Abstract
Increased oxidative stress with elevated levels of reactive oxygen and nitrogen species (ROS/RNS) plays an important role in the pathophysiology of many disease states. Increased ROS/RNS can modulate cellular macromolecules of DNA, lipids, and proteins, negatively affecting their normal functions. Numerous reports have described the properties and implications of oxidized DNA and lipids. However, oxidative modifications of proteins were not fully studied partially due to the requirement for specific reagents, the lack of the methods to detect, purify and identify oxidatively-modified proteins, and the relatively late development of highly-sensitive analytical instruments. This chapter describes the detailed procedure for systematically identifying oxidative-modified proteins in biological samples. Applications and other suggestions to this method are also described to understand the functional roles of oxidatively-modified proteins in promoting ER stress and mitochondrial dysfunction, which ultimately contribute to organ damage.
Keywords: Reactive Oxygen Species, Cysteine Oxidation, Functional Redox Proteomics
1. Introduction
It is generally accepted that increased oxidative/nitrosative stress plays an important role in promoting many disease states, including Alzheimer’s disease, alcoholism-related organ damage, cancer, cardiovascular diseases, chronic inflammation, diabetes, eye diseases, Huntington’s disease, kidney diseases, non-alcoholic liver diseases, Parkinson disease, sepsis, stroke, etc, although the etiological cause for each disease may be different. Exposure to potentially toxic chemicals, drugs and environmental contaminants or agents such as heavy metals, smoking and UV/irradiation can also produce increased levels of reactive oxygen/nitrogen species (ROS/RNS).
In most cases, disruption of the mitochondrial electron transport chain is known to produce large amounts of ROS (Lin and Beal, 2006). In addition to the ROS production through mitochondrial dysfunction, other enzymes are also known to produce ROS/RNS. These enzymes include: ethanol-inducible cytochrome P450 2E1 (CYP2E1), NADPH-oxidase, xanthine oxidase, and inducible nitric oxide synthase (iNOS) (Caro and Cederbaum, 2004; Kono et al., 2000; Purohit et al., 2009; Song et al., 1996). Increased ROS/RNS interact with cellular macromolecules such as DNA, lipid, and proteins to produce oxidized DNAs, lipid peroxides, and oxidized proteins, respectively, and usually negatively affect their physiological functions.
In the past, numerous investigators have reported the increased production of oxidized DNA and lipid peroxides in many pathological states (see reviews by Esterbauer et al., 1991; Minko et al., 2009; Rubbo and Radi, 2008; Wells et al., 2009). However, only a small number of reports systematically described the oxidized proteins under increased oxidative stress in many disease states. Part of the reason could be due to the requirement for specific reagents, the lack of suitable methods to systematically detect, purify, and identify oxidized proteins, and the relatively late development of highly-sensitive mass spectral instruments.
Several amino acids are known to be oxidized under increased oxidative/nitrosative stress. For instance, it is known that cysteine (Cys), glutamine (Glu), histidine (His), lysine (Lys), methionine (Met), tyrosine (Tyr), tryptophane (Trp), etc are oxidatively-modified and their physiological functions altered under oxidative/nitrosative stress (Berlett and Stadtman 1997; Stadtman et al., 2003). Oxidation of these amino acids in many enzymes usually leads to formation of carbonylated proteins (with His, Arg, Lys, Pro, Thr, etc) with concomitant irreversible inactivation of their catalytic activities (Hensley et al, 1995). In contrast, some oxidized cysteines (i.e., sulfenic acid and disulfides) and methionine-sulfoxides are known to be reversibly reduced to cysteine and methionine, respectively, under proper conditions (Stadtman et al., 2003) to regain their normal functions. Despite the possibilities of oxidative modifications of many other amino acids under increased oxidative/nitrosative stress, oxidation of Cys residues in various proteins was evaluated through redox-related Cys-targeted proteomics approaches (Baty et al., 2002; Kim et al., 2000; Sethuraman et al., 2004; Venkatraman et al., 2004), partly because of the availability of relatively specific agents [e.g., N-ethylmaleimide, iodoacetamide, or acid-cleavable isotope-coded affinity tag (ICAT) reagent] toward Cys residues and biotin-conjugated N-maleimide (biotin-NM) or biotin-iodoacetamide (BIAM).
