Abstract
Two different approaches can be used in phylogenomics: combined or separate analysis. In the first approach, different datasets are combined in a concatenated supermatrix. In the second, datasets are analyzed separately and the phylogenetic trees are then combined in a supertree. The supertree method is an interesting alternative to avoid missing data, since datasets that are analyzed separately do not need to represent identical taxa. However, the supertree approach and the corresponding consensus methods have been highly criticized for not providing valid phylogenetic hypotheses. In this study, congruence of trees estimated by consensus and supertree approaches were compared to model trees obtained from a combined analysis of complete mitochondrial sequences of 102 species representing 93 mammal families. The consensus methods produced poorly resolved consensus trees and did not perform well, except for the majority rule consensus with compatible groupings. The weighted supertree and matrix representation with parsimony methods performed equally well and were highly congruent with the model trees. The most similar supertree method was the least congruent with the model trees. We conclude that some of the methods tested are worth considering in a phylogenomic context.
Keywords: combined analysis, consensus, DNA sequences, phylogenomics, separate analysis, supermatrix
Introduction
The phylogenomic era has brought a shift from single to multiple datasets (or genes) to study phylogenetic relationships.1 While increasing the number of characters decreases stochastic errors, it also increases phylogenetic signal.2–4 However, phylogenomic studies present numerous methodological challenges (see review by Delsuc et al).5 Two opposite views have been proposed as to how to incorporate the growing amount of data to infer evolutionary relationships. Whereas the combined approach (sensu de Queiroz)6 concatenates different datasets in a supermatrix,7–10 the consensus approach (sensu de Queiroz)6 analyzes datasets separately and the resulting trees are combined with a consensus11–13 or a supertree method.14–16 The pros and cons of these competing approaches have been debated at length in the literature.10,17–25 When the combined approach is used, the concatenation of numerous genes from different species often results in a supermatrix with missing data. Indeed, a taxon bias has been observed in genetic databases, with a large number of genes (or whole genome) sequenced for a few key species thus resulting in large supermatrices dominated by missing data.2,23,24,26,27
An approach that can be applied to deal with incomplete matrices is the supertree method.14–16,22 In the likely event that some gene sequences are not available for all species, it is possible to estimate a phylogenetic tree for each gene separately and then combine the resulting trees with a supertree approach. Whereas separate datasets may not contain identical sets of taxa (i.e. only overlapping sets), the resulting supertree includes all taxa. Numerous supertree methods have been developed, the most familiar being the matrix representation with parsimony (MRP).28–30 They can be defined as a generalization of consensus methods, which only applies to trees defined on an identical set of taxa.6,11,12,20,31 Interestingly, it is possible to compare performances of classical consensus methods with that of supertree methods in a consensus setting, i.e. when all datasets have identical taxa.32 Since supertree methods are often developed from existing consensus methods, a setting that allows both types of methods to be directly compared is desirable to assess their relative accuracies.
The supertree strategy seems to be increasingly used in phylogenomics, where large amounts of data can be subdivided to facilitate phylogenetic analyses (i.e. divide-and-conquer strategy).22,33 Furthermore, supertree methods have been proposed as representing the optimal solution to reconstruct the Tree of Life.14,15,33,34 Consensus and supertree methods are similar in design, and both can be referred to as separate analyses, by opposition to a combined analysis (sensu de Queiroz)6 where sequence datasets are concatenated in a single supermatrix. A heated debate between those in favor and those opposed to the use of consensus has been raging for the last decade.18–20,35–37 The same debate has recently been extended to supermatrices and supertrees.7,10,14,16,17,23,25,38–40
The mammalian phylogeny has been extensively studied and the analysis of different sources of data leads to congruent phylogenies (see review by Springer and Murphy,41 Springer et al42 and Wildman et al.)43 Recent studies of mammal species have shown that mitochondrial phylogenies can be congruent to nuclear phylogenies when potential phylogenetic biases are removed. For example, additional taxa can be added to break long branches44 and compositional bias and heterogeneity in substitution rates can be appropriately handle.44–46 Another alternative is to use different substitution models depending on the codon position in order to account for a compositional bias of nucleotides among species.47 Numerous mitogenomic sequences of mammalian species are available and this data availability combined to accurate phylogenetic hypotheses is an ideal setting to test different phylogenetic approaches.
