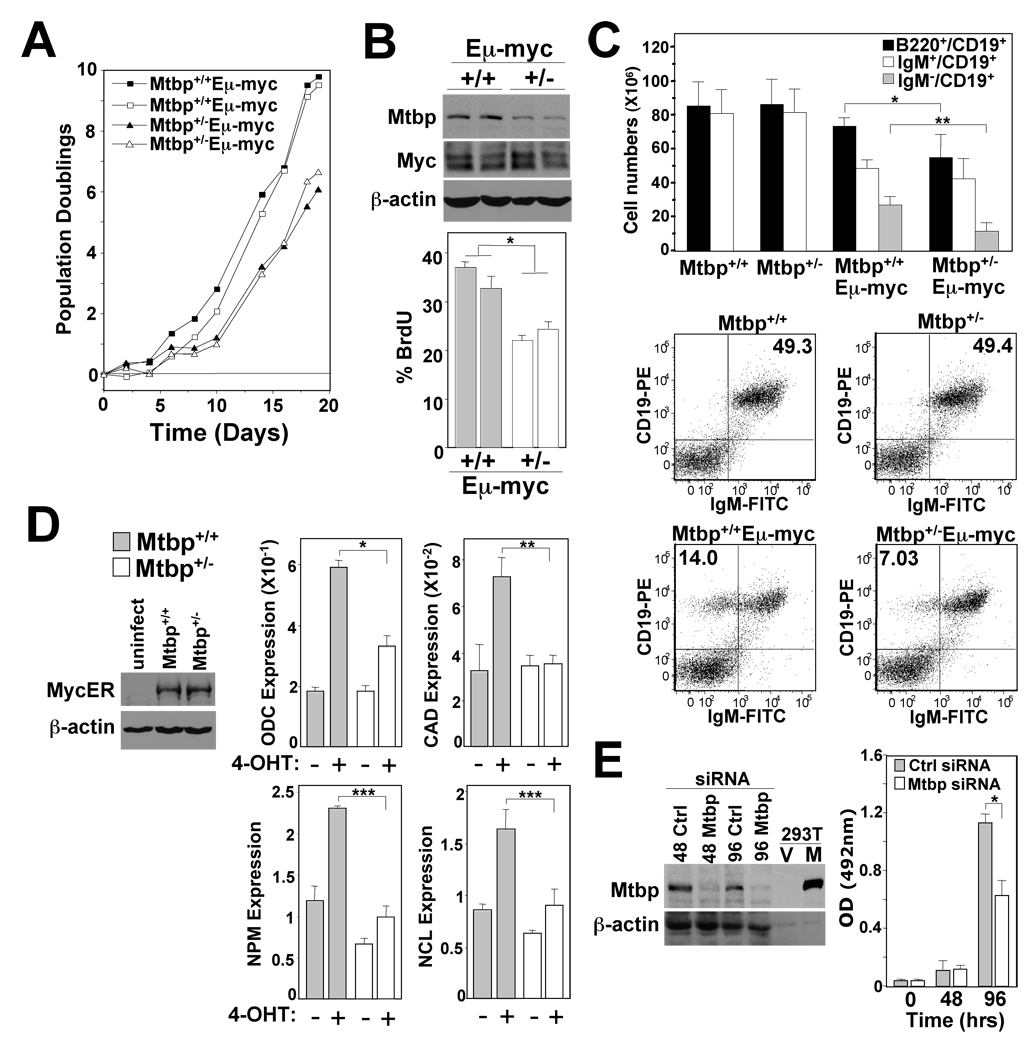
As an oncogene, increased levels of Myc force proliferation; consequently, we evaluated Myc-driven B cell growth. Bone marrows from Mtbp+/−Eµ-myc and Mtbp+/+Eµ-myc littermates prior to any detectable disease were placed into culture in IL-7 containing media, and viable cells were counted at intervals for 20 days. The Mtbp+/−Eµ-myc pre-B cells emerged from the cultures at a slower rate than the Mtbp+/+Eµ-myc pre-B cells (Fig. 6A). Once the pre-B cells dominated the cultures (7–10 days), the number of population doublings of the Mtbp+/−Eµ-myc pre-B cells was less than that of pre-B cells from Mtbp+/+Eµ-myc littermates. Mtbp+/−Eµ-myc pre-B cells expressed lower levels of Mtbp protein and incorporated decreased amounts of BrdU compared to Mtbp+/+Eµ-myc pre-B cells (Fig. 6B), illustrating fewer Mtbp haploinsufficient cells in S-phase. Therefore, the decreased rate of Myc-induced proliferation in Mtbp+/− B cells indicates that Mtbp is limiting when Myc is forcing proliferation.
