Abstract
Fibroblast activation protein (FAP) is a type II integral membrane glycoprotein belonging to the serine protease family. It is selectively expressed by tumor stromal fibroblasts and transiently in the fibroblasts of healing wounds. FAP has been shown to modulate growth, differentiation, adhesion, and metastasis of tumor cells. Despite the importance of FAP in cancer, the mechanisms that govern its expression have not been defined. In this study, we determined the transcription start site of the FAP gene and identified a 2-kb segment with promoter activity in cells expressing FAP. Truncation of this fragment revealed that the core promoter activity resided in a 245-bp fragment surrounding the transcription start site. Electrophoretic mobility shift assay showed that EGR1 binds to the FAP promoter. Mutation of the EGR1 site within this fragment significantly decreased the promoter activity of FAP and eliminated EGR1 binding. Down-regulation of EGR1 resulted in a significant reduction in endogenous FAP mRNA expression. These findings identify the basal transcriptional requirements of FAP gene expression and show EGR1 is an important regulator of FAP expression.
Keywords: Fibroblast activation protein, Promoter
2. INTRODUCTION
Fibroblast activation protein (FAP), also named “seprase”, is a cell-surface serine protease that was originally identified in 1986 in cultured fibroblasts (1). FAP is a member of post prolyl amino peptidases that are uniquely capable of cleaving the NH2-terminal dipeptides from polypeptides with penultimate L-prolines or L-alanines (2). Active FAP is a 170 kDa homodimer that contains two N-glycosylated 97 kDa subunits. Dipeptidyl peptidase IV (DPPIV or CD26) is the closest member to FAP and the most studied. Unlike DPPIV, FAP has both dipeptidyl peptidase and endopeptidase activity which are mediated by an active site serine624 (3). FAP is a type II transmembrane protein of 761 amino acids consisting of a large extracellular domain, transmembrane segment and a short cytoplasmic domain. Human FAP gene is located on chromosome 2q23 (4). The homologue of the FAP gene has been observed in several species including mouse, rat and xenopus (5). The mouse FAP gene, also located on chromosome 2, shares similar genomic organization and 89% amino-acid-sequence identity with human FAP including a perfectly conserved catalytic triad (2).
FAP has a unique expression pattern. Most normal adult tissues and benign epithelial tumors show little or no detectable FAP expression. However, FAP expression is detected in the stroma of over 90% of malignant breast, colorectal, lung, skin and pancreatic tumors, fibroblasts of healing wounds, soft tissue sarcomas, and some fetal mesenchymal cells (6-8).FAP has a potential role in cancer growth and metastasis through cell adhesion and migration processes, as well as rapid degradation of ECM components (9, 10). In invadopodia, FAP is associated with the fibronectin receptor, integrin alpha3beta1 (11), and forms a heterodimer complex with DPPIV or urokinase plasminogen activator receptor (uPAR) (12). The formation of FAP/DPPIV or FAP/uPAR protease complexes at the invadopodia of migratory fibroblasts (13), human endothelial cells (14) and invading membranes of malignant melanoma LOX cells (15) were shown to be critical for cell invasion and migration. Besides tumor associated fibroblasts, FAP expression was also found in reactive fibroblasts or fibroblast-like cells located at the tissue remodeling interface of healing wounds (6), rheumatoid arthritis synovium (16), cirrhotic liver (17), and fibrotic human lung (18). Overexpression of FAP has significant effects on cell adhesion, migration, proliferation and apoptosis in epithelial and fibroblastic cell lines (19).
Despite this growing knowledge of the enzymatic function of FAP and its role in cancer and many other diseases, the promoter elements of FAP have not yet been characterized. Our present study aims to unravel the transcription mechanism of FAP by identifying its minimal promoter and the transcriptional factors that are involved in the regulation of FAP mRNA expression.
3. MATERIALS AND METHODS
3.1. Cell culture and RNA extraction
Human epithelial carcinoma HeLa cells, V20, human embryonic kidney cell line (HEK 293), HT 1080 and mouse embryonic fibroblasts (MEFs) were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. HOS cells were grown in Minimum essential medium with 0.1 mM non-essential amino acid, 1 mM sodium pyruvate and 10% fetal bovine serum. HT-29 and MCF 7 cells were cultured in McCoy’s medium and RPMI, respectively, with 10% fetal bovine serum. All the cell lines and MEFs came from the cell line bank at Fox Chase Cancer Center. Total RNA was extracted according to the instructions of RNeasy® Mini Kit (Qiagen) before the analysis by qRT-PCR. The mRNA of MEFs was obtained according to the instructions of FastTrack® mRNA isolation Kit (Invitrogen).
3.2. Identification of transcription start site by 5′- Rapid Amplification of cDNA Ends (RACE)
To determine the transcription start site of FAP, 5′- RACE assay was performed according to the instruction of GeneRacer Kit (Invitrogen). Briefly, 250 ng of mFAP mRNA was treated with calf intestinal phosphatase to remove truncated mRNA. The treated full-length mRNA was decapped and subsequently ligated to a RNA oligo. Ligated mRNA was reverse-transcribed and 5′cDNA end was amplified by PCR with a nested FAP primer (592-567, 5′-ACA AAT TCC CCA TTC TGA AGG TCG TA-3′) and 5′GeneRacer primer provided by the kit. PCR product was sequenced and analyzed.
