Abstract
Objective(s)
An individual's risk of developing cardiovascular disease (CVD) is influenced by genetic factors. This study focussed on mapping genetic loci for CVD-risk traits in a unique population isolate derived from Norfolk Island.
Methods
This investigation focussed on 377 individuals descended from the population founders. Principal component analysis was used to extract orthogonal components from 11 cardiovascular risk traits. Multipoint variance component methods were used to assess genome-wide linkage using SOLAR to the derived factors. A total of 285 of the 377 related individuals were informative for linkage analysis.
Results
A total of 4 principal components accounting for 83% of the total variance were derived. Principal component 1 was loaded with body size indicators; principal component 2 with body size, cholesterol and triglyceride levels; principal component 3 with the blood pressures; and principal component 4 with LDL-cholesterol and total cholesterol levels. Suggestive evidence of linkage for principal component 2 (h2 = 0.35) was observed on chromosome 5q35 (LOD = 1.85; p = 0.0008). While peak regions on chromosome 10p11.2 (LOD = 1.27; p = 0.005) and 12q13 (LOD = 1.63; p = 0.003) were observed to segregate with principal components 1 (h2 = 0.33) and 4 (h2 = 0.42), respectively. Conclusion(s): This study investigated a number of CVD risk traits in a unique isolated population. Findings support the clustering of CVD risk traits and provide interesting evidence of a region on chromosome 5q35 segregating with weight, waist circumference, HDL-c and total triglyceride levels.
Key Words: Norfolk Island, Population isolate, Principal component, Linkage analysis, 5q35, CVD
Introduction
Cardiovascular disease is a leading cause of morbidity and mortality world-wide. The World Health Organisation reported a total of 16.7 million deaths globally in 2002 to be a direct result of CVD [1]. An individual's risk of developing CVD is influenced by multiple environmental and genetic factors. Major risk factors include tobacco use, physical inactivity, unhealthy diet, obesity, dyslipidemia, hypertension, diabetes mellitus and the metabolic syndrome. Combinations of these risk factors along with a positive family history significantly increase the likelihood of disease and its related effects on morbidity and mortality [2].
Genome-wide studies of cardiovascular-related phenotypes report linkage to various chromosomal regions, indicating that this disorder is genetically heterogeneous. To simplify dimensions of CVD risk, multivariate data reduction techniques such as factor and principal component analysis (PCA) have been employed to extract uncorrelated components from numerous inter-correlated phenotypes [3, 4]. This method has been used to identify loci linked to the clustering of CVD indicators, particularly those comprising the metabolic syndrome [5,6,7,8]. Of particular relevance is the use of a genetic isolate derived from Kosrae [5]. Isolated populations, like that of Kosrae, offer several advantages in gene mapping studies compared to outbred populations, with extreme geographical and cultural isolation reducing the effects of non-genetic variables by promoting a uniform lifestyle [9]. Genetic heterogeneity may also be reduced if the isolate is derived from a limited number of ancestors and has undergone population bottlenecks and endogamy during population expansion.
In the present study we tested the descendents of HMS Bounty mutineers and Tahitian population founders derived from the Norfolk genetic isolate [10]. Prior demographic analysis indicates this population possesses unique characteristics which may facilitate gene mapping studies of complex multifactorial diseases such as CVD [3]. Quantitative epidemiological data was available for related individuals on body size, blood pressure and lipid levels, all factors which contribute to the risk of CVD. To examine the relationship of these indicators, principal component analysis was performed to extract orthogonal components. Linear scores were calculated for each individual and used to determine the heritability of each factor in the population cohort. Quantitative trait loci segregating with individual factors were then identified by multipoint variance components linkage analysis using a genome-wide panel of STRs.
Methods
Sample Ascertainment
Norfolk is a small, isolated volcanic island situated in the South Pacific Ocean, approximately 1,700 kilometres northeast of Sydney. In 2001, the Island's permanent population totalled 1574 individuals of whom 756 claimed to be of Pitcairn decent [11]. The population supports itself from local produce, however as a result of both isolation and small land mass the population is highly dependent on imports of primary produce and manufactured goods [11]. The Islanders live a relatively homogeneous lifestyle due to their isolation, strict quarantine and immigration laws and community centred culture. Furthermore, a large proportion of the adult population are descended from 9 Isle of Man HMS Bounty mutineers, including acting lieutenant Fletcher Christian and 12 Tahitian women who relocated to Norfolk Island from Pitcairn Island in the 1850s [10].
Collection and phenotypic characterisation of the Norfolk Island cohort has been previously described in detail [3, 12]. Briefly, ethical clearance was granted prior to the commencement of the study by the Griffith University Human Research Ethics Committee. Study participants over 18 years of age were recruited via local media announcements. All participants signed informed consent statements prior to inclusion in the study. A detailed questionnaire was used to obtain specific information from study participants including ancestry, lifestyle habits and extensive medical history. Participants were extensively phenotyped for anthropometric measures, blood pressure, lipids, lipoproteins and blood chemistry. Blood samples were not taken after fasting at the time of collection. Responses to medical questionnaires indicated that 40 individuals reported anti-hypertensive use and 22 individuals reported use of lipid lowering therapy at the time of recruitment. Systolic and diastolic blood pressure values were corrected for anti-hypertensive use in accordance with the methods described by Tobin et al. (2005).
Pedigree Structure
A total of 377 individuals were determined to have familial links to the Tahitian (Polynesian) and HMS Bounty mutineer founders and were thus the focus of this investigation. These related individuals make up part of the current Norfolk pedigree (n = 6537) that extends through 11 generations to the original population founders [3]. To alleviate analysis burden imparted by the presence of multiple inbreeding and marriage loops in early generations and the large volume of missing data, the pedigree was trimmed (n = 978) using a peeling algorithm in the pedigree database management system PEDSYS [13]. A total of 285 genotyped and phenotyped individuals comprised the trimmed pedigree structure; the remaining 92 individuals became disjoint and though not included in the linkage analysis results, were retained for principal component analysis and covariate screening.