Figure 6.
Mtbp heterozygous pre-B cells have reduced Myc-driven proliferation. (A) Bone marrow from two Mtbp+/+Eµ-myc (squares) and two Mtbp+/−Eµ-myc (triangles) littermates prior to any detectable lymphoma was placed into IL-7 containing medium (day 0). Viable cells were counted at intervals. Data is representative of three independent experiments from separate litters of mice. (B) Western blots for the indicated proteins and the percentage of cells with BrdU incorporation in pre-B cells from two Mtbp+/−Eµ-myc transgenics (+/−) and two Mtbp+/+Eµ-myc transgenic littermates (+/+) was determined. Differences in BrdU incorporation between the two genotypes were significant (p=0.02, t-test). (C) Total numbers of B cells (B220+/CD19+) or B cell subsets in spleens of the indicated genotypes. Each bar represents four-six mice of each genotype. Asterisks denote statistically significant differences (*p=0.02; **p=0.0006, t-test). Representative dot plots of CD19-PE versus IgM-FITC gated on total lymphocytes from spleens. (D) Relative mRNA levels of the indicated gene transcripts were determined by qRT-PCR in Mtbp+/− and Mtbp+/+ MycER expressing MEFs following MycER activation with 4-OHT (*p=0.0005, **p=0.003, ***p=0.002; t-test). All data were generated in triplicate and normalized to β-actin levels. The levels of MycER were determined by Western blot. (E) p53−/− MEFs were transfected with control siRNA (Ctrl) or a pool of Mtbp-specific siRNAs. MTS assays were performed at intervals (hours) (*p=0.0003, t-test) and protein lysates were Western blotted. Protein lysates from 293T cells transfected with a vector encoding Mtbp (M) or empty vector (V) were controls (1/6 of the amount of protein of the other samples).
Myc overexpression in the B cell compartment of wild-type Eµ-myc mice leads to increased numbers of pre-B cells, which escape the bone marrow and are detected in the spleens of these mice (Langdon et al., 1986). If decreased levels of Mtbp inhibit Myc-induced proliferation, we postulated that B cell numbers would be altered in Mtbp+/−Eµ-myc mice. Evaluation of Mtbp+/− and Mtbp+/+ spleens from littermates showed normal numbers and percentages of total (B220+/CD19+) and mature (IgM+/CD19+) B cells (Fig. 6C). Although the percentage of total B cells in Mtbp+/−Eµ-myc spleens was similar to that in Mtbp+/+Eµ-myc littermates (data not shown), there were decreased numbers of total B cells in Mtbp+/−Eµ-myc transgenic spleens. This difference in total B cell numbers was due to significantly reduced numbers of pre-B cells (IgM−/CD19+) in the spleens in Mtbp+/−Eµ-myc mice, as immature/mature B cell (IgM+/CD19+) numbers were similar and not statistically different (Fig. 6C). Thus, B cell development was altered in Mtbp+/−Eµ-myc mice and not in Mtbp+/− mice in the absence of the Myc transgene. Therefore, these results combined with the data above suggest that an insufficient amount of Mtbp is present in Mtbp+/−Eµ-myc pre-B cells for Myc to effectively drive proliferation and expansion of this population.
To further investigate the decreased Myc-induced proliferation of Mtbp deficient cells, we evaluated the expression of four Myc transcriptional targets that are necessary for proliferation following Myc activation. MEFs from Mtbp+/− and Mtbp+/+ littermates were infected with the bicistronic retrovirus encoding MycER and GFP. MEFs were chosen for these experiments, since in the presence of serum, they proliferate when MycER is activated instead of undergoing apoptosis. Equal levels of MycER and GFP were detected in both genotypes of MEFs, and both grew at similar rates (Fig. 6D & data not shown). Activation of MycER with 4-OHT in the GFP+ wild-type MEFs resulted in the induction of ODC, carbamoyl-phosphate synthetase 2/aspartate transcarbamylase/dihydroorotase (CAD), nucleophosmin (NPM), and nucleolin (NCL) in three hours (Fig. 6D). In contrast, in the GFP+ Mtbp+/− MEFs, these Myc target genes showed a significantly reduced induction of expression following MycER activation. Similar results were obtained up to eight hours post MycER activation (data not shown). Therefore, decreased expression of Mtbp resulted in a reduction of Myc-mediated transcription of genes necessary for proliferation.