3.3. Generation of FAP promoter luciferase reporter constructs
The analysis of the 5′ flank region of mouse or human FAP gene by rVista ( http://rvista.dcode.org/) showed an area of approximately 3000 bp that had high homology between these two species. BAC clone RPCI23 was purchased from Children’s Hospital Oakland Research Institute (CHORI). 1991 bp upstream of the first exon of mouse FAP was amplified from RPCI23 with primers that had the Bgl II (forward, 5′-GCA GAT CTC CCG TAT ACT AGT ACT TTC AA-3′) and Hind III (reverse, 5′-GGG AAG CTT TTT TCC AGA TGT TTT TGC AAG-3′) restriction sites before cloned into pGL3 luciferase reporter construct (Promega). Further promoter bashing was done by generating pGL3 luciferase reporter constructs that contain various regions of conserved domains within 1991 bp 5′ of FAP gene. Fragments of FAP promoters (−1780, −1469, −1051, −452, −245, −133, −96 and −63) were amplified from pGL3-1991 with corresponding primers that included Bgl II site and subcloned into pGL3 Enhancer luciferase reporter construct. All fragments share the same Hind III site downstream of the FAP transcription start site and upstream of the initiating ATG. Deletion and mutation of pGL3-245 were created by GeneTailor Site-Directed Mutagenesis System (Invitrogen). The primers that include deletions or mutations are indicated in Figure. 4 or 5, respectively.
Figure 4.
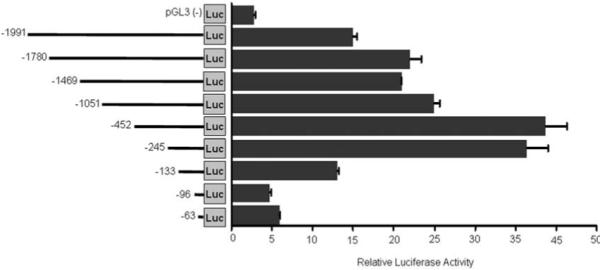
Identification of promoter sequences in FAP gene. Control pGL3 luciferase constructs or pGL3 contains various regions of FAP 5′ flank regions indicated in the graph were transiently co-transfected with Renilla control into HOS cells and luciferase activity was measured 48 hrs after transfection. The ratio of luciferase activity versus renilla control is shown in the graph. Values are mean +/− SE representing at least three independent experiments. The constructs are numbered on the left relative to the initiate ATG.
Figure 5.
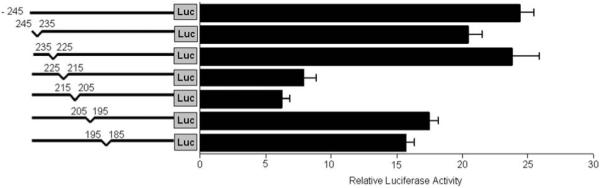
Deletion analysis of the minimal FAP promoter. A series of pGL3-245 internal deletion luciferase constructs were transiently co-transfected with Renilla control into HOS cells and luciferase activity was measured 48 hrs after transfection. The ratio of luciferase activity versus renilla control is shown in the graph. Values are mean +/− SE representing at least three independent experiments.
3.4. Transient transfection and luciferase assay
HOS cells were transiently transfected with various FAP-promoter luciferase constructs by Fugene 6 (Roche) according to the manufacturer’s instructions. In brief, 500 ng of various plasmid DNAs was transfected to cells plated in 12-well plate (40,000/well). pRL-TK vector (Promega) containing renilla luciferase was co-transfected at 1:100 ratio as internal transfection control. Luciferase activities were detected 48 hrs after transfection by Dualglo™ Luciferase Assay System (Promega) and the luminescence was measured by Envision Multilabel Plate Readers (PerkinElmer). The relative luciferase activity was presented as ratio of the firefly luciferase activity versus that of renilla.
3.5. Electrophoretic mobility-shift assay (EMSA)
Complementary oligonucleotides were synthesized by Integrated DNA Technologies: wild type FAP promoter fragment −225 ~ −205: 5′-CAAGAACGCCCCCAAAATCT-3′ and mutant early growth response 1 (EGR1) oligos: 5′-CAAGAATAGTACTCAAATCT-3′. Wild type FAP promoter oligo (8.75 pmol) was labeled with [gamma-32P] ATP (3000 Ci/mmol) and 10 units of T4 polynucleotide kinase for 10 min at 37°C and purified by G-50 microcolumn (GE healthcare). Recombinant EGR1 protein (Sigma) was incubated with 1 microgram poly d (I,C) in DNA binding buffer (10 mM Tris pH 7.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, and 5% (v/v) glycerol) for 15 min at room temperature. The 32P-labeled oligo (75 fmol) was then added to the reaction mixture and incubated for another 15 min at room temperature. DNA/protein complexes were separated from free oligonucleotide by electrophoresis on native 5% polyacrylamide gels in 0.5 X TBE buffer. Competition experiments to determine the specificity of the DNA binding complexes were performed by incubating the protein with a 100-fold molar excess of either the unlabeled consensus or mutant oligonucleotide for 20 min prior to the addition of labeled oligo. For supershift assays, antibodies against EGR1 was added to the reaction mixtures after the DNA-binding incubation and incubated for an additional 30 min at room temperature.
3.6. Small interfering RNA transfection and quantitative RT-PCR
The ON-TARGETplus SMARTpool small interfering RNAs (siRNA) against EGR1, E2F1 and HOXA4 were purchased from Dharmacon. RNA was extracted from HOS cells after transfected with 100 nM on-target or non-target siRNAs for 24 hrs. All the Quantitative RT-PCR was performed by the core facility in Fox Chase Cancer Center (FCCC). Real-time Taqman PCR assays were run using an ABI 7900 HT instrument. The sequences for detecting human FAP cDNA were F: TCA AAG AAG TAT CCC TTG CTA ATT CA; R: GCA ATG ACC ATC CCT TCC TTA C; P: (6FAM) TGG TGG TCC CTG CAG TCA GAG TGT AAG (BHQ1). The primers for EGR1, E2F1 and HoxA4 were purchased from Applied Biosystems. Actin was used as the reference gene. A 4 points 4-fold dilutions standard curve established with a calibrator sample was used to convert the Ct values into quantities.