Although a few Cys-targeted proteomics approaches have been reported (Kim et al., 2000; Sethuraman et al., 2004; Venkatraman et al., 2004), these methods using BIAM, 4-iodobutyl-triphenylphosphonium, or ICAT reagent may have disadvantages (or limitations) in detecting subtle increments in the amounts of oxidatively-modified proteins, primarily because the levels of oxidized proteins labeled with these reagents (e.g., BIAM, 4-iodobutyl-triphenylphosphonium, or ICAT) are inversely related to the increased levels of oxidative stress. To overcome these technical limitations (inconvenience), we have developed a simple, sensitive Cys-targeted biotin-switch method of using biotin-NM as a specific probe to systematically identify the oxidatively-modified protein thiols. The levels of the oxidized proteins were positively correlated with increased oxidative stress in alcohol-exposed E47 HepG2 hepatoma cells with over-expressed human CYP2E1 (Suh et al., 2004) and rodent tissues (Kim et al., 2006a; Moon et al., 2006).
As illustrated in Figure 1, we initially labeled the free Cys thiols with N-ethylmaleimide (NEM). After removing excess NEM by spinning through mini-spin Sephadex-G25 columns (Amersham Biosciences-GE Healthcare), we reduced the oxidized cysteines (sulfenic acid, disulfides, mixed disulfides with glutathione, S-nitrosylated Cys, etc) to free Cys thiols with dithiothreitol (DTT). The newly reduced sulfhydryl groups were then labeled with biotin-NM. After removing excess biotin-NM with the second Sephadex G25 mini-spin columns, we detected or affinity-purified biotin-labeled oxidized proteins with either streptavidin-agarose or monoclonal antibody to biotin-conjugated agarose. After washing the non-specifically bound proteins, agarose-bound biotin-NM-labeled oxidized proteins were dissolved and analyzed by 1-D SDS-polyacrylamide gel electrophoresis (PAGE) or 2-D PAGE for protein display and identification by mass spectrometric analysis (Suh et al., 2004). We believe that this Cys-targeted proteomics method for positively detecting oxidized proteins has a significant advantage over the previously described methods (Kim et al., 2000; Sethuraman et al., 2004; Venkatraman et al., 2004) in detecting small increments in oxidized proteins under increased oxidative stress.
Figure 1.
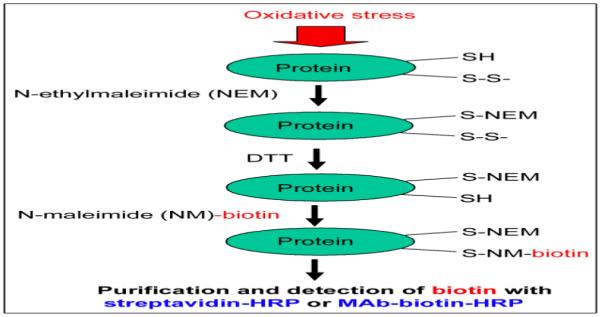
Schematic diagram to positively identify oxidized proteins by using a Cys-targeted biotin-switch method.
2. Materials
2.1 Chemicals and other materials
NEM, biotin-NM, DTT, CHAPS, and agarose-bound monoclonal antibody to biotin were obtained from Sigma Chemical (St. Louis, MO) in highest purity. Horse radish peroxidase (HRP)-conjugated streptavidin, streptavidin-agarose, and monoclonal antibody against biotin (MAb-biotin) were purchased from Molecular Probe (Eugene, OR). Protease inhibitor cocktail and phosphatase inhibitor cocktail were purchased from Calbiochem (San Diego, CA). Sephadex G25 mini-spin columns and immobilized pH gradient IEF gel strips (usually pH 3-10) were obtained from Amersham–Biosciences (Immobiline DryStrip from GE-Healthcare, Piscataway, NJ). Mass spectrometry-compatible silver stain was obtained from BioRad (Silver Stain Plus, Hercules, CA). Porcine sequencing grade modified trypsin was purchased from Promega (Madison, WI). Other reagents not mentioned here were the same as described (Kim et al., 2006a; Moon et al., 2006; 2008a; 2008b; Suh et al., 2004).
3. Methods
Actual procedures for identifying oxidatively-modified Cys residues with biotin-NM
Prepare proper buffer solutions freshly pre-equilibrated with nitrogen or argon gas for at least 30 min to remove oxygen dissolved in the extraction buffer. For isolating mitochondria, 250 mM sucrose should be included in the homogenizing buffer. However, no reducing agent such as DTT should be contained in the extraction buffer since it will interfere with the following biotin-switch method.
After removing the culture media, briefly rinse E47 HepG2 hepatoma cells with oxygen-free PBS buffer for 10 min 3 times before cell harvest by spinning for 5 min at 1,500 x g at 4 °C.