Norfolk Island represents an admixed population of Caucasian (Isle of Man) and Polynesian (Tahitian) ancestry, which had expanded 11 generations since at the time of collection. Recent admixture is an important consideration in genetic studies. Depending on the ethnic origin, trait alleles can display markedly different frequencies and may confer varied risks in the case of disease. The haplotype surrounding the trait (or disease) allele may extend longer distances in admixed populations, leading to extended regions of linkage disequilibrium (LD) between loci. The presence of long range LD means that lower density STR maps should be sufficient to identify regions segregating with traits of interest. Due to these facts, admixture mapping has been used to localise loci for numerous disorders, including multiple sclerosis, prostate cancer and hypertension in populations with African American ancestry [14,15,16]. Though the extent of admixture is yet to be characterised in the Norfolk population, the extent of LD across a well-characterised region of the X chromosome and also between variants in the NOS2A gene on chromosome 17q11.2-q12 has been previously assessed [12]. This study revealed LD to extend approximately 9.5–11.5Mb across the Xq13.3 region in the Norfolk Island population, a result comparable to those reported in other known population isolates [17].
Genome-Wide Scan
DNA was isolated from lymphocytes using a standard salting out procedure [18]. Samples were genotyped at the Australian Genome Research Facility (AGRF) using the Applied Biosystems PRISM Human Linkage Mapping Set version 2.5. The linkage mapping set comprised of 382 highly polymorphic dinucleotide microsatellite markers, spaced at an average distance of 10 cM throughout the human genome. Markers were individually amplified by PCR using fluorescently labelled primer pairs. Markers were then pooled into panels for capillary separation on the ABI3730 DNA Analyser. Genotyping results were analyzed using ABI GeneMapper software version 4.0.
Statistical Methods
Data was screened using SPSS version 14.0. Measurements greater than or equal to 4 standard deviations from the mean were assessed and any data entry errors or extreme outliers were excluded. Components with a high kurtosis were log transformed. Only subjects with measurements for all phenotypes were included in the principal component analysis. Differences in the sex-specific means of the quantitative phenotypes were investigated by one-way analysis of variance (ANOVA).
Principal components analysis was used to extract orthogonal components from cardiovascular and obesity related measurements. Obesity related traits included body mass index (BMI) calculated as kg/m2, hip circumference, waist circumference, percentage body fat and weight. Cardiovascular related traits included blood pressures (systolic and diastolic), lipids (total cholesterol and triglycerides) and HDL-cholesterol and LDL-cholesterol levels. The initial solution, principal component 1 explained the maximum variance, while successive components explained progressively smaller portions of the total variance. Principal components were simplified by orthogonal rotation (varimax). This minimised the number of variables with high loadings on each component. Principal components with eigenvalues greater or equal to 1 were retained (principal components with variances less than 1 contain less information than one of the original variables and hence are not worth retaining). Relationships between components are explained by factor loadings, values greater than or equal to 0.4 were used to indicate meaningful correlations between the component and the variable. PCA has 4 statistical assumptions; (1) random, independent sampling; (2) interval level measurements; (3) linearity, and (4) normality. In order to assess the violation of random, independent sampling, PCA component matrices were calculated 100 times for n = 75 randomly selected individuals (20% of the total cohort). Coefficients of congruence were computed for the entire dataset and each random sample [19].
A regression method was used to estimate factor score coefficients for the retained principal components and formed the basis of linkage phenotypes for each individual. Factor scores had a mean of 0 and a standard deviation of 1, skewness and kurtosis were less than 0.6, satisfying the assumption of normality. The 4 factor scores were screened for the covariate effects of age, sex, age2 and their interactions, prior to calculating heritability estimates using SOLAR.
Genotypic data was analysed for discrepancies, including mendelian inheritance violations using the PEDSYS program INFER and Simwalk2 [13, 20]. The Pedigree RElationship Statistical Test (PREST) was used to verify the pedigree structure and detect relationship misspecification [21]. Discrepant genotypes were blanked prior to analysis. Identity-by descent (IBD) matrices were computed using the program Loki at points throughout the genome for every relative pair [22]. Chromosomal marker maps were sex-averaged and obtained from the Marshfield Centre for Medical Genetics (http://research.marshfieldclinic.org/genetics). Multipoint variance component linkage methods were used to assess linkage between the 382 autosomal markers and quantitative phenotypes using the statistical program SOLAR 4.0.6 [23]. Additionally, each quantitative phenotype was simulated under the null hypothesis of no linkage. In this process, a fully-informative marker, unlinked to the trait, was simulated and trait linkage was tested at that marker 10,000 times for each quantitative linkage phenotype. This information was used to calculate empirical p values for LOD scores. For LOD score analysis in human pedigrees it has been proposed that a point-wise p value of 4.9 × 10–5 (LOD ∼ 3.3) is indicative of significant linkage, while a point-wise p value of 1.7 × 10–3 (LOD ∼ 1.9) is suggestive at the genome-wide level [39]. As these values do not take into account the effects of multiple independent tests, a Bonferroni correction was applied to adjust the significance thresholds for this study. Adjusted values of p = 4.3 × 10–4 for suggestive linkage and p = 1.2 × 10–5 for significant linkage were used to interpret results.
Results
Table 1 displays the population and sex-specific means and standard deviations of the original measurements used in the PCA for related Norfolk Island Individuals. The cohort used in this study consisted of 171 men and 206 women. The mean age of both male and female were 48.9 and 49.5 years, respectively, with little deviation in the variance between genders (p > 0.05). The remaining 11 quantitative phenotypes indicated significant (p < 0.05) sex-specific differences in trait variance. Males were observed to have significantly higher values pertaining to hip and waist circumference, BMI, weight, total triglycerides, total cholesterol, LDL-cholesterol and diastolic and systolic blood pressures compared to females. Females had significantly higher values of percentage body fat and HDL-cholesterol levels than men.
Table 1.
