Abstract
Background
It is widely appreciated that genomewide association studies often yield overestimates of the association of a marker with disease when attention focuses upon the marker showing the strongest relationship. For example, in a case-control setting the largest (in absolute value) estimated odds ratio has been found to typically overstate the association as measured in a second, independent set of data. The most common reason given for this observation is that the choice of the most extreme test statistic is often conditional upon first observing a significant p value associated with the marker. A second, less appreciated reason is described here. Under common circumstances it is the multiple testing of many markers and subsequent focus upon those with most extreme test statistics (i.e. highly ranked results) that leads to bias in the estimated effect sizes.
Conclusions
This bias, termed ranking bias, is separate from that arising from conditioning on a significant p value and may often be a more important factor in generating bias. An analytic description of this bias, simulations demonstrating its extent, and identification of some factors leading to its exacerbation are presented.
Key Words: Estimation bias, Multiple comparisons
Introduction
As high dimensional assay technology has become more available for genomic investigations there has been a corresponding increase in methodology to limit false positive findings [1, 2]. Until recently, less attention has been given to the effect sizes of those comparisons that are determined to reach statistical significance. In many instances this may be appropriate as effect sizes may not be of great importance per se; however in other circumstances it may be important to accurately assess the magnitude of the associated effects.
As an example, there has been increased recognition that some form of replication is desirable to support a claim of association found as a single result among a large panel of genetic markers [3]. However, using estimates of effect size derived from an initial study of as many as a million markers can lead to an underpowered evaluation as the initial estimate is likely biased toward more extreme results than are true.
A common rationale given for this bias is that it results from conditioning on the initial estimate meeting p value criteria for declaring significance – often referred to as selection or truncation bias. The term ‘significance bias’ will be used here to stress the conditioning upon a significant p value. Garner [4] describes this in the context of genomewide association scans; Allison et al. [5] describe it in the context of quantitative trait loci of paired siblings. This conditioning on a significant result has the effect of truncating the distribution of the marker's sampling distribution and creating an associated bias. For example, given a normally distributed variable X with mean μ and standard deviation ν, one can show the expected value of X conditional upon X > c (where c may be chosen to generate a significant p value) is given by
| (1.1) |
where φ(·) denotes the probability density function of normally distributed variable with mean μ and standard deviation ν. The second term on the right hand side of (1.1) captures the bias from conditioning. This description of bias does not recognize the effect multiple testing may exert on creating bias beyond its role in requiring a p value threshold less than 0.05. In this paper multiple testing and focusing on extreme results (i.e. highly ranked results) is shown to generate overestimation in situations where significance bias may have little effect. The terms ‘significance bias’ and ‘ranking bias’ are not in common use – the phrases ‘truncation bias’ and ‘selection bias’ are used more often but are insufficiently specific and fail to draw the distinction made in this paper.
Though a detailed description is given in the next section the idea of ranking bias is briefly described here. Given a number of test statistics, each one is composed of a trend term (related to true underlying effect size) and some random variation. When looking at the test statistic with the largest positive observed effect size it is likely (under many circumstances) that the random variation component is positive as well – because the large, positive observed effect is the sum of a trend and the random component. This random component captures the difference between the true and observed effect size and positive values correspond to a type of overestimation bias – called ranking bias here.
Allison et al. [5] briefly describe ranking bias but focus the work on significance bias. Göring et al. [6] discuss both significance bias and ranking bias. They make an analogy between the ranking bias discussed here with that corresponding to bias in model selection procedures.
Though they discuss ranking bias their modeling and analytical results are derived solely from models of significance bias. Zöllner and Pritchard [7] use the term ‘Winner's Curse’ to describe significance bias in a single marker and present an analytical approach toward correcting this bias. Sun and Bull [8] explicitly describe significance bias and more implicitly describe ranking bias in the context of a mathematical model with a number of non-informative markers and 1 informative marker. In the present work we present more general models with a number of informative markers and the relationship between bias (both significance and ranking) and the pattern of the markers' effect sizes is explicitly explored.