Homogenize the cells or tissue samples with cold STE buffer (250 mM sucrose, 50 mM Tris-Cl, pH 7.4 and 1 mM EDTA) with protease inhibitor cocktail and phosphatase inhibitor cocktail for 40 strokes with a glass-plastic homogenizer.
Spin whole cell extracts from control and ethanol-treated E47 HepG2 hepatoma cells or tissue homogenates from ethanol-exposed rodent livers for 10 min at 500 x g at 4 °C to collect plasma membrane, cell debris and nuclear fractions as pellets. Transfer the soluble fractions and spin at 9,000 x g for 15 min at 4 °C to prepare mitochondrial fractions (pellets) or cytoplasm (supernatant).
Transfer the soluble fractions into clean tubes and use them as cytosolic fraction (cytoplasm).
Rinse the crude nuclear fraction (#4) and mitochondrial fraction (#4) with at least three times with fresh STE buffer and then spin again at 13,000 x g for 10 min to remove contaminating any cytosolic proteins.
Incubate the nuclear and mitochondrial proteins with the buffer (40 mM Hepes, 50 mM NaCl, 1% CHAPS, 1 mM EDTA, 1 mM EGTA, protease and phosphatase inhibitor cocktails) for 15 min and then spin for 13, 000 x g to obtain the soluble proteins from the nuclear and mitochondrial fractions, respectively, as described (Kim et al., 2006b). Determine the protein concentration for each sample group.
For the biotin-switch labeling procedure, treat the same amounts of solubilized mitochondrial proteins (from each sample group) with 30 mM NEM for 20 min to block reduced thiols.
Gently spin NEM-treated protein samples (e.g., cytosolic or solubilized mitochondrial proteins from each group) through Sephadex G25 mini-spin columns pre-equilibrated with the 1% CHAPS containing buffer. Sephadex G25 mini-spin columns are pre-spun at 300 x g for 20 seconds to remove the equilibrium buffer. After removing the equilibrium buffer from the mini-spin columns, load the NEM-treated protein samples carefully onto the center of the Sephadex G25 beads (try to avoid loading proteins onto the side of the Sephadex beads in the mini-spin columns). Then spin the Sephadex G25 mini-spin columns with protein samples at 1,000 x g for 1 min to efficiently collect the proteins as column eluates without NEM, which should be retained on the Sephadex G25 beads.
Determine the protein concentration of the eluted proteins from the first Sephadex G25 mini-spin columns. Treat the same amount of eluted cytosolic or solubilized mitochondrial proteins with 5 mM DTT for 10 min to reduce any oxidized Cys residues (including sulfenic acid, disulfides and mixed disulfides with glutathione or nitrosylated thiols) to reduced thiols.
Incubate the protein samples with 7 mM biotin-NM for another 20 min to label freshly-reduced Cys residues with biotin-NM.
Purify the same amount of biotin-NM labeled proteins (from each sample group) through the second Sephadex G25 mini-spin columns to remove excess amounts of biotin-NM, as carefully as described above (#9).
For quick analysis, separate the biotin-NM labeled oxidized proteins on 1-D SDS-PAGE and subject them to immunoblot analysis with the specific monoclonal antibody against biotin or streptavidin-conjugated HRP.
To further characterize the identities of oxidatively-modified proteins, purify the biotin-NM labeled oxidized proteins with streptavidin-agarose beads (or agarose-bound monoclonal antibody to biotin). Wash the agarose-bound biotin-NM labeled proteins with the elution buffer at least twice to remove non-specifically bound proteins. Then dissolve biotin-NM labeled oxidized proteins in 2-D PAGE buffer (8 M urea, 20 mM DTT, 2% CHAPS, 0.5% IPG buffer, pH 3-10) for 30 min before isoelectrofocusing analysis on dry IPG strips for 24 h at 50,000 Vh, as recommended by the manufacturer. Stain the oxidized proteins resolved on 2-D gels with mass-spectrometry compatible Coomassie-blue or silver.
Pick the Coomassie-blue stained protein spots with new razor blades and analyze the protein identities by in-gel trypsin-digestion followed by mass-spectral analysis, as described (Blonder et al., 2004; Suh et al., 2004).
Confirm the presence of oxidized proteins with immunoblot analysis and activity measurements (Moon et al., 2005; 2007; Suh et al., 2004).
4. Discussion
By using the redox-related, Cys-targeted proteomics method, we expect to detect a greater number and intensity of biotin-NM-labeled oxidized proteins under increased oxidative stress compared to those found in control tissues, as exemplified in alcohol-exposed cells/tissues (Figure 2). This prediction was actually confirmed by data in ethanol-treated CYP2E1-containing E47 HepG2 hepatoma cells and rodent tissues (Suh et al., 2004). In fact, significant increases in the number and intensity of oxidatively-modified proteins were detected in the cytoplasm and mitochondria from ethanol-exposed mice or rats compared to that detected in pair-fed control rodents (Kim et al., 2006a; Moon et al. 2006; Song et al., 2008).