Phenotypic characteristics of participants (mean ± SD)
| Trait | Both sexes (n = 377) | Males (n = 171) | Females (n = 206) |
|---|---|---|---|
| Age, years | 49.2±16.6 | 48.9±16.9 | 49.5±16.4 |
| BMI, kg/m2 | 26.3±4.6 | 27.5±4.1 | 25.4±4.7 |
| DBP, mm Hg | 75.9±13.3 | 79.3±12.6 | 73.0±13.2 |
| DBP with adjustment, mm Hg | 76.8±13.6 | 80.5±13.9 | 73.6±12.6 |
| HDLc, mmol/l | 1.4±0.4 | 1.3±0.3 | 1.5±0.4 |
| Hip circumference, cm | 101.9±9.6 | 103.3±7.3 | 100.9±11.0 |
| LDLc, mmol/l | 2.8±1.0 | 3.0±0.9 | 2.7±1.0 |
| Percent body fat, % | 30.3±8.7 | 24.4±6.6 | 35.3±6.9 |
| SBP, mm Hg | 127.4±22.3 | 134.0±19.6 | 121.8±22.9 |
| SBP with adjustment, mm Hg | 129.2±24.0 | 135.7±21.2 | 123.7±24.9 |
| TC, mmol/l | 5.6±1.1 | 5.8±1.1 | 5.5±1.1 |
| TG, mmol/l | 2.0±1.2 | 2.3±1.3 | 1.7±1.1 |
| Waist circumference, cm | 87.3±13.7 | 95.1±11.1 | 81.2±12.4 |
| Weight, kg | 76.0±16.2 | 85.5±14.6 | 68.3±13.0 |
BMI = Body Mass Index; DBP = Diastolic Blood Pressure; HCLc = High Density Lipoprotein cholesterol; LDLc = Low Density Lipoprotein Cholesterol; SBP = Systolic Blood Pressure; TC = Total Cholesterol; TG = Total Triglycerides.
Analysis of BMI revealed Norfolk Islanders, particularly males were on average slightly overweight. The accepted ranges for BMI were as follows; values of 25–29 kg/m2 were classified as overweight, 30–34 kg/m2 as obese and 35 or above morbidly obese. Of particular interest to CVD, trait break down of BMI indicated that 39.3% of adults were overweight, 16.1% were obese and 3.2% were morbidly obese. Women had a higher percentage of body fat than men, however this difference is expected.
Waist circumference is an indicator of abdominal obesity, which according to the World Health Organisation (WHO) increases the risk of CVD in males and females with measurements exceeding 101 and 89 cm respectively [1]. The average waist circumference of males (95.1 cm) and females (81.2 cm) was within the recommended range. According to World Health Organisation guidelines 24.6% of males and 22.6% of females had values of waist circumference exceeding the recommended range and therefore at an increased risk of developing CVD.
The average systolic (127.4) and diastolic (75.9) blood pressure, a strong indicator of hypertension and CVD, was within expected normal limits. Analysis of blood pressure revealed 27.7% of related individuals to be hypertensive based on a systolic value ≥140 mm Hg and/or a diastolic reading ≥90 mm Hg. A total of 40 individuals reported using anti-hypertensive medications at the time of collection. As medication affects blood pressure measurements and may impact upon the reliability of the resulting test statistics, both diastolic blood pressure (DBP) and systolic blood pressure (SBP) measurements were adjusted using a correction devised by Tobin et al. (2005). This required adding 15 and 10 mm Hg to SBP and DBP scores for those individuals using antihypertensive medications, respectively [24]. The original and adjusted SBP and DBP are reported in table 1. A slight increase (29.6%) in the number of hypertensive individuals was observed when this correction was taken into account.
Lipid analysis was compared to published WHO European guidelines [1]. Mean total cholesterol (5.6 mmol/l) exceeded the WHO European recommendation of less than 5.0 mmol/l. Mean total triglycerides (2.0 mmol/l) exceeded the recommended less than 1.7 mmol/l. Mean LDL-cholesterol levels (2.8 mmol/l) were in the healthy range of less than 3.0 mmol/l. While HDL-cholesterol levels in men (1.3 mmol/l) and women (1.5 mmol/l) were within the recommended limits (equal or greater than 1.0 mmol/l in men and 1.2 mmol/l in females).
Bivariate Pearson's correlation coefficients and significance levels between the 11 variables are shown in table 2. Examination of the component matrix indicated that there was a high degree of correlation among the variables. In total, 41 pairs of variables were significant below p = 0.001 level. The high level of correlations between the 11 variables supported the use of PCA.
Table 2.
Pearson's correlation coefficient matrix of the traits after normalisation and blood pressure adjustments
| BMI | DBP adjusted | HDLc | Hip circumference | LDLc | Percent bodyfat | SBP adjusted | TC | TG∗ | Waistcir-cumference | Weight | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BMI | 1.00 | ||||||||||
| DBPadjusted | 0.43d | 1.00 | |||||||||
| HDLc | –0.36d | –0.11 | 1.00 | ||||||||
| Hip circumference | 0.86d | 0.36 d | –0.23d | 1.00 | |||||||
| LDLc | 0.20d | 0.29 d | –0.06 | 0.18c | 1.00 | ||||||
| Percent body fat | 0.48 d | 0.13 b | 0.06 | 0.50 d | 0.03 | 1.00 | |||||
| SBP adjusted | 0.44d | 0.76 d | –0.14b | 0.37d | 0.26d | 0.06 | 1.00 | ||||
| TC | 0.26d | 0.35 d | 0.04 | 0.23 d | 0.92d | 0.11a | 0.35d | 1.00 | |||
| TG∗ | 0.44 d | 0.34 d | –0.48d | 0.33d | 0.25d | 0.12a | 0.38d | 0.41d | 1.00 | ||
| Waistcircumference | 0.81d | 0.43 d | –0.39d | 0.77d | 0.26d | 0.17b | 0.48d | 0.31d | 0.46d | 1.00 | |
| Weight | 0.84d | 0.39 d | –0.42d | 0.75d | 0.15b | 0.14b | 0.36d | 0.18b | 0.38d | 0.82d | 1.00 |
Log transformed
p < 0.05
p < 0.01
p < 0.001
p < 0.0001.
PCA extracted 4 components, which explained nearly 83% of the total variation of the 11 original quantitative traits (table 3). Principal component 1 had high loadings of traits that reflect body size, particularly adiposity (hip circumference, BMI, Percentage body fat, weight and waist circumference) and explained the largest portion of the total variance (44%). Principal component 1 is a strong indicator of atherosclerosis. Principal Component 2 was loaded predominantly with HDL-cholesterol, total triglycerides, weight and waist circumference, all of which are associated with obesity and atherogenic dyslipidemia. Comparably, principal component 3 contained high loadings of blood pressure, reflecting the risk of essential hypertension and lastly, principal component 4 was loaded with LDL-cholesterol and total cholesterol, both strong indicator of ischemic stroke and heart attack risk.