4. RESULTS
4.1. Sequence analysis of putative FAP promoter region and identification of transcription start site in FAP gene
The cDNAs of human or mouse FAP were cloned more than a decade ago (7, 20). It has been reported that both mouse and human FAP genes are located on chromosome 2 (2, 4). Clustal alignment of the human and mouse FAP gene demonstrated a high degree of conservation of proximal putative promoter regions (Figure. 1). The sequence of −500 upstream of ATG is 78% conserved between mouse and human FAP genes. Sequence analysis by rVista program revealed the absence of a TATA box in the 5′ flanking region of the FAP gene.
Figure 1.
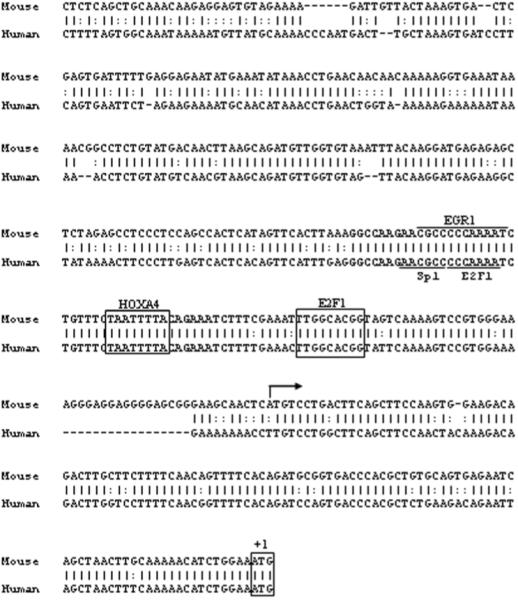
Promoter sequence analysis of FAP genes. Cluster alignment of the proximal promoter regions of the human and mouse FAP genes. Conserved transcription factor binding sites between human and mouse are boxed or underlined. A 40-bp region with 100% homology between human and mouse is highlighted in bold letters. Arrow indicates the position of the transcription start site.
To identify the transcription start site in mouse FAP gene, 5′- RACE was performed with the mRNA from mouse embryonic fibroblasts which contains endogenous FAP. After amplification with generacer oligo and nested FAP gene-specific primer, a dominant DNA product was visualized on agarose gel (Figure. 2), subcloned, and sequenced. 5′- RACE of mRNA from MEF indicated FAP transcription initiates at 119 nt upstream of ATG translation initiation site. 5′- RACE using total RNA from human HeLa cells indicated the transcription start site of human FAP is −154 nt upstream of ATG (Figure. 2). To be consistent, the numbering of all the sequences in the manuscript is relative to ATG.
Figure 2.
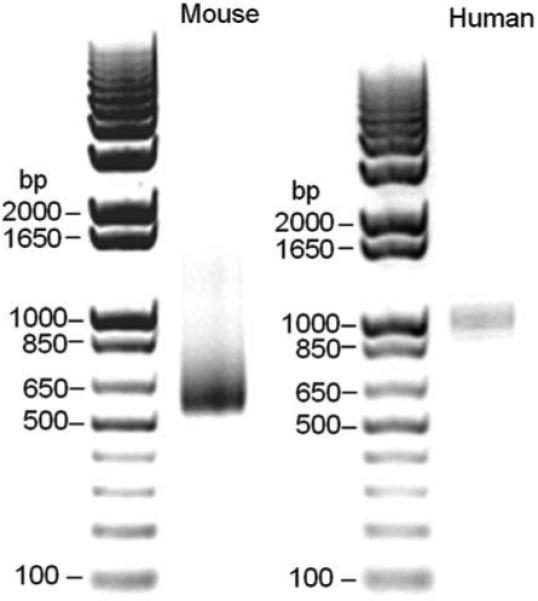
Transcription start site of FAP genes. 5′- RACE assay was performed with RNAs from mouse or human FAP positive cells. Photograph of ethidium bromide-stained agarose gels showing one dominant 5′- RACE product.
4.2. Identification and characterization of FAP promoter sequence
In vitro, FAP is expressed in sarcoma cell lines but not in most cancer epithelial cells (7, 21). To find an in vitro cell model to study the transcriptional regulation of FAP, the levels of FAP mRNA were first evaluated by qRT-PCR among several cell lines with various origins. Consistent with previous reports, human sarcoma cell lines HOS and V20, as well as human cervical carcinoma cell line HeLa, have high levels of endogenous FAP expression. In contrast, FAP is undetectable in the cancer epithelial cells such as HT-29, HEK 293 and MCF 7 (Figure. 3A).
Figure 3.
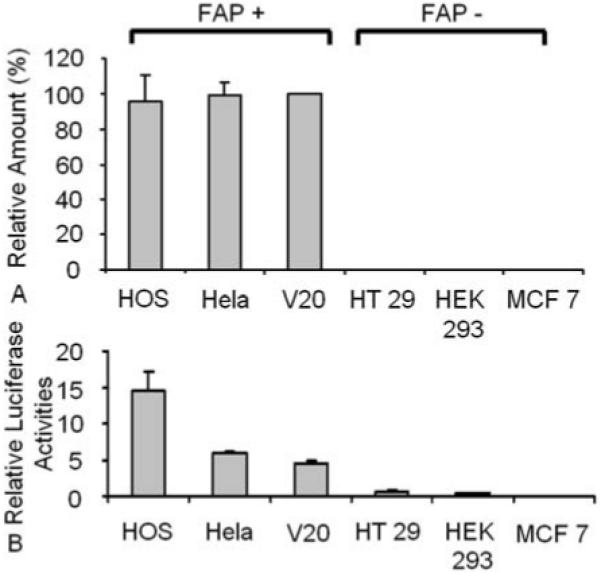
Putative FAP promoter activity is specific. (A) The mRNA level of FAP was detected in various cancer cell lines by qRT-PCR. The percent of FAP mRNA was compared with that of V20 after normalized with actin. (B) The luciferase construct that contains ~2kb 5′ flank region of FAP gene and the renilla control vector were transfected into cell lines with different levels of FAP. 48 hrs later, the luminescence was measured and the ratio of luciferase activity versus renilla control is shown in the graph. Values are mean +/− E representing at least three independent experiments.