As a separate independent measure of Mtbp’s role in proliferation and to further test the contribution of p53 to Mtbp function, we knocked down Mtbp expression in p53−/− MEFs with Mtbp-specific siRNAs. Within 48 hours, MEFs transfected with a pool of four different Mtbp-specific siRNAs showed an approximately 90% reduction in Mtbp expression, a larger decrease in Mtbp to that in Mtbp+/− cells, compared to control (Fig. 6E). Control siRNA did not impact cell growth, whereas MEFs with the Mtbp-specific siRNAs showed a significant reduction in proliferation 96 hours post transfection. There was no detectable difference in apoptosis between the samples (data not shown). Therefore, loss of Mtbp expression suppressed proliferation in a p53-independent manner.
Mtbp is overexpressed in lymphomas
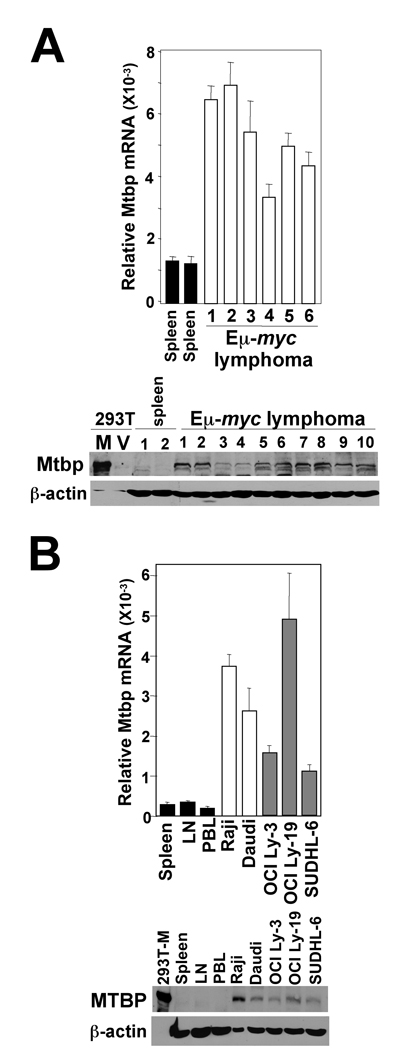
Our results suggest that Mtbp expression contributes to Myc functions necessary for growth and transformation, and is induced by Myc, and thus, we postulated Mtbp expression would be increased in lymphomas that overexpress Myc. Mtbp mRNA levels as determined by qRT-PCR were significantly elevated in Eµ-myc lymphomas compared to Mtbp levels in untransformed Eµ-myc splenocytes (Fig. 7A). Similarly, protein levels of Mtbp were also increased in Eµ-myc lymphomas. There were also significantly elevated levels of MTBP mRNA and protein in human B cell lymphoma cell lines, as compared to three normal human lymphoid tissue controls (Fig. 7B). Additionally, there were increased levels of MTBP in human Burkitt lymphomas that have MYC translocations, as well as in the human diffuse large B cell lymphoma (DLBCL) cell lines, which frequently overexpress Myc. Therefore, B cell lymphomas express increased levels of MTBP, possibly due to its role in proliferation, and thus, may contribute to lymphomagenesis.
Figure 7.
Increased Mtbp expression in murine and human lymphomas. (A, B) qRT-PCR and Western blots were performed with splenocytes (prior to detectable disease) and B cell lymphomas from wild-type Eµ-myc transgenic mice (A) and human B cell lymphoma lines (Burkitt lymphomas, white bars; DLBCL, grey bars) and normal human lymphatic tissues (spleen; lymph node, LN; peripheral blood lymphocytes, PBL, black bars) (B). Protein lysates from 293T cells transfected with an Mtbp expressing vector (M) or empty vector (V) were controls (1/6 of the amount of protein of the other samples). qRT-PCR data were generated in triplicate and normalized to β-actin levels.