Figure 2.
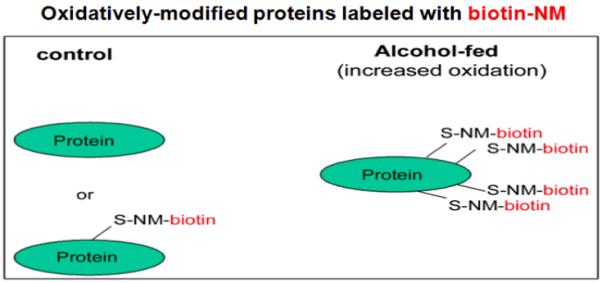
Comparison of biotin-NM labeled oxidatively-modified proteins in biological samples. The increased number of biotin-NM labeled oxidized proteins under elevated oxidative stress (in alcohol-exposed cells/tissues than control cells/tissues).
Furthermore, the increased number and intensity of oxidatively-modified proteins were also observed in non-alcohol induced liver damage such as ischemia-reperfusion hepatic injury (Moon et al., 2008a) or following exposure to 3,4-methylenedioxymethamphetamine (MDMA, ecstasy) (Moon et al., 2008b). We would thus expect to detect increased levels of oxidized proteins in many other tissues such as brain (Moon et al., unpublished observation) in various disease states.
4.1 Advantages of the simple redox-based Cys-targeted proteomics method
The redox-based, Cys-targeted approach exhibits multiple advantages over other existing methods:
1) Positive correlation between the levels of oxidized proteins and increased oxidative stress
It is known that oxidized Cys residues do not react with the sulfhydryl reagents such as BIAM and ICAT. Therefore, decreased efficiencies of labeling oxidized proteins with these methods would be expected. Unlike the other methods reported (Kim et al., 2000; Sethuraman et al., 2004; Venkatraman et al., 2004), where the number of oxidized proteins was inversely related to increased oxidative stress, the current redox-based proteomics method allows the positive identification of oxidized proteins. In fact, we observed a positive correlation between the increased number of the oxidatively-modified proteins and increased oxidative stress (Kim et al., 2006a; Moon et al., 2006; 2008a; Suh et al., 2004).
2) No requirements for special reagents
Instead of requiring a specific antibody to 4-iodobutyltriphenyl-phosphonium (Venkatraman et al., 2004) or a cysteine-specific ICAT reagent (Sethuraman et al., 2004), this biotin-switch method described here does not need special reagents. All reagents used in this method are available commercially and easy to obtain (Suh et al., 2004).
3) Functional proteomics analysis
The main objective of many proteomics approaches including 2-D Fluorescence Difference Gel Electrophoresis (2-D DIGE) system is to detect alterations in the expressed levels of many proteins in two different samples (e.g., treated and untreated controls). However, many proteins can be inhibited without significant quantitative differences, suggesting post-translational modifications including oxidative modifications of Cys residues (Kim et al., 2006a; Moon et al., 2006; 2008a). Identification of the oxidatively-modified proteins detected with the Cys-targeted redox proteomics approach allows us to predict functional implications (e.g., inhibition) of the oxidized proteins/enzymes even in the absence of any changes in protein contents. We can simply search for Cys residues in the active site(s) of each oxidized protein in the literature. For instance, mitochondrial 3-ketoacyl-CoA thiolase, the last enzyme in the mitochondrial β-oxidation pathway of fatty acids, contains two Cys residues and one His residue in its active site (Zeng and Li, 2004). Although its protein level was unchanged, we expected its inactivation through oxidative modifications of catalytic Cys residues under increased oxidative stress in alcohol-exposed animals (Moon et al., 2006; Song et al., 2008) and non-alcoholic liver injury models (Moon et al., 2008a; 2008b). In fact, the inhibition of 3-ketoacyl-CoA thiolase activity was correlated with fat accumulation measured by biochemical measurement of triglyceride levels (Moon et al., 2008a; 2008b; Song et al., 2008) and histological fat staining with oil-red O dye (Moon et al., 2006). The conservation of the active site Cys and His residues between mitochondrial and peroxisomal 3-ketoacyl-CoA thiolases can be used in predicting the inhibition of the peroxisomal enzyme under increased oxidative stress. Similarly, we also expect inhibition of the oxidatively-modified aldehyde dehydrogenase (ALDH) isozymes such as ALDH5 and ALDH7, although expressed in low levels, since all ALDH isozymes contain a highly conserved Cys residue in their active sites (as discussed in Moon et al., 2007).