A fragment of DNA sequence upstream of the FAP ATG translation initiation site (−1991 nt) was inserted in a pGL3 luciferase reporter construct and analyzed for luciferase activity in all six cell lines that are either FAP positive or negative. Transfection of the luciferase construct containing 1991 nucleotides of the putative FAP promoter region produced a 5 to 15 fold increase in luciferase activity among FAP positive cell lines HOS, V20 and HeLa, whereas very low or no luciferase activity was detected in the FAP negative cell lines tested (Figure. 3B). This data indicates that the 1991-kb putative FAP promoter directs significant cell type-restricted expression in vitro. The FAP positive sarcoma cell line HOS was chosen as a model for further study because of its high endogenous FAP level and high transfection efficiency in luciferase assay.
To delineate the functional elements within the promoter region, a series of promoter fragment deletions were fused to the luciferase reporter gene in pGL3, and the resulting constructs were transiently transfected into HOS cells (Figure. 4). Transfection of the pGL3-FAP construct containing 1991 nucleotides of the FAP promoter region produced a 15 fold increase in luciferase activity compared to empty pGL3 vector control. Further deletion to −452 produced even higher luciferase activity (38 fold) implying the presence of inhibitory elements between nucleotides −1991 and −452. The luciferase activities of pGL3-452 and −245 are similar, however deletion from −245 to −133 caused a significant decrease in promoter activity and further deletions beyond the transcription start site abolished the promoter activities (Figure. 4). These data indicate that the promoter fragment contained in pGL3-245 is sufficient to drive FAP promoter activity. The minimal promoter of FAP appears located between −245 to −119, or 126 nt upstream of the transcription starting site.
To identify which region in the FAP minimal promoter is critical for the transcription of FAP, the promoter activities of six pGL3-245 internal deletion constructs were analyzed in transfected HOS cells. As shown in Figure. 5, the internal deletion constructs were created by sequentially removing 10 bp from the 5′ end of −245. Comparable promoter activity to pGL3-245 was seen with the two deletions targeting region −245 to −225. In contrast, removal of either −225 ~ −215 or −215 ~ −205 significantly decreased the promoter activities to 33% or 26%, respectively. Moderate decreases of promoter activity to 60 ~ 70% were observed after deleting −205 ~ −195 or −195 ~ −185. Taken together, these data suggest that the FAP promoter fragment −225 to −185, particularly the 20 bp between −225 to −205, contains the binding sites of the primary transcription factors that are responsible for FAP transcription. Interestingly, this region was shown to have 100% sequence homology between human and mouse as seen in Figure. 1.
4.3. EGR1 binds to FAP promoter and participates in FAP gene transcription
Based on the analysis by bioinformatics software, four transcription factor binding sites were predicted between the −225 to −185 region of the FAP promoter consisting of EGR1, E2F1, Sp1 and HOXA4. Among them, the EGR1 site located at −219 was the only one that is identical with the known consensus sequence (22) and was consistently predicted by two bioinformatics software, independently, rVista and ConSite (http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite). Moreover, the binding sites of E2F1 and Sp1 overlapped with that of EGR1. To test if the binding sites containing EGR1, E2F1 and Sp1 are important in the transcription of FAP, the predicted EGR1 motif from −219 to −211 in the pGL3-245 plasmid was mutated according to previously published sequence (22) which is shown as Mut 1 in Figure 6. Two other mutations (Mut 2 or Mut 3) were introduced in pGL3-245 plasmids as indicated in Figure 6 to disrupt the binding of HOXA4 or to serve as a negative control, respectively. Transfection of Mut 1 containing EGR1 mutations resulted in 40% loss of promoter activity (p less than 0.05) while Mut 2 and 3 led to slightly increased promoter activity. This mutation analysis revealed that the integrity of the EGR1 binding motif which overlaps with E2F1 and Sp1 is critical for FAP transcription.
Figure 6.
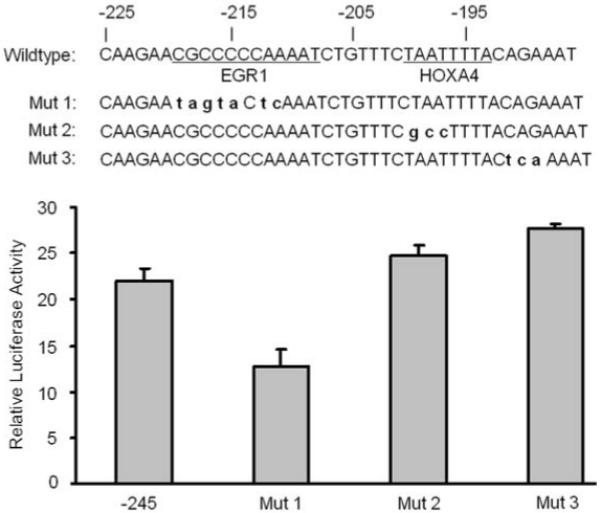
EGR1 is a potential transcription factor that regulates FAP transcription. Luciferase constructs pGL3-245 and its mutant as indicated were transiently co-transfected with Renilla control into HOS cells and luciferase activity were measured 48 hrs after transfection. The ratio of luciferase activity versus renilla control is shown in the graph. Values are mean +/− SE representing at least three independent experiments.