Table 3.
Coefficients and variances of factors satisfying the eigen-value ≥1 criterion
| Variable | Principal component |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| BMI | 0.83 | 0.39 | 0.28 | 0.09 |
| DBPadjusted | 0.18 | 0.07 | 0.89 | 0.17 |
| HDLc | –0.05 | –0.88 | 0.03 | 0.05 |
| Hip circumference | 0.87 | 0.26 | 0.22 | 0.08 |
| LDLc | 0.05 | 0.08 | 0.11 | 0.95 |
| Percent body fat | 0.78 | –0.31 | –0.08 | 0.07 |
| SBP adjusted | 0.14 | 0.14 | 0.90 | 0.15 |
| TC | 0.11 | 0.05 | 0.20 | 0.96 |
| TG∗ | 0.15 | 0.63 | 0.23 | 0.33 |
| Waistcircumference | 0.61 | 0.54 | 0.35 | 0.14 |
| Weight | 0.63 | 0.57 | 0.27 | 0.01 |
| Eigenvalue | 4.88 | 1.86 | 1.29 | 1.09 |
| Totalvariance, % | 44.35 | 16.87 | 11.73 | 9.92 |
| Accumulative variance, % | 44.35 | 61.21 | 72.94 | 82.86 |
Log transformed. Factor loadings in bold type are > 0.4.
To address the violation of independent random sampling, 20% (n = 75) of the cohort was randomly sampled 100 times. The first 4 principal components were assessed using the coefficient of congruence [19]. This coefficient is used to compare the factor loadings of two separate groups by measuring the cosine of the angle between the two vectors. As the coefficient approaches 1, vector similarity increases. For the current study, the mean coefficient of congruence between the entire sample and the 100 random samples were as follows; 0.99 (95% CI = 0.99–1.00) for principal component 1; 0.91 (95% CI = 0.89–0.94) for principal component 2; 0.72 (95% CI = 0.66–0.77) for principal component 3; and 0.75 (95% CI = 0.70–0.79) for principal component 4. These results indicate that the structures of principal component 1 and 2 were very close to the structure of the whole data, while principal component 3 and 4 were a poorer fit. The first 4 derived components for the 100 random samples accounted for 83.82% (95% CI = 83.50–84.14) of the total trait variance. The total variance of each individual component were; 45.07% (95% CI = 44.32–45.82) for principal component 1; 17.08% (95% CI = 16.68–17.48) for principal component 2; 12.19% (95% CI = 11.95–12.42) for principal component 3; and 9.48% (95% CI = 9.26–9.70) for principal component 4.
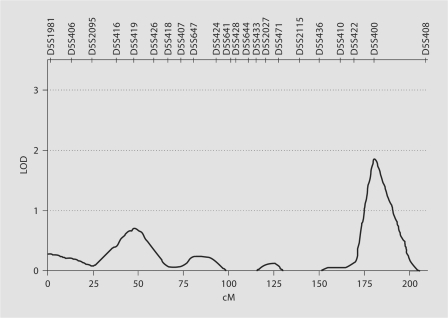
After assessing the coefficient of congruence we performed heritability screening for the 4 components. Three of the four components were significantly heritable after covariate correction. The heritabilities of principal components 1, 2 and 4 were 0.33 ± 0.14 (p = 0.007), 0.35 ± 0.17 (p = 0.02) and 0.42 ± 0.12 (p = 0.0002). Principal component 3 was not significantly heritable (h2 = 0.12 ± 0.19, p = 0.14). Table 4 details the PCA phenotypes and markers that produced maximum LOD scores in regions throughout the genome. The highest LOD score detected in this study was linked to principal component 2. Suggestive linkage for this phenotype was observed on chromosome 5q35 with a maximum LOD score of 1.85 for marker D5S400 (p = 0.0008) (fig. 1). The 1-LOD support interval extended from 174 to 193 cM. Principal component 2 was observed to segregate with additional regions on chromosomes 1, 7, 14, 15, 17, 18 and 20 (p < 0.01, LOD >1). Analysis of principal components 1 and 4 identified peak linkage signals on chromosomes 10 (LOD = 1.27; p = 0.005) and 12 (LOD = 1.63; p = 0.003), respectively. Additional linkage peaks for principal component 1 were localised to chromosomes 1, 7, 12, 17 and 20, while only one other peak was observed on chromosome 21 for principal component 3.
Table 4.
Summary of PCA genome scan results∗
| Principal component | Chromosome | cM | Nearest marker (cM) | LOD | Empirical p value |
|---|---|---|---|---|---|
| 1 | 1 | 237 | D1S213 (232.69) | 1.10 | 0.008 |
| 7 | 30 | D7S507 (32.20) | 1.22 | 0.006 | |
| 10 | 67 | D10S208 (61.08) | 1.27 | 0.005 | |
| 12 | 74 | D12S83 (75.06) | 1.02 | 0.01 | |
| 17 | 56 | D17S798 (56.73) | 1.00 | 0.01 | |
| 20 | 24 | D20S115 (25.13) | 1.19 | 0.006 | |
| 2 | 1 | 46 | D1S234 (45.44) | 1.11 | 0.01 |
| 5 | 181 | D5S400 (180.40) | 1.85 | 0.0008 | |
| 7 | 56 | D7S484 (55.58) | 1.01 | 0.01 | |
| 14 | 113 | D14S985 (118.30) | 1.11 | 0.01 | |
| 15 | 15 | D15S1002 (15.51 | 1.09 | 0.01 | |
| 17 | 76 | D17S1868 (75.89) | 1.20 | 0.008 | |
| 18 | 79 | D18S474 (74.33) | 1.02 | 0.01 | |
| 20 | 3 | D20S117 (2.94) | 1.03 | 0.01 | |
| 4 | 12 | 65 | D12S368(67.52) | 1.63 | 0.003 |
| 21 | 17 | D21S1256(13.20) | 1.10 | 0.01 | |
Only chromosomal regions producing LOD Scores ≥1 are displayed.
Fig. 1.
Multipoint linkage results for principal component 2 on chromosome 5.