In this study, three different approaches that are commonly used in phylogenomics to analyze DNA sequence matrices were studied in a consensus setting (sensu Bininda-Emonds32): (1) topological consensus methods, (2) topological supertree methods, and (3) weighted supertree methods that account for branch lengths. Congruence among these competing approaches was compared with respect to model trees that were obtained from a complete matrix of mitogenomic mammalian sequences (i.e. a phylogenetic tree obtained from concatenated gene sequences) of 102 species representing 93 mammal families.
Methods
Model tree
DNA sequence alignments
In February 2009, 96 mammal families had at least one species with a complete mitochondrial (mt) sequence in GenBank. In this study, one taxon per family was chosen. When more than one taxon was available, only one species was selected per family (with a few exceptions, see below) and the entire mt sequence was downloaded (see Appendix 1 for Gen-Bank accession numbers). The twelve mitochondrial genes of the H-strand were aligned using ClustalX 2.0.1048 and the alignment was further verified by eye in SeAl 2.0a11. Ambiguous sites and overlapping regions of the ATP6–ATP8 and NAD4-NAD4L were removed.
Stationarity of base frequencies across taxa was tested on the complete alignment using the chi-square test of homogeneity of base frequencies implemented in PAUP* 4.0.49 A test of congruence among distance matrices (CADM)50 was used to determine the congruence among the twelve mt genes using R 2.9.0,51,52 and Ape 2.3 package,53,54 with 9999 permutations for significance testing.
A well-supported tree, compatible with current consensus of mammal molecular phylogeny,41,55 was required to represent interfamilial mitogenomic relationships. However, systematic errors, mainly caused by reconstruction artifacts, can produce a biased tree topology.5 Among potential systematic errors, heterogeneity in base composition47,56–59 and different evolutionary rates among species60–65 have often been cited as confounding factors affecting the inference of mammalian mitogenomic relationships. Indeed, preliminary analyses of our dataset revealed the presence of systematic biases and different strategies were used to reduce their effect.46,58 For one, the third codon position was removed since it evolves more rapidly, especially in the mitochondrial genome,56,66 and is often saturated for higher-level relationships.47 Also, the first codon position of leucine (C and T) was recoded as pyrimidine (Y). Additionally, three problematic species were removed (Anomalurus sp., Anomaluridae, Erinaceus europaeus, Erinaceidae and Manis tetradactyla, Manidae). These species are known to be affected by either reduced or accelerated evolutionary rates which can lead to long-branch attraction or positional uncertainty due to short branches.46,62,66–68 Finally, nine extra species were added to break long branches46 within the following six families: Chrysochloridae, Elephantidae, Macroscelidae, Procaviidae, Soricidae and Talpidae. Consequently, a total of 102 complete mt sequences, representing 93 mammalian families, were included (see Appendix 1).
Phylogenetic inference
Modeltest 3.7 was used to identify the best model of nucleotide substitution.69 Both the hierarchical likelihood ratio tests (hLRTs) and Akaike information criterion (AIC) suggested a general time-reversible model (GTR)70–72 following a gamma distribution (Γ)73 with invariant sites (I). The equilibrium frequencies of nucleotides A, C, G, and T were: gA = 0.3452, gC = 0.2054, gG = 0.0901, gT = 0.3593, the relative substitution rates were: rAC = 1.1083, rAG = 6.7749, rAT = 1.1934, rCG = 1.3020, rCT = 3.9717, rGT = 1.0000, and parameters α and I were 0.6762 and 0.4437 respectively. Phylogenetic trees were estimated using two different methods: maximum likelihood (ML)74,75 and Bayesian maximum likelihood (BML).76,77 ML analysis was performed with PhyML 3.0,78 with a GTR + Γ + I model, where base frequencies, proportion of invariable sites and gamma shape distribution parameters were estimated from the data. The number of categories for the gamma distribution was set to six. A subtree pruning and regrafting (SPR) algorithm was selected, starting from a BioNJ tree, and ten additional random starting trees. Non-parametric bootstrap support (BS) was assessed using identical settings in PhyML for 100 replicates. BML was performed with MRBAYES 3.1.276 on a shared-memory multiprocessor computer (Altix 4700). Two MCMC analyses were run for 5,000,000 generations each, using the same GTR + Γ + I model. The Metropolis coupling used eight chains, starting from a random tree and eight swaps with Markov chains sampled every 100th generation, and with a burn-in of 10%. The majority-rule consensus tree and Bayesian posterior probabilities (BPP) were obtained from the tree distribution.