In order to investigate if EGR1 binds to the putative EGR1 site on FAP promoter region (−225 ~ −205), electrophoretic mobility shift assay (EMSA) was performed. Recombinant EGR1 protein was incubated with 32P-labeled FAP promoter oligonucleotides (−225 ~ −205) and resulted in the formation of one single DNA-binding complex whose intensity was depleted by pre-incubation with unlabeled wild type oligonucleotides and specific anti-EGR1 antibody, but not by mutant oligos (Figure 7).
Figure 7.
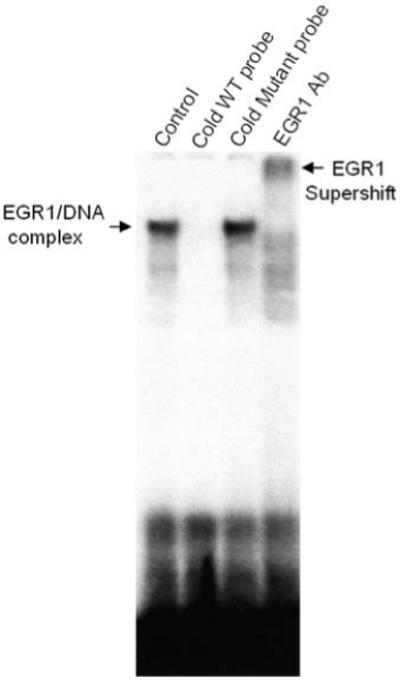
EGR1 binds to the putative EGR1 binding site in the FAP promoter. [γ-32P] ATP-labeled oligonucleotides covering −225 to −185 region of FAP promoter fragment was incubated with recombinant EGR1 protein (“Control”). Competition experiment was performed by pre-incubating EGR1 protein with a 100-fold excess of unlabeled consensus (“cold WT probe”) or EGR1 mutant oligos (“cold mutant probe”) prior to the addition of the 32P-labeled oligo. EGR1 antibody was used for supershift analysis. Arrows point at the DNA/protein complex of EGR1 and supershift band identified by the EGR1 antibody.
4.4. siRNA-mediated knockdown of EGR1 inhibits the mRNA expression of FAP
To determine the functional importance of EGR1 in mediating the endogenous expression of FAP, small interfering RNAs (siRNAs) against EGR1 were transfected into HOS cells. The siRNAs against E2F1 and HOXA4 were also tested as controls since they were also possible transcription factors within the FAP minimal promoter region. As shown in Figure 8, siRNA against EGR1 knocked down the expression of EGR1 mRNA to 25%, which lead to 50% inhibition of FAP mRNA expression as detected by qRT-PCR. In contrast, no significant change of FAP was observed in HOS cells when transfected with siRNAs of E2F1 and HoxA4, although the siRNAs almost completely knocked down the expression of both genes. This provides the direct evidence that the transcription of FAP is partially mediated by EGR1.
Figure 8.
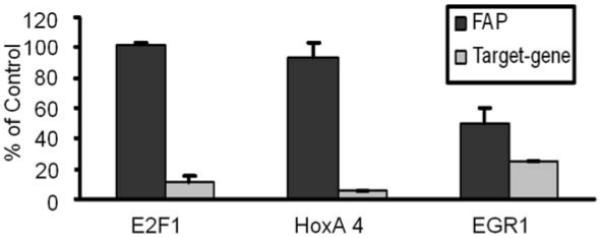
The expression of FAP mRNA is partially decreased by knockdown of EGR1 in HOS. The siRNAs of E2F1, HOXA4 and EGR1 or non-target control siRNAs were transfected into HOS cells at 100 nM for 24 hrs before the RNA was harvested. The mRNA level of FAP and genes of target were detected by qRT-PCR. Data is shown as relative amount comparing with non-target siRNA controls after normalized with actin. Values are mean +/− SE.
5. DISCUSSION
As an inducible tumor stromal antigen, FAP has been shown to play an important role in cancer cell growth and metastasis (10). The preclinical studies by our group and others have shown that overexpression of FAP in HEK 293 human embryonic kidney cells (23) and MDA-MB 231 breast cancer cells (24) lead to more rapid tumor growth compared to FAP negative control cells in mouse models. An in vivo study involving colon cancer patients reported that FAP expression in tumor stroma was inversely correlated with the stages of colon cancer (25). In contrast, decreased tumorigenicity was seen in mouse melanoma cells when FAP was re-introduced (26) although it was independent of FAP enzymatic activity. These findings suggest that the role of FAP in cancer growth is cell type dependent and the control of FAP expression at the transcriptional level may be important in regulating FAP function. Up-regulation of FAP has been demonstrated in cultured melanocytes (27), fibroblast cell lines (28) and primary chondrocytes (29) after stimulation with growth factors and pro-inflammatory cytokines such as transforming growth factor-beta (TGF-beta) or IL-1. However, there have been no previous reports about the promoter elements of FAP, and nothing is known about the mechanisms that lead to inducible transcription. For these reasons, in the present work we have focused our attention to the identification of the DNA sequences necessary for basal transcription of the FAP gene and the possible cis-elements involved in the differential expression of the gene. To the best of our knowledge, this is the first identification and characterization of the promoter of the FAP gene.