Description of Bias
The modeling proposed here is relatively simple both because it is easy to see how to apply this situation to others and because it allows for a transparent partitioning of a statistic into trend and random error terms that facilitates understanding ranking bias. Let N denote the common number of cases and controls drawn in population-based study, G denote the number of biallelic markers under consideration, G1 the number of markers with different allele frequencies between cases and controls, and G0 the number with no true difference between cases and controls, i.e. G = G1 + G0. For simplicity's sake a recessive pattern is assumed to influence the likelihood of disease for the informative markers. For each of the G markers the log odds ratio will be computed from an associated 2 × 2 table (table 1).
Table 1.
2 × 2 table for each biallelic marker
| Case | Control | Total | |
|---|---|---|---|
| 2 minor alleles | a | b | a + b |
| 0 or 1 minor alleles | c | d | c + d |
| N | N | 2N |
For the j-th marker let θj denote the log odds ratio and its estimate, log(ad/bc). Then
| (2.1) |
where
and ∼ N(0,1) indicates an approximate Gaussian distribution with mean 0 and variance 1. Given the observed log odds and associated test statistic
| (2.2) |
one can use the relation in (2.1) to obtain p values for the hypothesis that θj = 0 as well as to generate confidence intervals
where 1 – α is confidence associated with the interval and is the associated percentile from a N(0,1) distribution.
To investigate how bias arises it is useful to rewrite the relationship between (2.1) and (2.2). First, write
| (2.3) |
where
| (2.4) |
denote the empirically observed probability of two minor alleles in the case and control populations, respectively. The term is an estimate of σ, which is defined by equation (2.3) after replacing the estimated probabilities of having both minor alleles by the true unknown proportions of having both minor alleles. Then we can decompose the test statistic Tj as
| (2.5) |
| (2.6) |
The decomposition shows the test statistic Tj is composed of a trend term,
and a mean 0 random fluctuation term, . Note that and Tj differ in that has an approximately normal distribution centered about 0 while for Tj this is only true when θj = 0. The term captures the difference between estimated log odds and true log odds and its distribution is of primary interest in assessing overestimation bias.
For each j, has approximately an unconditional N(0,1) distribution. A critical question concerns the distribution of when j corresponds to most extreme observed Tj statistic. We will focus upon the case of largest Tj – the case for smallest, i.e. most negative, statistic is similar and generalizations involving it and other extreme statistics are discussed below.
Let r1,r2, …, rG order the T test statistics where
Typically, the distribution of
| (2.7) |
is not that of a normal distribution with mean 0 – this will be demonstrated with simulations below. The reason for the bias is that if a number of markers have similar sized trend components
then the random components are likely to play a larger role in determining which marker has the maximal observed value, . Under these conditions of multiple markers with similar trend values the contribution of for that with largest value is likely to be positive. From the definition of in (2.7) we see a positive value corresponds to overestimation of the underlying log odds ratio. The expected value of the difference between and is what is meant by ranking bias, i.e.
As discussed further below, it is important to note that rG, the index of the marker with greatest observed effect size, is random and can change in repeated sampling from the relevant populations.
Simulation Studies Demonstrating Bias
To illustrate the ranking bias a simple set of simulations are presented. In this case-control study there are G = 500 markers and there are N = 500 cases and 500 controls. In these simulation studies the markers are generated as independent variables. Populations of cases and controls were generated with 480 of the markers having no differences in allele frequencies. For the remaining 20 markers the odds ratios for diseased relative to healthy were 1.05, 1.10, …, 1.95, and 2.0. The probability of having 2 minor alleles in the control group was randomly chosen for each marker to lie between 0.05 and 0.25 (i.e. the minor allele frequencies varied between 0.224 and 0.5).