4) Proteomics analysis for different sub-cellular organelles
By using this simple biotin-switch method, we can systematically identify oxidatively-modified proteins in different sub-cellular organelles (e.g., cytoplasm, mitochondria, ER, nuclear fractions, etc) to theoretically study the underlying mechanisms of redox-related cellular metabolism, ER stress, mitochondrial dysfunction and modulation of transcription factors, respectively. By studying the time-dependent oxidative modifications of various proteins and cell/tissue damage, we can generate many interesting hypotheses toward tissue damage. For instance, oxidative modifications and inactivation of endoplasmic reticulum (ER)-resident chaperone proteins including protein disulfide isomerase (PDI) may lead to unfolded client proteins of PDI and other chaperone proteins, leading to unfolded protein responses and ER stress (Kim et al., 2006a). In addition, by studying the time-dependent oxidation of mitochondrial proteins during ischemia-reperfusion liver injury, we reported that oxidative modifications of many mitochondrial proteins take place at much earlier than the actual tissue damage observed later (Moon et al., 2008a). These results suggest that mitochondrial dysfunction through oxidative inactivation of many mitochondrial proteins contributes to tissue damage.
5) Proteomics analysis for different tissues
In addition, we can identify oxidatively-modified proteins in different organs/tissues (e.g., liver, brain, kidney, heart, intestine, etc), depending on the target organs of interest (Moon et al., 2008b and unpublished data). By comparing the patterns of oxidative protein modifications in different tissues, we can estimate the role of specific proteins in each organ.
6) Proteomics analysis for different disease states
Oxidatively-modified proteins in different disease states (e.g. such as ischemia-reperfusion hepatic injury, diabetes, etc) or following exposure to potentially toxic drugs/chemicals (e.g., MDMA-exposed rat liver or brain tissues) or environmental contaminants where increased oxidative stress plays a major role in cellular toxicity (Moon et al., 2008a; 2008b), can be studied.
7) Proteomics analysis for detecting mixed disulfides
Increased production of peroxynitrate (PN) in the presence of ROS/RNS can actively react with free Cys residues to form nitrosothiols as well as nitrate Tyr residues (3-nitroTyr) of many proteins and affect their functions (Lane et al., 2001; Ottesen et al., 2001). By using mild reducing agents such as ascorbate (Asc) or glutathione (GSH) (Jaffrey et al., 2001; Kashiba-Iwatsuki, et al., 1997) instead of DTT in the second step in Figure 1, proteins with mixed disulfides (e.g., glutathionylation, succinylation, or S-nitrosylation) can be identified, as reported (Moon et al., 2006; 2008a).
8) Application in translational research
Finally, this method can be employed in translational studies by evaluating the effectiveness or progress of treatment with a certain beneficial agent (e.g., anti-oxidants or cell protective agents). This can be accomplished by monitoring the levels of oxidatively-modified proteins in the biological specimens before, during, and after treatment with a beneficial agent. For instance, a polyunsaturated fatty acid diet containing physiological levels of arachidonic and docosahexaenoic acids effectively prevented protein oxidation, mitochondrial dysfunction and ultimately alcoholic fatty liver (Song et al., 2008). Based on our data, it is expected that the beneficial effects of other anti-oxidants against many disease states can be demonstrated in future studies.
4.2 Limitations of the redox-based Cys-targeted proteomics and alternative approaches
1) Detection of oxidized proteins expressed in low levels
Despite many advantages, this Cys-targeted proteomics approach also has some limitations. One of the major disadvantages of this method is that it depends on the amount of target proteins expressed in a given cell/tissue. Common to all proteomics methods, the Cys-targeted redox proteomics approach only allows detection of oxidatively-modified proteins expressed in abundance, as we originally described (Suh et al. 2004). For instance, it is unlikely that this systematic approach could be successfully used in directly detecting oxidation of many DNA repair enzymes including O6-methylguanine-DNA-methyltransferase, which contains Cys in its active site and can be inhibited via S-nitrosylation (Laval and Wink, 1994). Although oxidation of O6-methylguanine-DNA-methyltransferase or other DNA repair enzymes such as OGG1 was not observed in our studies (Kim et al., 2006a; Moon et al., 2006; Suh et al., 2004), the failure to detect these proteins could be due to low expression levels of these proteins relative to other proteins in the liver.