Discussion
Isolates, such as Norfolk are unique populations to study complex multi-factorial disorders. The combination of geographical and cultural isolation leads to individuals sharing a common environment, minimising differences in lifestyle factors such as diet, exercise and sanitation compared to out bred populations. This has been observed particularly in Amish and Hutterite populations [25, 26]. The homogeneous environment shared by individuals is of great significance in studies of complex disorders, particularly those of cardiovascular origin where there appears to be a threshold effect influenced by lifestyle factors. Norfolk is also of interest in genetic studies as a large number of individuals in this population can trace their heritage back to a small number of families derived from the original Bounty mutineer and Polynesian founders. The limited number of ancestors minimizes genetic heterogeneity. This can reduce the number of susceptibility genes underlying disease. As a result of the expected reduction in genetic and non-genetic variables, population isolates with known founder effect have been exploited in numerous gene mapping studies of complex disorders [25,26,27,28,29,30,31].
This current study focussed on a large complex family from the Norfolk Isolate to dissect the genetic and environmental variables underlying CVD risk. We performed PCA with orthogonal rotation to reduce 11 inter-correlated variables into groups of independent (un-correlated) components. This data reduction method identified principal components that explained 83% of the variation in the original quantitative traits. PCA identified four distinct components underlying CVD risk in this family. The component accounting for the largest portion of variation was strongly loaded by variables relating to obesity, which has long been known to be an independent predictor of CVD-related morbidity and mortality [32]. The second largest component, reflected traits of obesity loaded with HDL-cholesterol and triglyceride levels. Increased waist circumference, elevated total triglycerides and reduced HDL-cholesterol are 3 indicators of metabolic syndrome and a strong predictor of CVD risk [33]. Principal component 3 reflected the risk of essential hypertension. Lastly, principal component 4 was composed of serum LDL-cholesterol and total cholesterol levels, which when elevated promote arteriosclerosis, increasing the risk of chronic heart disease (CHD), ischemic stroke and heart attack [34].
Previous PCA studies have focused primarily on the metabolic syndrome, a major risk factor for CVD characterised by the clustering of traits of insulin resistance, hypertension, dyslipidemia and obesity [5,6,7,8, 31, 35]. Unfortunately, as values of serum levels of fasting glucose and fasting insulin were not available for our study participants, insulin resistance could not be directly assessed. However, our findings are consistent with other studies as obesity and lipid levels displayed high loadings in the derived components [3,4,5,6,7, 36]. In particular, the loading patterns of weight and waist circumference in both the first and second components suggest a close relationship between traits of obesity and CVD risk in Norfolk Islanders. This is supported by demographic studies, which report a higher prevalence of obesity and dyslipidemia in individuals of Polynesian ancestry [37, 38].
This study is an extension of an epidemiological cardiovascular study of Norfolk Islanders, which reported heritability estimates for eight of the eleven phenotypes presented in the current study [3]. Heritability estimates in the original paper ranged from 0.19 for DBP to 0.63 for LDL-cholesterol. In the current study we detected a substantial genetic contribution for 3 of the 4 components, with estimates ranging from 0.27 to 0.43. The third component, which reflected blood pressure, was not significantly heritable. Heritability estimates for the 4 components presented in this study are within the range of those reported by Bellis et al (2005). In the previous study, DBP was not significantly heritable (h2 = 0.19, p = 0.06). Likewise in this study, the blood pressure component was not significantly heritable.
The genetic predisposition of these phenotypes was investigated by means of non-parametric variance component linkage analysis. The most significant linkage signal in this study was observed on chromosome 5q35. This locus was linked to the second component comprising weight, waist circumference, HDL-cholesterol and total triglyceride levels. A LOD score of 1.85 (p = 0.0008) was detected at marker D5S400. This locus was suggestive of linkage at the genome-wide threshold (p = 1.7 × 10–3). However, after adjustment for the effects of multiple testing across the four principle components, the 5q35 locus was no longer suggestive of linkage. However, this linkage peak is of potential interest as it has been reported with a range of traits related to CVD risk.
Linkage analysis in founder isolates of Kosraen and Amish origin have reported the 5q35 locus to segregate with serum leptin levels and BMI, respectively [31, 40]. Additional family studies have reported segregation with traits of BMI, HDL-cholesterol levels, LDL-cholesterol levels, serum leptin levels, lean mass, body fat measures and type II diabetes mellitus within a 10 cM interval of D5S400 [8,41,42,43,44,45,46,47]. Linkage results support this chromosome 5 locus to be strongly influenced by traits of obesity. Interestingly, the 5q34-qter chromosomal region contains numerous obesity gene candidates. One such gene, the Beta-2-Adrenergic Receptor (ADRB2; OMIM 109690) spanning 5q32–34, a member of the G protein-coupled receptor superfamily, functions as a major lypolytic receptor in human adipocytes [48]. Variations of this gene have been reportedly associated with nocturnal asthma, arteriosclerosis, hypertension, obesity, metabolic syndrome and type 2 diabetes [49,50,51,52,53,54]. However, despite the number of positive associations there appears to be inconsistencies between studies, for instance a candidate gene study conducted in 7,808 middle-aged white subjects was unable to demonstrate any consistent associations between ADRB2 variants and obesity, hypertension or type 2 diabetes [55].
In addition to the 5q35 locus, we identified peak LOD scores linked to principal components 1 and 4 on chromosome 10p11.2 near marker D10S208 and 12q13 near marker D12S368, respectively. Though neither suggestive nor significant at the genome-wide threshold for linkage, these two regions have been reported to segregate with CVD-related risk traits. The chromosome 10 marker, D10S208 has been reported to be linked with obesity in 2 large studies in European and African Americans [56, 57]. The second strongest signal in this study (LOD = 1.63; p = 0.003) observed on chromosome12q13. The component segregating with this locus was loaded with LDL-cholesterol and total cholesterol and is in the same vicinity of a region reported to segregate with LDL-cholesterol and LDL-apoB levels [58]. In addition to these peak regions for the 3 heritable principal components, linkage peaks (LOD >1; p < 0.01) were identified on chromosomes 1, 7, 12, 14, 15, 17, 18, 20 and 21. Though these genetic loci were neither suggestive nor significant at the genome-wide threshold for linkage, their presence supports the underlying heterogeneous nature of cardiovascular-related phenotypes.