Given a simulated sample of cases and controls the following steps were performed:
For each of G = 500 markers find
Because the true log odds ratios (θj) are known, one can calculate a realization of
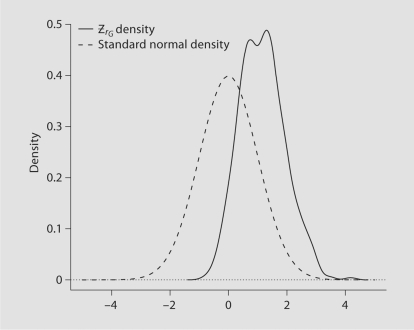
After making many such samples (1000 simulations) the resulting distribution of is shown along with the standard normal distribution in figure 1. Of note are the facts that the distribution is not centered about 0 (indicating typically overestimates ) and that the distribution appears bimodal. Also, the distribution is relatively narrow compared to the standard normal density – this is perhaps surprising given asymptotic efficiency properties of normal approximations. However, the standard normal distribution is not relevant here as it neglects to take into account that rG, the index of the maximal T statistic, is random and changes in different samples drawn from the underlying populations. For example, in the 1000 simulations approximately 34% of the time rG corresponded to that marker with the odds ratio of 2.0, 32% of the time rG was associated with the 1.95 odds ratio, and 8% of the time with that associated with 1.90, and so forth. Because different markers are chosen as the simulated datasets change a mixture distribution arises as suggested by the non-unimodal character of the distribution.
Fig. 1.
Distribution of and a standard normal distributions; maximum odds ratio of 2.0.
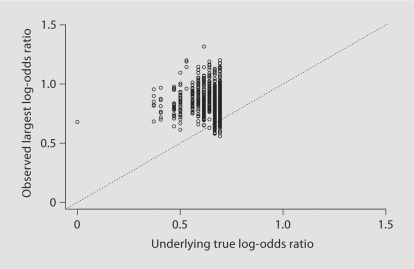
The observed and true log-odds values associated with the largest test statistic (i.e. and ) for each of the 1000 simulations are shown in figure 2. The discrete vertical bands correspond to the 12 distinct values of that were obtained: {0, log(1.45), log(1.50), …, log(2.0) = 0.69}. The degree to which the points lie above the diagonal 45 degree line indicate the amount of bias in each simulation (fig. 2).
Fig. 2.
Comparison of δrG (X-axis) and δˆrG (Y-axis) in 1000 simulations.
Ranking Bias vs. Significance Bias
Thus far there has been no reliance upon a p value threshold as a means for creating the observed bias. Truncation bias related to conditioning on a significant p value (i.e. significance bias) is a common rationale given for overestimation bias in marker studies and is an independent contributing factor. The bias demonstrated thus far is not conditioned on the largest T statistic first exceeding some threshold, e.g. one that corresponds to a p value = 0.025/G. Hence it cannot be significance bias. To clarify the difference significance bias is now further discussed.
Recent work involving significance bias focuses upon the effect in a single marker [4, 7]. In our setting we illustrate the idea using the marker with largest true effect size. Let r0G designate this marker, i.e.
Because r0G is fixed (though unknown) it is the case that for large sample size N
| (4.1) |
and hence However, a bias is introduced if one conditions on exceeding a thres hold value so that the associated p value is sufficiently small. For instance, if then the associated p value will be less than 0.025/500 and in our simulations the marker would be judged showing significant association using a Bonferroni criterion. However, using standard derivations concerning conditional expectations of normally distributed random variables one can show that
| (4.2) |
| (4.3) |
where φ and Φ denote the density and cumulative distribution function of a standard normal distribution. Consequently it follows from (4.1) that
| (4.4) |
| (4.5) |
The approximation arises because has been replaced by and the asymptotic normal distribution was used. From the result in (4.5) one can show how changes in sample size, odds ratio, allele frequency (affecting the σ term), and p value threshold change the significance bias. Such an assessment was performed by Garner [4].