A few other enzymes, that were not detected by our systematic Cys-targeted redox-proteomics approaches (Kim et al., 2006a; Moon et al., 2006) but confirmed for oxidative-modifications of active site Cys residues, may include: Rpn2, which is a subunit of 26S proteasome complex system (Zmijewski et al., 2009), methionine adenosyltransferase (Avila et al., 1998; Ruiz et al., 1998), mitogen-activated protein kinase phosphatases (Heneberg and Draber, 2005; Kim et al., 2003), and tumor suppressor protein PTEN (Lee et al., 2002). Oxidative-modifications of these proteins may not be easily detected by the Cys-targeted redox-proteomics approach described here due to low expression levels of these proteins. However, it is possible to successfully demonstrate oxidative modifications of these or other key enzymes/proteins (including some transcription factors) by immunoprecipitation with a specific antibody against each target protein and then immunoblot analysis with a specific antibody against Cys-S-NO, 3-nitrotyrosine, or glutathione. This alternative approach should be correlated with the activity measurement to further confirm functional implication of the oxidative modifications of the critical Cys or other amino acid residues.
2) Detection of covalent modifications of Cys residues
Another point for consideration is that Cys residues can undergo many different covalent modifications such as conjugation with carbonyl compounds such as acetaldehyde, acrolein, croton aldehyde, malondialdehyde, and 4-hydroxynonenal (4-HNE), all of which can be produced through lipid peroxidation under oxidative stress (Esterbauer et al., 1991; Catala, 2009). For instance, Cys residues of mitochondrial ALDH2 are known to directly interact with 4-HNE (Doorn et al., 2006) as well as conjugation with the metabolites of disulfiram, daunorubicin, and acetaminophen (discussed in Moon et al., 2009) with concomitant ALDH2 inactivation. The ALDH2 activity is also decreased in many pathological conditions such as alcoholic fatty liver, hepatic cancer, aging, and hepatic ischemia-reperfusion injury (as discussed in Moon et al., 2009). In most cases, the ALDH2 protein levels might not be altered, suggesting that the catalytic and other critical Cys residues of ALDH2 are likely oxidatively-modified and/or conjugated with 4-HNE or other reactive metabolites such as acetaminophen and disulfiram. The covalent modifications of Cys residues by the latter cases can be demonstrated by measuring the enzyme activity after incubation with a strong reducing agent DTT, which can not reverse the cysteine-carbonyl conjugates or drug-conjugation adducts. If the suppressed enzyme activities are recovered by the addition of DTT, the target proteins are likely to be oxidatively modified to sulfenic acid, S-nitrosylation, or disulfides (including mixed disulfides with glutathione). If the activities are not restored even after incubation with DTT, this likely represents the irreversible, covalent modifications (i.e., conjugation adducts) or hyper-oxidation of Cys residues to sulfinic/sulfonic acids. Alternatively, other amino acid residues of the target proteins may also be oxidatively-modified and thus contribute to irreversible inactivation of the target protein (Moon et al., 2009). Therefore, it is advised to consider many possible routes of oxidative modifications during interpretation of the data.
Acknowledgement
This research was supported by the intramural Program fund at the National Institute on Alcohol Abuse and Alcoholism. Part of this research was also supported by a grant for the Chronic Liver Disease Project (to B.J. Song) from the Center for Biological Modulators in South Korea. The authors are grateful to Drs. Sue-Goo Rhee and Klaus Gawrisch for their advice and support.