Both located on chromosome 2, mouse and human FAP gene have similar genomic organization (2). Mouse FAP gene spans approximately 60 kb and contains 26 exons ranging in size from 46 bp to 195 bp. We have identified the transcription starting sites of mouse and human FAP gene and cloned the mouse FAP promoter. Consistent with their similarities in chromosomal location and structure, both human and mouse FAP gene have only one dominant transcription start site at −154 and −119 (relative numbering to ATG), respectively. The unique expression pattern of FAP has prompted us to compare the activity of FAP promoter constructs in either FAP positive or negative cell lines. Using a transient luciferase reporter assay, we have demonstrated that the luciferase activity of a 2-kb putative mouse FAP promoter construct correlates with the expression levels of FAP in different cell lines. This suggests that FAP promoter constructs can only be transactivated by factors present in FAP-expressing cells. Further analysis within the 2-kb FAP promoter fragment demonstrated that minimal transcription of the mouse FAP gene requires a proximal promoter region of 126 nt upstream of the transcription start site. This region has approximately 80% sequence homology between human and mouse FAP gene, indicating the probability of important conserved cis-elements regulating FAP transcription. Indeed, deletion analyses revealed a 20-bp positive regulatory element within minimal promoter of FAP which contains three adjacent or overlapping binding sites of transcription factors including EGR1, E2F1 and Sp1. Using a series of mutational analyses, EMSA and siRNA-mediated knockdown experiments, we have demonstrated that EGR1, but not Sp1 or E2F1, is required in governing expression of FAP.
EGR1 or the early growth response gene product is a zinc-finger transcription factor of 59 kDa that preferentially binds GC-rich regulatory elements (30). EGR1 has been localized in endothelial cells, macrophages, and smooth muscle cells (31). EGR1 is a master regulator that controls the expression of a wide variety of genes such as transcription factors, growth factors and cytokines (TGF-beta, platelet-derived growth factors, IL-2), cell-cycle regulators (cyclin D1), matrix proteins (fibronectin, collagen) et al (30). Similar to FAP, EGR1 is also involved in wound healing (32). The roles of EGR1 in tumor cell phenotype are paradoxical. While the majority of studies define EGR1 as having a tumor suppressor function (30), EGR1 is oncogenic in prostate cancer (33). Elevated expressions of TGF-beta, collagen I, as well as FAP, were detected in the reactive stroma of prostate cancer (34). The expressions of TGF-beta and collagen driven by EGR1 contribute to the recruitment of tumor-associated vasculature and stromal components (31). Interestingly, it has also been shown that FAP mRNA is up-regulated by endothelial cells undergoing reorganization and capillary morphogenesis (35). FAP promotes growth of breast cancer tumors partly through enhanced angiogenesis (24). In ovarian carcinoma, EGR1 is critical in collagen-induced membrane type 1 matrix metalloproteinase (MT1-MMP) up-regulation and cellular invasion (36). More recently, it was reported that type I collagen triggers the expression of FAP in ovarian carcinoma cell line and initiates tumor invasion and metastatic cascades (37). In our study, we have demonstrated that truncation and mutation of the EGR1 binding sequence in the FAP promoter resulted in decreased promoter activity. The interaction of EGR1 with the putative EGR1-binding motif in the FAP promoter is dependent on the integrity of the core-binding sequence as shown by EMSA studies. Moreover, functional analysis using EGR1 siRNA-mediated knockdown experiments showed that the expression of FAP mRNA is partially blocked by down-regulating EGR1. Given our evidence showing that EGR1 regulates FAP transcription, it will be worth studying if EGR1 is involved in collagen-induced expression of FAP in ovarian cancer or FAP-induced angiogenesis in prostate cancer.
Although our data demonstrates the important role of EGR1 in FAP transcription, we can not exclude the possibilities that other unidentified transcription factors might be involved. This is highlighted by the fact that the interference of EGR1 binding or expression did not completely block FAP transcription. Further studies are required to investigate the identities of other possible transcription factors that may occupy the region −225 to −205 of the FAP promoter.
Taken together, our study provides the first insight into the basal transcriptional mechanism of the FAP gene and the importance of transactivation by EGR1 in this process. The developments of therapeutic antibodies and small molecule inhibitors against FAP have been tested in clinical trials (38, 39). Our discovery of EGR1 as one of the transcriptional regulators of FAP provides a new rationale to disrupt the function of FAP.
6. ACKNOWLEDGEMENTS
We thank Emmanuelle Nicolas and the staff of Real-Time PCR Facility, Translational Research Facility, DNA sequencing and synthesis facility at Fox Chase Cancer Center for their services. We thank Lisa A. Vanderveer and Dr. Andrew Godwin for their assistance in EMSA analysis. This work was supported in part by Grants CA 090468 and CA 122301 and an Appropriation from the Commonwealth of Pennsylvania.