However, in most circumstances r0G is not known, i.e. in a study with a large number of markers and little a priori knowledge of which markers have the strongest effects only rG is observed and the preceding development based upon r0G is not really applicable in practice. What is of practical interest is However analytical evaluation of this expression is more complicated than assessing bias for r0G because rG is random. Consequently, simulations may be used to explore the following types of overestimation bias significance bias related to ranking bias related to and the combination of ranking bias and significance bias related to
Sets of simulations were performed to examine the relative size of biases across varying conditions. Each design has 500 markers with a smaller number of informative markers (G1) ranging between 20 and 1; the results are shown in table 2. Scenario 1 corresponds to the simulations generating figure 1, the maximum true odds ratio is 2 (log odds ratio of 0.69) and the maximum effect size is given by θr0G/σr0G = 0.69/3.465 where 3.465 is the associated σ value derived from the underlying allele probabilities (the probability of 2 minor alleles is 0.1685 in the control group) and odds ratio. An estimate of σ is given by in equation (2.3). From equation (4.5) an analytical estimate of the significance bias applied to this particular marker is 0.072 as shown in the first column of table 2 (all biases in this section are reported on a log odds ratio scale). In the 1000 simulations the observed bias for this marker arising from conditioning on its T statistic exceeding 3.90 was 0.075 – in good agreement with the analytic result. The simulations' estimate of ranking bias related to is about 0.191 in this case – approximately 2.5 times larger than the significance bias of . As an aside it is worth noting that other markers with smaller true effect sizes will have larger significance bias, but smaller likelihood of reaching the threshold. Finally we can get an estimate of the combined effects of ranking bias and significance bias for by examining just those simulations in which TrG > 3.90. In these 1000 simulations TrG > 3.90 in 99% of the occasions so the additional significance bias induced by conditioning on this common event is essentially negligible as it raises the bias only from 0.191 to 0.192 (table 2).
Table 2.
Ranking and significance bias on a log odds ratio scale
| Scenario 1: n = 500, G1 = 20 informative ORs {1.05, …, 2.0} | 0.072 | 0.191 | 0.192 | |||
| Scenario 2: n = 500, G1 = 40 {1.025, …, 2. 0} | 0.072 | 0.230 | 0.230 | |||
| Scenario 3: n = 500, G1 = 5 {1.20, …, 2.0} | 0.072 | 0.113 | 0.129 | |||
| Scenario 4: n = 500, G1 = 1 {2.0} | 0.072 | 0.069 | 0.085 | |||
| Scenario 5: n = 500, G1 = 1 {4.0} | 0.000 | 0.000 | 0.000 | |||
| Scenario 6: n = 1000, G1 = 20 {1.05, …, 2.0} | 0.002 | 0.112 | 0.112 | |||
| Scenario 7: n = 500, G1 = 10 {1.04, …,1.40} | 0.330 | 0.448 | 0.471 | |||
| Scenario 8: n = 500, G1 = 10 {1.30, …, 1.30} | 0.395 | 0.476 | 0.543 |
Next we investigate how results change when the number of informative markers change from the design in Scenario 1. Scenario 2 has a similar structure but has 40 informative markers taking values 1.025, 1.05, …, 1.975, and 2.0. Because the sample size, standard deviations, and maximum odds ratio of 2 stay the same the significance bias associated with remains the same at 0.072. However, the changes in informative effect sizes induce changes in the ranking bias and combined ranking and significance bias associated with . These figures have increased to 0.230 for both bias measures. Conversely, when the of informative markers are reduced to only 5 as in Scenario 3, taking values 1.20, …, 1.80, 2.0, then the two bias measures associated with are reduced to 0.113 and 0.129 though the significance bias associated with remains unchanged. These three scenarios show how bias in depends upon the distribution of all markers, not just the conditions associated with the single marker having the largest true effect size.
In Scenario 4 the more extreme case of a single informative marker with an odds ratio of 2 is considered. Here the ranking bias of is considerably less because there is a relatively low probability that any other marker will be associated with rG – in 92.7% of the simulations the marker with an odds ratio of 2 generated the largest T statistic. Therefore, the significance bias of and are quite comparable as shown in the first and third column. In Scenario 5 the effect size of the single informative marker is increased so that this marker produces the smallest p value in all the simulations so there is essentially no ranking bias. Further, the sample size is sufficiently large so that the associated T statistic will exceed the 3.90 cutoff with an extremely high probability so there is no detected significance bias. Scenario 6 changes Scenario 1 by increasing the sample size. As in Scenario 5 this essentially eliminates the significance bias associated with the marker having an odds ratio of 2. However, there is still considerable bias associated with indicating ranking bias is a more important factor than significance bias in this case.