In conclusion, principal component analysis reduced 11 interrelated CVD risk traits to 4 newly defined components. As these components are uncorrelated, each one can be interpreted to represent a distinct phenotype underlying CVD risk in Norfolk Islanders. Heritability screening indicated that these components are influenced by a strong genetic element in this family. Linkage analysis identified a suggestive locus underlying CVD risk on chromosome 5, which has been reported to segregate with similar phenotypes. Our findings support the clustering of CVD risk factors in Norfolk Islanders and report a potentially interesting latent cardiovascular risk variable loaded with obesity and lipid levels on chromosome 5q35. Further investigations are warranted in this population to characterise the nature and involvement of this locus in CVD risk.
Acknowledgements
This research was supported by funding from the National Health and Medical Research Council (NHMRC) of Australia, from a Medical Bioinformatics Genomics Proteomics Program (MGBPP) grant and the National Heart Foundation (NHF). Also Hannah Cox was supported by a NHMRC Biomedical Postgraduate Scholarship. Genotyping was performed by the Australian Genome Research Facility (AGRF). The SOLAR statistical genetics computer package is supported by a grant from the US National Institute of Mental Health (MH059490). We would like to acknowledge the Research Computing Services team at Griffith University, particularly Paul Jardine for providing support on Griffith's Sun Solaris HPC cluster. Lastly, our appreciation to the Norfolk Islanders who volunteered for this study.
References
- 1.Mackay J, Mensah GA. The Atlas of Heart Disease and Stroke. Geneva: WHO; 2004. [Google Scholar]
- 2.Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults Executive summary of the third report of the national cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 3.Bellis C, Hughes RM, Begley KN, Quinlan S, Lea RA, Heath SC, Blangero J, Griffiths LR. Phenotypical characterisation of the isolated Norfolk Island population focusing on epidemiological indicators of cardiovascular disease. Hum Hered. 2005;60:211–219. doi: 10.1159/000090545. [DOI] [PubMed] [Google Scholar]
- 4.Goodman E, Dolan LM, Morrison JA, Daniels SR. Factor analysis of clustered cardiovascular risks in adolescence: obesity is the predominant correlate of risk among youth. Circulation. 2005;111:1970–1977. doi: 10.1161/01.CIR.0000161957.34198.2B. [DOI] [PubMed] [Google Scholar]
- 5.Shmulewitz D, Auerbach SB, Lehner T, Blundell ML, Winick JD, Youngman LD, Skilling V, Heath SC, Ott J, Stoffel M, Breslow JL, Friedman JM. Epidemiology and factor analysis of obesity, type II diabetes, hypertension, and dyslipidemia (syndrome X) on the island of Kosrae, Federated States of Micronesia. Hum Hered. 2001;51:8–19. doi: 10.1159/000022953. [DOI] [PubMed] [Google Scholar]
- 6.Cai G, Cole S, Freeland-Graves JH, Maccluer JW, Blangero J, Comuzzie AG. Principal component for metabolic syndrome risk maps to chromosome 4p in Mexican Americans: The San Antonio Family Heart Study. Hum Biol. 2004;76:651–655. doi: 10.1353/hub.2005.0001. [DOI] [PubMed] [Google Scholar]
- 7.Tang W, Miller MB, Rich SS, North KE, Pankow JS, Borecki IB, Myers RH, Hopkins PN, Leppert M, Arnett DK. Linkage analysis of a composite factor for the multiple metabolic syndrome: the National Heart, Lung, and Blood Institute Family Heart Study. Diabetes. 2003;52:2840–2847. doi: 10.2337/diabetes.52.11.2840. [DOI] [PubMed] [Google Scholar]
- 8.He LN, Liu YJ, Xiao P, Zhang L, Guo Y, Yang TL, Zhao LJ, Drees B, Hamilton J, Deng HY, Recker RR, Deng HW. Genomewide linkage scan for combined obesity phenotypes using principal component analysis. Ann Hum Genet. 2008;0:1–8. doi: 10.1111/j.1469-1809.2007.00423.x. [DOI] [PubMed] [Google Scholar]
- 9.Varilo T, Peltonen L. Isolates and their potential use in complex gene mapping efforts. Curr Opin Genet Dev. 2004;14:316–323. doi: 10.1016/j.gde.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Hoare M. Norfolk Island: A Revised and Enlarged History 1774–1998. Rockhamptom: Central Queesland University; 1999. [Google Scholar]
- 11.Matthews SP: Norfolk Island Census of Population and Housing 7 August 2001 – Statistical Report on Characteristics of Population and Dwellings. Norfolk Island, Photopress International, 2001.