Abbreviations
- FAP
fibroblast activation protein
- DPPIV
dipeptidyl peptidase IV
- uPAR
urokinase plasminogen activator receptor
- EMSA
electrophoretic mobility shift analysis
- siRNA
small interfering RNAs
- EGR1
early growth response 1
- TGF-beta
transforming growth factor-beta
- MT1-MMP
membrane type 1 matrix metalloproteinase
7. REFERENCES
- 1.Rettig WJ, Chesa PG, Beresford HR, Feickert HJ, Jennings MT, Cohen J, Oettgen HF, Old LJ. Differential expression of cell surface antigens and glial fibrillary acidic protein in human astrocytoma subsets. Cancer Res. 1986;46(12 Pt 1):6406–12. [PubMed] [Google Scholar]
- 2.Niedermeyer J, Enenkel B, Park JE, Lenter M, Rettig WJ, Damm K, Schnapp A. Mouse fibroblast-activation protein--conserved Fap gene organization and biochemical function as a serine protease. Eur J Biochem. 1998;254(3):650–4. doi: 10.1046/j.1432-1327.1998.2540650.x. [DOI] [PubMed] [Google Scholar]
- 3.Park JE, Lenter MC, Zimmermann RN, Garin-Chesa P, Old LJ, Rettig WJ. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J Biol Chem. 1999;274(51):36505–12. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 4.Mathew S, Scanlan MJ, Mohan Raj BK, Murty VV, Garin-Chesa P, Old LJ, Rettig WJ, Chaganti RS. The gene for fibroblast activation protein alpha (FAP), a putative cell surface-bound serine protease expressed in cancer stroma and wound healing, maps to chromosome band 2q23. Genomics. 1995;25(1):335–7. doi: 10.1016/0888-7543(95)80157-h. [DOI] [PubMed] [Google Scholar]
- 5.Brown DD, Wang Z, Furlow JD, Kanamori A, Schwartzman RA, Remo BF, Pinder A. The thyroid hormone-induced tail resorption program during Xenopus laevis metamorphosis. Proc Natl Acad Sci U S A. 1996;93(5):1924–9. doi: 10.1073/pnas.93.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garin-Chesa P, Old LJ, Rettig WJ. Cell surface glycoprotein of reactive stromal fibroblasts as a potential antibody target in human epithelial cancers. Proc Natl Acad Sci U S A. 1990;87(18):7235–9. doi: 10.1073/pnas.87.18.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scanlan MJ, Raj BK, Calvo B, Garin-Chesa P, Sanz-Moncasi MP, Healey JH, Old LJ, Rettig WJ. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci U S A. 1994;91(12):5657–61. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rettig WJ, Garin-Chesa P, Beresford HR, Oettgen HF, Melamed MR, Old LJ. Cell-surface glycoproteins of human sarcomas: differential expression in normal and malignant tissues and cultured cells. Proc Natl Acad Sci U S A. 1988;85(9):3110–4. doi: 10.1073/pnas.85.9.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng JD, Weiner LM. Tumors and their microenvironments: tilling the soil. Commentary re: A. M. Scott et al., A Phase I dose-escalation study of sibrotuzumab in patients with advanced or metastatic fibroblast activation protein-positive cancer. Clin. Cancer Res. 2003;9:1639–1647. [PubMed] [Google Scholar]; Clin Cancer Res. 2003;9(5):1590–5. [PubMed] [Google Scholar]
- 10.Kelly T. Fibroblast activation protein-alpha and dipeptidyl peptidase IV (CD26): cell-surface proteases that activate cell signaling and are potential targets for cancer therapy. Drug Resist Updat. 2005;8(1-2):51–8. doi: 10.1016/j.drup.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Mueller SC, Ghersi G, Akiyama SK, Sang QX, Howard L, Pineiro-Sanchez M, Nakahara H, Yeh Y, Chen WT. A novel protease-docking function of integrin at invadopodia. J Biol Chem. 1999;274(35):24947–52. doi: 10.1074/jbc.274.35.24947. [DOI] [PubMed] [Google Scholar]
- 12.Chen WT, Kelly T. Seprase complexes in cellular invasiveness. Cancer Metastasis Rev. 2003;22(2-3):259–69. doi: 10.1023/a:1023055600919. [DOI] [PubMed] [Google Scholar]
- 13.Ghersi G, Dong H, Goldstein LA, Yeh Y, Hakkinen L, Larjava HS, Chen WT. Regulation of fibroblast migration on collagenous matrix by a cell surface peptidase complex. J Biol Chem. 2002;277(32):29231–41. doi: 10.1074/jbc.M202770200. [DOI] [PubMed] [Google Scholar]
- 14.Ghersi G, Zhao Q, Salamone M, Yeh Y, Zucker S, Chen WT. The protease complex consisting of dipeptidyl peptidase IV and seprase plays a role in the migration and invasion of human endothelial cells in collagenous matrices. Cancer Res. 2006;66(9):4652–61. doi: 10.1158/0008-5472.CAN-05-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artym VV, Kindzelskii AL, Chen WT, Petty HR. Molecular proximity of seprase and the urokinase-type plasminogen activator receptor on malignant melanoma cell membranes: dependence on beta1 integrins and the cytoskeleton. Carcinogenesis. 2002;23(10):1593–601. doi: 10.1093/carcin/23.10.1593. [DOI] [PubMed] [Google Scholar]
- 16.Bauer S, Jendro MC, Wadle A, Kleber S, Stenner F, Dinser R, Reich A, Faccin E, Godde S, Dinges H, Muller-Ladner U, Renner C. Fibroblast activation protein is expressed by rheumatoid myofibroblast-like synoviocytes. Arthritis Res Ther. 2006;8(6):R171. doi: 10.1186/ar2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy MT, McCaughan GW, Abbott CA, Park JE, Cunningham AM, Muller E, Rettig WJ, Gorrell MD. Fibroblast activation protein: a cell surface dipeptidyl peptidase and gelatinase expressed by stellate cells at the tissue remodelling interface in human cirrhosis. Hepatology. 1999;29(6):1768–78. doi: 10.1002/hep.510290631. [DOI] [PubMed] [Google Scholar]