In Scenario 7 more modest effect sizes are considered. Here there are 10 informative ORs between 1.04 and 1.40. In this case 10,000 simulations were considered as there was low power (7.1%) for any of the 10 markers with informative ORs to exceed the Bonferroni threshold. Because the power is low, one might expect higher significance bias and this is in fact the case. The marker with OR = 1.4 had significance bias of 0.330 – indicating the observed log-odds ratio was typically twice as high as the true effect (log(1.4) = 0.336) when the criterion was met. However, in this case the ranking bias was extensive as well with values of 0.448 (unconditional) and 0.471 (conditional upon a significant statistic). Ranking bias remains high because those conditions that create large significance bias (e.g. a large Bonferroni threshold relative to true effect size) also tend to create large ranking bias (many different markers could potentially be highest ranked, with large bias for any one marker being highest ranked). In general it is difficult to find conditions in which significance bias will appreciably exceed the rank bias – either unconditional or conditional upon the highest ranked test statistic exceeding a threshold. Indeed, as it is usually the case that and one would not expect significance bias, to exceed the conditional ranking bias,
Finally, Scenario 8 shows the case when all the informative markers have the same effect size. As described in the Discussion and Appendix sections one should expect high ranking bias and this is in fact the case.
Among the conclusions to be drawn from these comparisons of different types of bias are (1) significance bias associated with a single, fixed marker may not be particularly relevant; (2) ranking bias is distinct from significance bias; (3) ranking bias is likely the greater of the two sources when many variables are considered. Finally, as shown by comparing the first 4 scenarios in table 2 that have many common design elements, the effect sizes of all markers play a role in determining ranking bias – efforts to understand bias associated with highly ranked markers must take this into account.
Discussion
Appendix A shows that ranking bias (scaled by the marker's standard deviation and sample size) for independent markers is given approximately by
| (5.1) |
where
| (5.2) |
for j in 1, …, G. In words, M–j represents the largest of all Tk (besides Tj) minus the trend term corresponding to the j-th marker. As discussed further in the appendix, some understanding of the bias can be deduced from relation (5.1). First, and perhaps most importantly, it is clear the bias depends in principle on the effect sizes, θj/σj, for all G of the markers. In practice, perhaps a few or even just one marker is relevant, but in principle one needs to take into account the true effect sizes of all.
As the sample size increases then each of the M–j increase toward infinity in absolute value (with probability 1 under common conditions) so that the bias tends to zero as we expect assuming there are some informative markers and one is more informative than the others. On the other hand, bias is considerably worse when the effect sizes, θj/σj, are the same for all j = 1, …, G as would be the case is there were no informative markers. If the markers are independent then in this case
An important related question that underlies equation (5.1) is: given repeated samples of size N from the underlying populations, how many different markers could reasonably be selected as rG? If one marker has an effect size that is much larger than all the rest then in repeated samples we would expect rG to almost always correspond to that marker and no appreciable bias should result – empirically this is seen in Scenario 5 of table 2. Analytically this is demonstrated in the appendix when γ → ∞. If, on the other hand, there are a number of markers that have non-trivial probabilities of producing the largest test statistic in a given sample then in these cases the corresponding terms play a larger role in determining which marker is chosen and we would expect a correspondingly greater degree of bias given the relationship between bias and . One way of thinking about these factors is to see that the distribution of is a mixture of the conditional distributions given rG = j with the weights of the mixture given by the probability that rG = j. These conditional distributions are typically not centered about 0 – in fact they are subject to a kind of truncation bias similar in form to that arising from conditioning on a significant p value, but instead arising from conditioning on rG = j. When there are a number of markers with non-negligible Prob[rG = j] this mixture leads to non-negligible bias for . Appendix A provides technical details.