Discussion
A previous overexpression study has implicated Mdm2, and consequently p53, in mediating Mtbp functions (Boyd et al., 2000; Brady et al., 2005). However our unbiased genetic approach has revealed that Mdm2 appears to have little to do with Mtbp function. Utilizing the Eµ-myc transgenic mouse model, which has well-established pathways of tumor development, we have demonstrated that Mtbp is a novel regulator of tumor development and that decreased levels of Mtbp significantly inhibited lymphoma development in a manner different from that when Mdm2 or p53 levels are altered. Although both Mtbp and Mdm2 heterozygous Eµ-myc mice had profound delays in tumor development, here we have made the novel finding that the extended tumor latency of both mice was caused by two distinct mechanisms. Specifically, cells from Mtbp+/− mice were impaired in their ability to respond to Myc overexpression, resulting in decreased proliferation and delayed tumor development. In contrast, Mdm2+/− B cells are hyper-responsive to Myc overexpression and undergo p53-dependent apoptosis causing a delay in tumorigenesis (Alt et al., 2003). If reduced levels of Mtbp lead to decreased levels of Mdm2 as was reported (Brady et al., 2005), then Mtbp+/−Eµ-myc mice should have had increased B cell apoptosis and a high frequency of p53 mutations in the lymphomas that arose. However, there was no significant increase in apoptosis of Mtbp+/− cells when Myc was overexpressed, and the frequency of p53 mutations and deletions in Mtbp+/−Eµ-myc lymphomas was similar to that of wild-type Eµ-myc lymphomas. In addition, loss of one allele of ARF, which restored lymphoma latency in Mdm2+/−Eµ-myc mice to that of wild-type Eµ-myc mice (Eischen et al., 2004), only accelerated lymphoma latency in Mtbp+/−Eµ-myc mice to the rate observed for ARF+/−Eµ-myc mice. Notably, deletion of p53, which rescues the Mdm2-null lethality, did not rescue Mtbp−/− embryos (Iwakuma et al., 2008). Moreover, Mtbp+/− thymocytes had a normal p53 response following gamma irradiation (Iwakuma et al., 2008). Combined, the data demonstrate that Mtbp does not function through Mdm2, but it is clearly a regulator of Myc-induced tumorigenesis.
Determining the role of Mtbp in mediating the delay in lymphoma development was challenging, since it did not appear to function through Mdm2 or impact apoptosis. However, since Mtbp expression was increased by pro-proliferative signals from growth factors and oncogenes and decreased when cells were arrested, this suggested Mtbp may participate in proliferation. The decreased proliferation we detected following knockdown of Mtbp expression supported this concept. We also observed that under conditions of hyperproliferative signals from the oncogene Myc, decreased Mtbp expression had significant effects on Myc function. Specifically, cells from Mtbp+/− mice were impaired in their ability to respond to Myc overexpression, resulting in decreased proliferation and transcriptional activation of Myc target genes necessary for cell growth. This was reflected in Mtbp+/−Eµ-myc mice by a significantly reduced expansion of pre-B cells normally observed in Eµ-myc mice. Previously, B-cells with a decreased ability to proliferate from Myc overexpression have impaired Myc-induced lymphomagenesis. For example, deletion of E2F1 or loss of one allele of ODC led to an inhibition of Myc-induced proliferation of MEFs or B cells, respectively (Baudino et al., 2003; Nilsson et al., 2005). Eµ-myc mice lacking E2F1 or heterozygous for ODC had a delay in B cell lymphomagenesis that was attributed to reduced proliferation. We observed a decrease in the ability of Myc to upregulate ODC and other genes necessary for proliferation when Mtbp was heterozygous, which could explain the reduced rates of Myc-mediated proliferation in Mtbp+/− Eµ-myc B cells. Therefore, a decrease in Myc-induced proliferation should account for the significant delay in B cell lymphoma development in Mtbp+/−Eµ-myc mice. However, we cannot rule out the possibility that Mtbp contributes to transformation in other ways as well. In addition, the biochemical mechanism by which Mtbp impacts Myc proliferative and transformation functions is unresolved. If Mtbp is a direct transcriptional target of Myc, as our data suggests, Mtbp could influence proliferation downstream of Myc. We have also detected Mtbp in the nucleus and associated with chromatin (CME unpublished data); thus, Mtbp could directly or indirectly affect Myc transcriptional functioning. Although additional studies are needed to define the biochemical mechanism of Mtbp function, our results do show that Mtbp is regulated by Myc and has a critical role in proliferation mediated by Myc.