- 18.Acharya PS, Zukas A, Chandan V, Katzenstein AL, Pure E. Fibroblast activation protein: a serine protease expressed at the remodeling interface in idiopathic pulmonary fibrosis. Hum Pathol. 2006;37(3):352–60. doi: 10.1016/j.humpath.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 19.Wang XM, Yu DM, McCaughan GW, Gorrell MD. Fibroblast activation protein increases apoptosis, cell adhesion, and migration by the LX-2 human stellate cell line. Hepatology. 2005;42(4):935–45. doi: 10.1002/hep.20853. [DOI] [PubMed] [Google Scholar]
- 20.Niedermeyer J, Scanlan MJ, Garin-Chesa P, Daiber C, Fiebig HH, Old LJ, Rettig WJ, Schnapp A. Mouse fibroblast activation protein: molecular cloning, alternative splicing and expression in the reactive stroma of epithelial cancers. Int J Cancer. 1997;71(3):383–9. doi: 10.1002/(sici)1097-0215(19970502)71:3<383::aid-ijc14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 21.Dolznig H, Schweifer N, Puri C, Kraut N, Rettig WJ, Kerjaschki D, Garin-Chesa P. Characterization of cancer stroma markers: in silico analysis of an mRNA expression database for fibroblast activation protein and endosialin. Cancer Immun. 2005;5:10. [PubMed] [Google Scholar]
- 22.Halvorson LM, Ito M, Jameson JL, Chin WW. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone beta-subunit gene expression. J Biol Chem. 1998;273(24):14712–20. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- 23.Cheng JD, Dunbrack RL, Jr., Valianou M, Rogatko A, Alpaugh RK, Weiner LM. Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model. Cancer Res. 2002;62(16):4767–72. [PubMed] [Google Scholar]
- 24.Huang Y, Wang S, Kelly T. Seprase promotes rapid tumor growth and increased microvessel density in a mouse model of human breast cancer. Cancer Res. 2004;64(8):2712–6. doi: 10.1158/0008-5472.can-03-3184. [DOI] [PubMed] [Google Scholar]
- 25.Henry LR, Lee HO, Lee JS, Klein-Szanto A, Watts P, Ross EA, Chen WT, Cheng JD. Clinical implications of fibroblast activation protein in patients with colon cancer. Clin Cancer Res. 2007;13(6):1736–41. doi: 10.1158/1078-0432.CCR-06-1746. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez-Montagut T, Blachere NE, Sviderskaya EV, Bennett DC, Rettig WJ, Garin-Chesa P, Houghton AN. FAPalpha, a surface peptidase expressed during wound healing, is a tumor suppressor. Oncogene. 2004;23(32):5435–46. doi: 10.1038/sj.onc.1207730. [DOI] [PubMed] [Google Scholar]
- 27.Rettig WJ, Garin-Chesa P, Healey JH, Su SL, Ozer HL, Schwab M, Albino AP, Old LJ. Regulation and heteromeric structure of the fibroblast activation protein in normal and transformed cells of mesenchymal and neuroectodermal origin. Cancer Res. 1993;53(14):3327–35. [PubMed] [Google Scholar]
- 28.Rettig WJ, Su SL, Fortunato SR, Scanlan MJ, Raj BK, Garin-Chesa P, Healey JH, Old LJ. Fibroblast activation protein: purification, epitope mapping and induction by growth factors. Int J Cancer. 1994;58(3):385–92. doi: 10.1002/ijc.2910580314. [DOI] [PubMed] [Google Scholar]
- 29.Milner JM, Kevorkian L, Young DA, Jones D, Wait R, Donell ST, Barksby E, Patterson AM, Middleton J, Cravatt BF, Clark IM, Rowan AD, Cawston TE. Fibroblast activation protein alpha is expressed by chondrocytes following a pro-inflammatory stimulus and is elevated in osteoarthritis. Arthritis Res Ther. 2006;8(1):R23. doi: 10.1186/ar1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13(2):115–24. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khachigian LM. Early growth response-1 in cardiovascular pathobiology. Circ Res. 2006;98(2):186–91. doi: 10.1161/01.RES.0000200177.53882.c3. [DOI] [PubMed] [Google Scholar]
- 32.Braddock M. The transcription factor Egr-1: a potential drug in wound healing and tissue repair. Ann Med. 2001;33(5):313–8. doi: 10.3109/07853890109002083. [DOI] [PubMed] [Google Scholar]
- 33.Abdulkadir SA. Mechanisms of prostate tumorigenesis: roles for transcription factors Nkx3.1 and Egr1. Ann N Y Acad Sci. 2005;1059:33–40. doi: 10.1196/annals.1339.018. [DOI] [PubMed] [Google Scholar]
- 34.Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res. 2002;8(9):2912–23. [PubMed] [Google Scholar]
- 35.Aimes RT, Zijlstra A, Hooper JD, Ogbourne SM, Sit ML, Fuchs S, Gotley DC, Quigley JP, Antalis TM. Endothelial cell serine proteases expressed during vascular morphogenesis and angiogenesis. Thromb Haemost. 2003;89(3):561–72. [PubMed] [Google Scholar]
- 36.Barbolina MV, Adley BP, Ariztia EV, Liu Y, Stack MS. Microenvironmental regulation of membrane type 1 matrix metalloproteinase activity in ovarian carcinoma cells via collagen-induced EGR1 expression. J Biol Chem. 2007;282(7):4924–31. doi: 10.1074/jbc.M608428200. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy A, Dong H, Chen D, Chen WT. Elevation of seprase expression and promotion of an invasive phenotype by collagenous matrices in ovarian tumor cells. Int J Cancer. 2009;124(1):27–35. doi: 10.1002/ijc.23871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott AM, Wiseman G, Welt S, Adjei A, Lee FT, Hopkins W, Divgi CR, Hanson LH, Mitchell P, Gansen DN, Larson SM, Ingle JN, Hoffman EW, Tanswell P, Ritter G, Cohen LS, Bette P, Arvay L, Amelsberg A, Vlock D, Rettig WJ, Old LJ. A Phase I dose-escalation study of sibrotuzumab in patients with advanced or metastatic fibroblast activation protein-positive cancer. Clin Cancer Res. 2003;9(5):1639–47. [PubMed] [Google Scholar]
- 39.Narra K, Mullins SR, Lee HO, Strzemkowski-Brun B, Magalong K, Christiansen VJ, McKee PA, Egleston B, Cohen SJ, Weiner LM, Meropol NJ, Cheng JD. Phase II trial of single agent Val-boroPro (Talabostat) inhibiting Fibroblast Activation Protein in patients with metastatic colorectal cancer. Cancer Biol Ther. 2007;6(11):1691–9. doi: 10.4161/cbt.6.11.4874. [DOI] [PubMed] [Google Scholar]