The results thus far presented only focus upon the marker with largest test statistic. A natural question is how much bias may be present if attention is paid to all markers that exceed a significance threshold. This corresponds to the common strategy of following-up on all markers that meet a stringent significance threshold. For each simulation under Scenario 1 in table 2 the number of markers with T scores > 13.90 was recorded and the difference between the observed δˆ and the true underlying θ was recorded. Table 3 provides information about all highly ranked markers that exceed the threshold. In 99% of the simulations at least one marker met the threshold requirement. The bias associated with the highest ranking marker exceeding the threshold is 0.192 when measured on a log odds scale – this is essentially the same information as reported in the first row of table 2. Again it is worth noting that this is not reporting the results for the marker with true log-odds ratio of 2.0, but rather the average bias for the marker with the highest observed T statistic – in 34% of the simulations this did correspond to the marker with odds ratio of 2.0, in 32% of occasions the marker with odds ratio of 1.95, etc…. The other rows of table 3 provide new information: in 92.5% of the simulations the second highest ranked marker met the Bonferroni criteria and the bias for these instances was estimated as 0.143 log-odds units. Table 3 gives information for the 3rd, 4th, 5th, 6th, and 7th mostly highly ranked markers (no more than 7 markers ever met the criteria) and we see the bias conditional upon exceeding the threshold remains considerable. This table indicates that the ranking bias is not just a problem for the most highly ranked marker – it can be present and substantial for all those markers meeting significance criteria.
Table 3.
Bias of markers exceeding threshold under scenario 1
| Marker rank | ||||
|---|---|---|---|---|
| 1st highest ranked: | 0.990 | 0.192 | ||
| 2nd highest ranked: | 0.925 | 0.143 | ||
| 3rd highest ranked: | 0.741 | 0.124 | ||
| 4th highest ranked: | 0.438 | 0.128 | ||
| 5th highest ranked: | 0.193 | 0.132 | ||
| 6th highest ranked: | 0.054 | 0.131 | ||
| 7th highest ranked: | 0.007 | 0.129 | ||
Conclusions
In this report the focus has been on measuring bias associated with the largest effect size. Ranking bias is potentially present in any study with multiple tests where attention is drawn to outcomes associated with the most extreme observed effect sizes. Here we have seen that focus upon highly ranked results, rather than conditioning on significant p values, can be responsible for much of overestimation bias. Some authors have put forth methods to correct for both ranking and significance bias. The bootstrap/cross-validation approach put forth by Sun and Bull [8] can accommodate both types of bias. A similar bootstrap approach by Jeffries [9, 10] has been employed to correct for ranking bias in microarray and diagnostic modeling contexts. Analytic approaches have been put forth by Zöllner and Pritchard [7], Ghosh et al. [11], and Zhong and Prentice [12] however these appear to address significance bias but not ranking bias. The more extensive dependence of rank bias on potentially all effect sizes (as opposed to significance bias that depends only upon one marker's effect size) complicates analytical solutions. It is likely that analytic approaches to biased overestimation from genomic studies that ignore the need to consider all markers' effect sizes may be missing an important aspect of the problem, i.e. the distribution of the most extreme test statistic is a mixture distribution depending, in principle, upon all markers' parameters.
The modeling in this paper was very simple: a single stage study (i.e. no two-stage or higher stage designs) with a recessive genetic model and Wald test approach to evaluating significance. The simple approach allowed for a simple analytic description of the problem – more complicated designs are likely prone to the same types of biases and work remains for examining the role of ranking bias in these circumstances.
Appendix A
Calculation of the Bias
Here an effort is made to sketch the degree of bias that may be expected and link this magnitude to some factors such as sample size, distribution of true effect sizes, and the number of markers. Simplifying assumptions will be employed as necessary. Recall that the j-th test statistic Tj may be decomposed into a trend and random fluctuation component as
where and is approximated by a standard normal distribution. Then we may calculate the expected value of (and hence obtain a measure of the bias of ) as
| (A.1) |
and
| (A.2) |
| (A.3) |
| (A.4) |
where is the conditional distribution of given rG = j and f (zj,rG = j) describes the joint distribution of and the event rG = j.