Our data indicate that Mtbp regulates Myc-induced lymphoma development in a manner that is consistent with an oncogene rather than a tumor suppressor. Specifically, decreased expression of Mtbp inhibited Myc-induced proliferation and tumor development. Mtbp was not deleted in wild-type Eµ-myc lymphomas, nor was there LOH of Mtbp in Mtbp+/−Eµ-myc lymphomas. In another study, Mtbp+/−p53+/−mice had a similar tumor latency as p53+/− mice (Iwakuma et al., 2008). Therefore Mtbp does not appear to be a classic tumor suppressor, as was originally proposed (Boyd et al., 2000). Instead, Mtbp levels were elevated in B cell lymphomas from mice and humans. These data are consistent with the observation that the region that encodes MTBP is frequently amplified in human colorectal cancers and multiple myeloma (Carrasco et al., 2006; Martin et al., 2007). Moreover, public microarray databases show increased MTBP mRNA in many human cancers as compared to normal tissues. Therefore, mounting data indicate Mtbp overexpression is selected for and likely contributes to tumorigenesis, and thus Mtbp may be oncogenic. However, there is also data suggesting that decreased levels of Mtbp contribute to tumor metastasis in p53 heterozygous mice (Iwakuma et al., 2008). Although the biochemical function of Mtbp in metastasis in p53+/− mice is unresolved, the requirements of Mtbp in metastasis are likely to be different than that for Mtbp in Myc-induced proliferation and tumorigenesis. It will be important in the future to determine whether Mtbp is oncogenic in its own right or a critical partner of an oncogene such as Myc, and its precise role in metastasis.
Acknowledgments
The authors thank Dr. Silvia Plaza, Brandon Metge, Jane Kennedy, and Dr. Chris Carbone for expert technical assistance, Dr. Xavier Graña for assistance with the adenoviruses and the H1299 cell cycle studies, and Dr. William Dupont and Dale Plummer for Kaplan-Meier analysis. We also thank the following individuals for providing recombinant adenoviruses: Dr. Joseph Nevins (Duke University)-E2F1 and c-Myc, Dr. Juan Fueyo (MD Anderson Cancer Center)-p16, Dr. Wafik El-Deiry (University of Pennsylvania)-p21, and Dr. Ruiz-Lozano (Burnham Institute)-GFP. This work was supported by NCI grants R01CA098139, R01CA117935, and P30CA068485 (CME), the Leukemia & Lymphoma Society (CME), and P01CA095569 (DH and JG).
Footnotes
Conflict of Interest
There are no competing financial interests with the research described.
References
- Alt JR, Greiner TC, Cleveland JL, Eischen CM. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J. 2003;22:1442–1450. doi: 10.1093/emboj/cdg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudino TA, Maclean KH, Brennan J, Parganas E, Yang C, Aslanian A, Lees JA, Sherr CJ, Roussel MF, Cleveland JL. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol Cell. 2003;11:905–914. doi: 10.1016/s1097-2765(03)00102-3. [DOI] [PubMed] [Google Scholar]
- Boyd MT, Vlatkovic N, Haines DS. A novel cellular protein (MTBP) binds to MDM2 and induces a G1 arrest that is suppressed by MDM2. J Biol Chem. 2000;275:31883–31890. doi: 10.1074/jbc.M004252200. [DOI] [PubMed] [Google Scholar]
- Brady M, Vlatkovic N, Boyd MT. Regulation of p53 and MDM2 activity by MTBP. Mol Cell Biol. 2005;25:545–553. doi: 10.1128/MCB.25.2.545-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, Protopopov A, Sukhdeo K, Hanamura I, Stephens O, Barlogie B, Anderson KC, Chin L, Shaughnessy JD, Jr, Brennan C, Depinho RA. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Alt JR, Wang P. Loss of one allele of ARF rescues Mdm2 haploinsufficiency effects on apoptosis and lymphoma development. Oncogene. 2004;23:8931–8940. doi: 10.1038/sj.onc.1208052. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Woo D, Roussel MF, Cleveland JL. Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol. 2001;21:5063–5070. doi: 10.1128/MCB.21.15.5063-5070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakuma T, Tochigi Y, Van Pelt CS, Caldwell LC, Terzian T, Parant JM, Chau GP, Koch JG, Eischen CM, Lozano G. Mtbp haploinsufficiency in mice increases tumor metastasis. Oncogene. 2008;27:1813–1820. doi: 10.1038/sj.onc.1210827. [DOI] [PubMed] [Google Scholar]
- Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- Martin ES, Tonon G, Sinha R, Xiao Y, Feng B, Kimmelman AC, Protopopov A, Ivanova E, Brennan C, Montgomery K, Kucherlapati R, Bailey G, Redston M, Chin L, DePinho RA. Common and distinct genomic events in sporadic colorectal cancer and diverse cancer types. Cancer Res. 2007;67:10736–10743. doi: 10.1158/0008-5472.CAN-07-2742. [DOI] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW, Cleveland JL. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, Felsher DW. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res. 2006;66:6598–6605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2008;27:1590–1598. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]