References
- Avila MA, Mingorance J, Martínez-Chantar ML, Casado M, Martin-Sanz P, Boscá L, Mato JM. Regulation of rat liver S-adenosylmethionine synthetase during septic shock: role of nitric oxide. Hepatology. 1997;25:391–396. doi: 10.1002/hep.510250222. [DOI] [PubMed] [Google Scholar]
- Baty JW, Hampton MB, Winterbourn CC. Detection of oxidant sensitive thiol proteins by fluorescence labeling and two-dimensional electrophoresis. Proteomics. 2002;2:1261–1266. doi: 10.1002/1615-9861(200209)2:9<1261::AID-PROT1261>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Blonder J, Conrads TP, Yu LR, Terunuma A, Janini GM, Issaq HJ, Vogel JC, Veenstra TD. A detergent- and cyanogen bromide-free method for integral membrane proteomics: application to Halobacterium purple membranes and the human epidermal membrane proteome. Proteomics. 2004;4:31–45. doi: 10.1002/pmic.200300543. [DOI] [PubMed] [Google Scholar]
- Caro AA, Cederbaum AI. Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu. Rev. Pharmacol. Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
- Catalá A. Lipid peroxidation of membrane phospholipids generates hydroxyalkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem. Phys. Lipids. 2009;157:1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Doorn JA, Hurley TD, Petersen DR. Inhibition of human mitochondrial aldehyde dehydrogenase by 4-hydroxynon-2-enal and 4-oxonon-2-enal. Chem. Res. Toxicol. 2006;19:102–110. doi: 10.1021/tx0501839. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- Heneberg P, Dráber P. Regulation of cys-based protein tyrosine phosphatases via reactive oxygen and nitrogen species in mast cells and basophils. Curr. Med. Chem. 2005;12:1859–1871. doi: 10.2174/0929867054546636. [DOI] [PubMed] [Google Scholar]
- Hensley K, Hall N, Subramaniam R, Cole P, Harris M, Aksenov M, Aksenova M, Gabbita SP, Wu JF, Carney JM, Lovell M, Markesbery WR, Butterfield DA. Brain regional correspondence between Alzheimer’s disease histopathology and biomarkers of protein oxidation. J. Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:L1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- Kashiba-Iwatsuki M, Kitoh K, Kasahara E, Yu H, Nisikawa M, Matsuo M, Inoue M. Ascorbic acid and reducing agents regulate the fates and functions of S-nitrosothiols. J. Biochem. (Tokyo) 1997;122:1208–1214. doi: 10.1093/oxfordjournals.jbchem.a021883. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Hood BL, Aragon RA, Hardwick JP, Conrads TP, Veenstra TD, Song BJ. Increased oxidation and degradation of cytosolic proteins in alcohol-exposed mouse liver and hepatoma cells. Proteomics. 2006a;6:1250–1260. doi: 10.1002/pmic.200500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Ryu SW, Song BJ. JNK- and p38 kinase mediated phosphorylation of Bax leads to its activation, mitochondrial translocation and apoptosis of human hepatoma HepG2 cells. J. Biol. Chem. 2006b;281:21256–21265. doi: 10.1074/jbc.M510644200. 2006. [DOI] [PubMed] [Google Scholar]
- Kim HS, Song MC, Kwak IH, Park TJ, Lim IK. Constitutive induction of p-Erk1/2 accompanied by reduced activities of protein phosphatases 1 and 2A and MKP3 due to reactive oxygen species during cellular senescence. J. Biol. Chem. 2003;278:37497–37510. doi: 10.1074/jbc.M211739200. [DOI] [PubMed] [Google Scholar]
- Kim JR, Yoon HW, Kwon KS, Lee SR, Rhee SG. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal. Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- Kono H, Rusyn I, Yin M, Gabele E, Yamashina S, Dikalova A, Kadiiska MB, Connor HD, Mason RP, Segal BH, Bradford BU, Holland SM, Thurman RG. NADPH oxidase-derived free radicals are key oxidants in alcohol-induced liver disease. J. Clin. Invest. 2000;106:867–872. doi: 10.1172/JCI9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane P, Hao G, Gross SS. S-nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation. Sci. STKE. 2001;2001:RE1. doi: 10.1126/stke.2001.86.re1. [DOI] [PubMed] [Google Scholar]
- Laval F, Wink DA. Inhibition by nitric oxide of the repair protein, O6-methylguanine-DNA-methyltransferase. Carcinogenesis. 1994;15:443–447. doi: 10.1093/carcin/15.3.443. [DOI] [PubMed] [Google Scholar]
- Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Minko IG, Kozekov ID, Harris TM, Rizzo CJ, Lloyd RS, Stone MP. Chemistry and biology of DNA containing 1,N(2)-deoxyguanosine adducts of the alpha,beta-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 2009;22:759–778. doi: 10.1021/tx9000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Abdelmegeed MA, Song BJ. Inactivation of cytosolic aldehyde dehydrogenase via S-nitrosylation in ethanol-exposed rat liver. FEBS Lett. 2007;581:3967–3972. doi: 10.1016/j.febslet.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Hood BL, Kim BJ, Hardwick JP, Conrads TP, Veenstra TD, Song BJ. Inactivation of oxidized and S-nitrosylated mitochondrial proteins in alcoholic fatty liver of rats. Hepatology. 2006;44:1218–1230. doi: 10.1002/hep.21372. [DOI] [PubMed] [Google Scholar]