| (A.5) |
| (A.6) |
Here we have used the decomposition in (2.5) to show how the bias depends on the random fluctuation terms, and . To simplify we will approximate the sdˆj and sdˆk terms by σj and σk (the associated true population values). Then we obtain
| (A.7) |
| (A.8) |
where
| (A.9) |
and
| (A.10) |
To proceed further we condition the expectation on M–j and recall that is marginally distributed as a standard Gaussian random variable and thus, for independent markers,
| (A.11) |
| (A.12) |
| (A.13) |
fz denotes a Gaussian distribution and the expectation in (A.13) is necessary because M–j contains random elements . The integral may be explicitly rewritten as
| (A.14) |
From (A.14) one sees that bias is inversely related to the absolute value of the M–j terms. Some consequences of this derivation are as follows.
Consider the effect of increasing the sample size holding all else constant. We focus upon the case in which there exists a single true maximum effect size and designate that marker's index by r0G, i.e.
It is worthwhile to examine M–j for the case when j = r0G and j ≠ r0G separately where we assume only one variable (with index r0G) has the most positive effect size, i.e. there are no ties. Then
| (A.15) |
| (A.16) |
| (A.17) |
In either case we have that M2–j → ∞ so from (A.14) one sees
The case for expanding the differences among effect sizes is similar – at least for the simplified example below. For a given pattern of effect sizes among the G variables (again with no ties for the largest effect size), consider a new pattern of effect sizes given by multiplying each original effect by a constant γ > 0. Then if r0G designates the marker with the most positive true effect size
| (A.18) |
| (A.19) |
| (A.20) |
Consequently the same conclusion of no bias follows. If one reverses the limiting action of γ so that γ → 0 from above then
| (A.21) |
where the are standard normal statistics and the bias is then positive. This situation corresponds to the situation of no variables showing differential expression.
The case for increasing G, the number of variables is less clear cut as it depends upon the combination of effect sizes. Empirically it seems that adding variables with effect sizes at or near the size of the largest preexisting effect sizes exaggerates the bias effects for θrG In terms of figuring the change of M–j terms as above there is more ambiguity as some terms M2−j terms will likely increase, others decrease, and some new terms will be introduced.
References
- 1.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 2.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Editorial Freely associating. Nat Genet. 1999;22:1–2. doi: 10.1038/8702. [DOI] [PubMed] [Google Scholar]
- 4.Garner C. Upward bias in odds ratio estimates from genome-wide association studies. Genet Epidemiol. 2007;31:288–295. doi: 10.1002/gepi.20209. [DOI] [PubMed] [Google Scholar]
- 5.Allison D, Fernandez JR, Heo M, Zhu S, Etzel C, Beasley TM, Amos CI. Bias in estimates of quantitative-trait locus effect in genome scans: demonstration of the phenomenon and a methodof-moments procedure for reducing bias. Am J Hum Genet. 2002;70:575–585. doi: 10.1086/339273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Göring H, Terwilliger JD, Blanger J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet. 2001;69:1357–1363. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zöllner S, Pritchard JK. Overcoming the winner's curse: estimating penetrance parameters from casecontrol data. Am J Hum Genet. 2007;80:605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun L, Bull SB. Reduction of selection bias in genomewide studies by resampling. Genet Epidemiol. 2005;28:352–367. doi: 10.1002/gepi.20068. [DOI] [PubMed] [Google Scholar]
- 9.Jeffries N. Multiple comparisons distortions of parameter estimates. Biostatistics. 2007;8:500–504. doi: 10.1093/biostatistics/kxl025. [DOI] [PubMed] [Google Scholar]
- 10.Jeffries N. Performance of a genetic algorithm for mass spectrometry proteomics. BMC Bioinformatics. 2004;5:180. doi: 10.1186/1471-2105-5-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh A, Zou F, Wright FA. Estimating odds ratios in genome scans: an approximate conditional likelihood approach. Am J Hum Genet. 2008;82:1064–1074. doi: 10.1016/j.ajhg.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong H, Prentice RL: Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics 2008, in press. [DOI] [PMC free article] [PubMed]