- 12.Bellis C, Cox HC, Ovcaric M, Begley KN, Lea RA, Quinlan S, Burgner D, Heath SC, Blangero J, Griffiths LR: Linkage disequilibrium analysis in the genetically isolated Norfolk Island population. Heredity 2007. [DOI] [PubMed]
- 13.Dyke B. 2. San Antonio, Texas: Southwest Foundation for Biomedical Research; 1995. PEDSYS: A Pedigree Data Management System. User's Manual. Ed Tech Rep No. [Google Scholar]
- 14.Reich D, Patterson N, Jager PLD, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BAC, Hauser SL, Oksenberg JR, Hafler DA. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 15.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Charles Gu C, Tang H, Rao DC, Risch N, Weder A. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 17.Angius A, Bebbere D, Petretto E, Falchi M, Forabosco P, Maestrale G, Casu G, Persico I, Melis P, Pirastu M. Not all isolates are equal: linkage disequilibrium analysis on Xq13.3 reveals different patterns in Sardinian sub-populations. Hum Genet. 2002;111:9–15. doi: 10.1007/s00439-002-0753-z. [DOI] [PubMed] [Google Scholar]
- 18.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korth B, Tucker LR. The distribution of chance congruence coefficients from simulated data. Psychometrika. 1975;40:361–372. [Google Scholar]
- 20.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almasy L, Blangero J. Multipoint quantitative trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobin MD, Sheehan NA, Scurrah KH, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 25.Hsueh W-C, Mitchell BD, Schneider JL, St. Jean PL, Pollin TI, Ehm MG, Wagner MJ, Burns DK, Sakul H, Bell CJ, Shuldiner AR. Genome-wide scan of obesity in the Old Order Amish. J Clin Endocrinol Metab. 2001;86:1199–1205. doi: 10.1210/jcem.86.3.7358. [DOI] [PubMed] [Google Scholar]
- 26.Ober C, Cox NJ, Abney M, Di Rienzo A, Lander ES, Changyaleket B, Gidley H, Kurtz B, Lee J, Nance M, Pettersson A, Prescott J, Richardson A, Schlenker E, Summerhill E, Willadsen S, Parry R. Genome-wide search for asthma susceptibility loci in a founder population. The Collaborative Study on the Genetics of Asthma. Hum Mol Genet. 1998;7:1393–1398. doi: 10.1093/hmg/7.9.1393. [DOI] [PubMed] [Google Scholar]
- 27.Hovatta I, Varilo T, Suvisaari J, Terwilliger JD, Ollikainen V, Arajärvi R, Juvonen H, Kokko-Sahin M-L, Väisänen L, Mannila H, Lönnqvist J, Peltonen L. A genomewide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am J Hum Genet. 1999;65:1114–1124. doi: 10.1086/302567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsueh W-C, Mitchell BD, Schneider JL, Wagner MJ, Bell CJ, Nanthakumar E, Shuldiner AR. QTL Influencing blood pressure maps to the region of PPH1 on chromosome 2q31-34 in Old Order Amish. Circulation. 2000;101:2810–2816. doi: 10.1161/01.cir.101.24.2810. [DOI] [PubMed] [Google Scholar]
- 29.Laitinen T, Daly MJ, Rioux JD, Kauppi P, Laprise C, Petays T, Green T, Cargill M, Haahtela T, Lander ES, Laitinen LA, Hudson TJ, Kere J. A susceptibility locus for asthma-related traits on chromosome 7 revealed by genome-wide scan in a founder population. Nat Genet. 2001;28:87–91. doi: 10.1038/ng0501-87. [DOI] [PubMed] [Google Scholar]
- 30.Pajukanta P, Terwilliger JD, Perola M, Hiekkalinna T, Nuotio I, Ellonen P, Parkkonen M, Hartiala J, Ylitalo K, Pihlajamäki J, Porkka K, Laakso M, Viikari J, Ehnholm C, Taskinen M-R, Peltonen L. Genomewide scan for familial combined hyperlipidemia genes in Finnish families, suggesting multiple susceptibility loci influencing triglyceride, cholesterol, and apolipoprotein B levels. Am J Hum Genet. 1999;64:1453–1463. doi: 10.1086/302365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shmulewitz D, Heath SC, Blundell ML, Han Z, Sharma R, Salit J, Auerbach SB, Signorini S, Breslow JL, Stoffel M, Friedman JM. Inaugural article: linkage analysis of quantitative traits for obesity, diabetes, hypertension, and dyslipidemia on the island of Kosrae, Federated States of Micronesia. Proc Natl Acad Sci USA. 2006;103:3502–3509. doi: 10.1073/pnas.0510156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubert HB, Feinleib M, McNamara PM, Castelli WP. Obesity as an independent risk factor for cardiovascular disease: a 26-year follow-up of participants in the Framingham Heart Study. Circulation. 1983;67:968–977. doi: 10.1161/01.cir.67.5.968. [DOI] [PubMed] [Google Scholar]
- 33.Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) Final Report. Circulation. 2002;106:3143. [PubMed] [Google Scholar]
- 34.Stamler J, Daviglus ML, Garside DB, Dyer AR, Greenland P, Neaton JD. Relationship of baseline serum cholesterol levels in 3 large cohorts of younger men to long-term coronary, cardiovascular, and all-cause mortality and to longevity. JAMA. 2000;284:311–318. doi: 10.1001/jama.284.3.311. [DOI] [PubMed] [Google Scholar]
- 35.Arya R, Blangero J, Williams K, Almasy L, Dyer TD, Leach RJ, O'Connell P, Stern MP, Duggirala R. Factors of insulin resistance syndrome-related phenotypes are linked to genetic locations on chromosomes 6 and 7 in nondiabetic Mexican-Americans. Diabetes. 2002;51:841–847. doi: 10.2337/diabetes.51.3.841. [DOI] [PubMed] [Google Scholar]
- 36.Austin MA, Edwards KL, McNeely MJ, Chandler WL, Leonetti DL, Talmud PJ, Humphries SE, Fujimoto WY. Heritability of multivariate factors of the metabolic syndrome in nondiabetic Japanese Americans. Diabetes. 2004;53:1166–1169. doi: 10.2337/diabetes.53.4.1166. [DOI] [PubMed] [Google Scholar]
- 37.Simmons D, Thompson CF, Volklander D. Polynesians: prone to obesity and Type 2 diabetes mellitus but not hyperinsulinaemia. Diabet Med. 2001;18:193–198. doi: 10.1046/j.1464-5491.2001.00435.x. [DOI] [PubMed] [Google Scholar]
- 38.Hodge AM, Dowse GK, Toelupe P, Collins VR, Zimmet PZ. The association of modernization with dyslipidaemia and changes in lipid levels in the Polynesian population of Western Samoa. Int J Epidemiol. 1997;26:297–306. doi: 10.1093/ije/26.2.297. [DOI] [PubMed] [Google Scholar]