- Moon KH, Hood BL, Mukhopadhyay P, Rajesh M, Abdelmegeed MA, Kwon YI, Conrads TP, Veenstra TD, Song BJ, Pacher P. Oxidative inactivation of key mitochondrial proteins leads to dysfunction and injury in hepatic ischemia reperfusion. Gastroenterology. 2008a;135:1344–1357. doi: 10.1053/j.gastro.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Kim BJ, Song BJ. Inhibition of mitochondrial aldehyde dehydrogenase by nitric oxide-mediated S-nitrosylation. FEBS Lett. 2005;579:6115–6120. doi: 10.1016/j.febslet.2005.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Lee YM, Song BJ. Inhibition of hepatic mitochondrial aldehyde dehydrogenase by carbon tetrachloride through JNK-mediated phosphorylation. Free Radic. Biol. Med. 2009 Nov 13; doi: 10.1016/j.freeradbiomed.2009.11.008. 2009. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Upreti VV, Yu LR, Lee IJ, Ye X, Eddington ND, Veenstra TD, Song BJ. Mechanism of 3,4-methylenedioxymethamphetamine (MDMA, ecstasy)-mediated mitochondrial dysfunction in rat liver. Proteomics. 2008b;8:3906–3918. doi: 10.1002/pmic.200800215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen L,H, Harry D, Frost M, Davies S, Khan K, Halliwell B, Moore K. Increased formation of S-nitrothiols and nitrotyrosine in cirrhotic rats during endotoxemia. Free Radic. Biol. Med. 2001;31:790–798. doi: 10.1016/s0891-5849(01)00647-5. [DOI] [PubMed] [Google Scholar]
- Purohit V, Gao B, Song BJ. Molecular mechanisms of alcoholic fatty liver. Alcohol. Clin. Exp. Res. 2009;33:191–205. doi: 10.1111/j.1530-0277.2008.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbo H, Radi R. Protein and lipid nitration: role in redox signaling and injury. Biochim. Biophys. Acta. 2008;1780:1318–1824. doi: 10.1016/j.bbagen.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Ruiz F, Corrales FJ, Miqued C, Mato JM. Nitric oxide inactivates rat hepatic methionine adenosyltransferase in vivo by S-nitrosylation. Hepatology. 1998;28:1051–1057. doi: 10.1002/hep.510280420. [DOI] [PubMed] [Google Scholar]
- Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol. Cell Proteomics. 2004;3:273–278. doi: 10.1074/mcp.T300011-MCP200. [DOI] [PubMed] [Google Scholar]
- Song BJ, Koop DR, Ingelman-Sundberg M, Nanji A, Cederbaum AI. Ethanol-inducible cytochrome P450 2E1: Biochemistry, molecular biology, and clinical relevance: 1996 update. Alcohol. Clin. Exp. Res. 1996;20:138A–146A. doi: 10.1111/j.1530-0277.1996.tb01764.x. [DOI] [PubMed] [Google Scholar]
- Song BJ, Moon KH, Olsson NU, Salem N., Jr. Prevention of alcoholic fatty liver and mitochondrial dysfunction in the rat by long-chain polyunsaturated fatty acids. J. Hepatol. 2008;49:262–273. doi: 10.1016/j.jhep.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER, Moskovitz J, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid. Redox Signal. 2003;5:577–582. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
- Suh SK, Hood BL, Kim BJ, Conrad TP, Veenstra TD, Song BJ. Identification of oxidized mitochondrial proteins in alcohol-exposed human hepatoma cells and mouse liver. Proteomics. 2004;4:3401–3412. doi: 10.1002/pmic.200400971. [DOI] [PubMed] [Google Scholar]
- Venkatraman A, Landar A, Davis AJ, Ulasova E, Page G, Murphy MP, Darley-Usmar V, Bailey SM. Oxidative modification of hepatic mitochondria protein thiols: effect of chronic alcohol consumption. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G521–G527. doi: 10.1152/ajpgi.00399.2003. [DOI] [PubMed] [Google Scholar]
- Wells PG, McCallum GP, Chen CS, Henderson JT, Lee CJ, Perstin J, Preston TJ, Wiley MJ, Wong AW. Oxidative stress in developmental origins of disease: teratogenesis, neurodevelopmental deficits, and cancer. Toxicol. Sci. 2009;108:4–18. doi: 10.1093/toxsci/kfn263. [DOI] [PubMed] [Google Scholar]
- Zeng J, Li D. Expression and purification of his-tagged rat mitochondrial 3-ketoacyl-CoA thiolase wild type and His352 mutant proteins. Protein Expr. Purif. 2004;35:320–326. doi: 10.1016/j.pep.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Zmijewski J, Banerjee S, Abraham E. S-Glutathionlyation of the Rpn2 regulatory subunit inhibits 26S proteosomal function. J. Biol. Chem. 2009;284:22213–22221. doi: 10.1074/jbc.M109.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]