- 39.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 40.Platte P, Papanicolaou GJ, Johnston J, Klein CM, Doheny KF, Pugh EW, Roy-Gagnon MH, Stunkard AJ, Francomano CA, Wilson AF. A study of linkage and association of body mass index in the old order Amish. Am J Med Genet Part C: Semin Med Genet. 2003;121C:71–80. doi: 10.1002/ajmg.c.20005. [DOI] [PubMed] [Google Scholar]
- 41.Almasy L, Hixson JE, Rainwater DL, Cole S, Williams JT, Mahaney MC, VandeBerg JL, Stern MP, MacCluer JW, Blangero J. Human pedigree-based quantitative-trait–locus mapping: localization of two genes influencing HDL-cholesterol metabolism. Am J Hum Genet. 1999;64:1686–1693. doi: 10.1086/302425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elbein SC, Hasstedt SJ. Quantitative trait linkage analysis of lipid-related traits in familial type 2 diabetes: evidence for linkage of triglyceride levels to chromosome 19q. Diabetes. 2002;51:528–535. doi: 10.2337/diabetes.51.2.528. [DOI] [PubMed] [Google Scholar]
- 43.Feitosa MF, Borecki IB, Rich SS, Arnett DK, Sholinsky P, Myers RH, Leppert M, Province MA. Quantitative-trait loci influencing body-mass index reside on chromosomes 7 and 13: The National Heart, Lung, and Blood Institute Family Heart Study. Am J Hum Genet. 2002;70:72–82. doi: 10.1086/338144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, Guy-Grand B, Froguel P. A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat Genet. 1998;20:304–308. doi: 10.1038/3123. [DOI] [PubMed] [Google Scholar]
- 45.Pérusse L, Rice T, Chagnon YC, Després J, Lemieux S, Roy S, Lacaille M, Ho-Kim M, Chagnon M, Province MA, Rao DC, Bouchard C. A genome-wide scan for abdominal fat assessed by computed tomography in the Québec Family Study. Diabetes. 2001;50:614–621. doi: 10.2337/diabetes.50.3.614. [DOI] [PubMed] [Google Scholar]
- 46.Zhao L, Xiao P, Liu Y, Xiong D, Shen H, Recker RR, Deng H. A genome-wide linkage scan for quantitative trait loci underlying obesity related phenotypes in 434 Caucasian families. Hum Genet. 2007;121:145–148. doi: 10.1007/s00439-006-0286-y. [DOI] [PubMed] [Google Scholar]
- 47.Reynisdottir I, Thorleifsson G, Benediktsson R, Sigurdsson G, Emilsson V, Einarsdottir AS, Hjorleifsdottir EE, Orlygsdottir GT, Bjornsdottir GT, Saemundsdottir J, Halldorsson S, Hrafnkelsdottir S, Sigurjonsdottir SB, Steinsdottir S, Martin M, Kochan JP, Rhees BK, Grant SFA, Frigge ML, Kong A, Gudnason V, Stefansson K, Gulcher JR. Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34-q35.2. Am J Hum Genet. 2003;73:323–335. doi: 10.1086/377139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbe P, Millet L, Galitzky J, Lafontan M, Berlan M. In situ assessment of the role of the beta 1-, beta 2- and beta 3-adrenoceptors in the control of lipolysis and nutritive blood flow in human subcutaneous adipose tissue. Br J Pharmacol. 1996;117:907–913. doi: 10.1111/j.1476-5381.1996.tb15279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jocken JWE, Blaak EE, Schiffelers S, Arner P, van Baak MA, Saris WHM. Association of a beta-2 adrenoceptor (ADRB2) gene variant with a blunted in vivo lipolysis and fat oxidation. Int J Obes. 2006;31:813–819. doi: 10.1038/sj.ijo.0803499. [DOI] [PubMed] [Google Scholar]
- 50.Barbato E, Berger A, Delrue L, Van Durme F, Manoharan G, Boussy T, Heyndrickx GR, De Bruyne B, Ciampi Q, Vanderheyden M, Wijns W, Bartunek J. GLU-27 variant of [beta]2-adrenergic receptor polymorphisms is an independent risk factor for coronary atherosclerotic disease. Atherosclerosis. 2007;194:e80–e86. doi: 10.1016/j.atherosclerosis.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 51.Krushkal J, Xiong M, Ferrell R, Sing CF, Turner ST, Boerwinkle E. Linkage and association of adrenergic and dopamine receptor genes in the distal portion of the long arm of chromosome 5 with systolic blood pressure variation. Hum Mol Genet. 1998;7:1379–1383. doi: 10.1093/hmg/7.9.1379. [DOI] [PubMed] [Google Scholar]
- 52.Contopoulos-Ioannidis DG, Manoli EN, Ioannidis JPA. Meta-analysis of the association of [beta]2-adrenergic receptor polymorphisms with asthma phenotypes. J Allergy Clin Immunol. 2005;115:963–972. doi: 10.1016/j.jaci.2004.12.1119. [DOI] [PubMed] [Google Scholar]
- 53.Dallongeville J, Helbecque N, Cottel D, Amouyel P, Meirhaeghe A. The Gly16-> Arg16 and Gln27->Glu27 polymorphisms of {beta}2-adrenergic receptor are associated with metabolic syndrome in men. J Clin Endocrinol Metab. 2003;88:4862–4866. doi: 10.1210/jc.2003-030173. [DOI] [PubMed] [Google Scholar]
- 54.Large V, Hellström L, Reynisdottir S, Lönnqvist F, Eriksson P, Lannfelt L, Arner P. Human beta-2 adrenoceptor gene polymorphisms are highly frequent in obesity and associate with altered adipocyte beta-2 adrenoceptor function. J Clin Invest. 1997;100:3005–3013. doi: 10.1172/JCI119854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gjesing A, Andersen G, Burgdorf K, Borch-Johnsen K, J⊘rgensen T, Hansen T, Pedersen O. Studies of the associations between functional beta2-adrenergic receptor variants and obesity, hypertension and type 2 diabetes in 7,808 white subjects. Diabetologia. 2007;50:563–568. doi: 10.1007/s00125-006-0578-8. [DOI] [PubMed] [Google Scholar]
- 56.Dong C, Wang S, Li W-D, Li D, Zhao H, Price RA. Interacting genetic loci on chromosomes 20 and 10 influence extreme human obesity. Am J Hum Genet. 2003;72:115–124. doi: 10.1086/345648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price R, Li W, Bernstein A, Crystal A, Golding E, Weisberg S, Zuckerman W. A locus affecting obesity in human chromosome region 10p12. Diabetologia. 2001;44:363–366. doi: 10.1007/s001250051627. [DOI] [PubMed] [Google Scholar]
- 58.Feitosa M, Borecki I, Rankinen T, Rice T, Després J, Chagnon Y, Gagnon J, Leon A, Skinner J, Bouchard C, Province M, Rao D. Evidence of QTLs on chromosomes 1q42 and 8q24 for LDL-cholesterol and apoB levels in the HERITAGE Family Study. J Lipid Res. 2005;46:281–286. doi: 10.1194/jlr.M400252-JLR200. [DOI] [PubMed] [Google Scholar]