Model tree topology
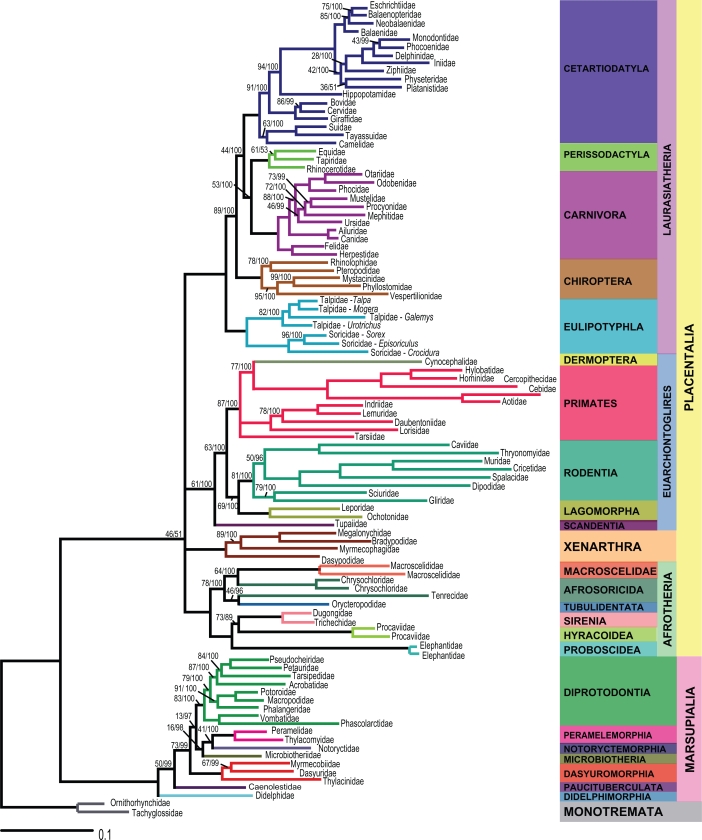
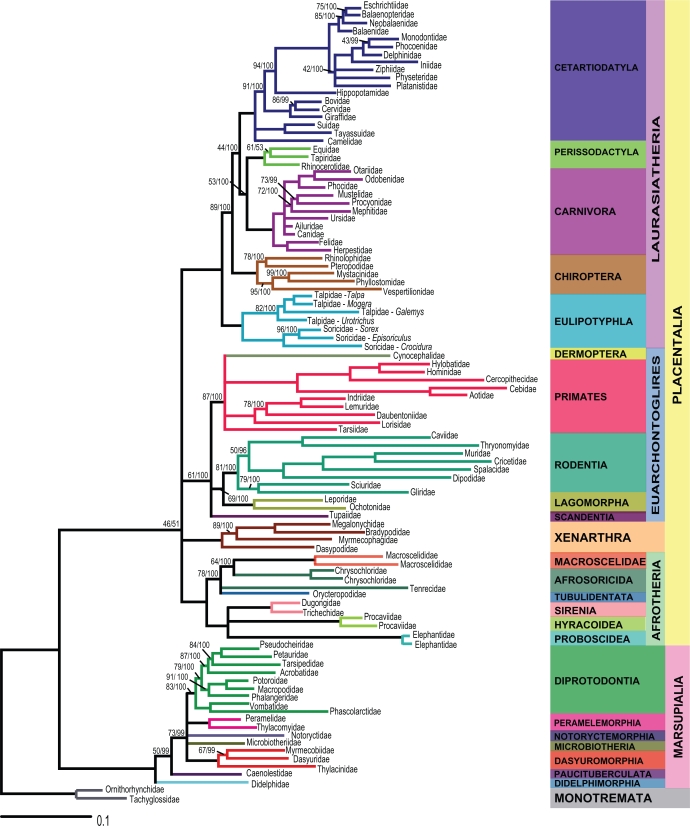
The ML and BML tree topologies were identical, except at two nodes. These incongruent clades were represented by polytomies in order to render the ML and BML trees completely congruent (Fig. 1). This topology was used as the first model tree (MT1). A second model tree (MT2) was then constructed by collapsing all branches that were not compatible to the current molecular consensus of mammal phylogeny. In this second tree (Fig. 2), eight extra polytomies were added to the first model tree to ensure that all clades were compatible with recent molecular studies.46,66,79–82
Figure 1.
First model tree (MT1) representing mitogenomic relationships among 93 mammalian families. Bootstrap values (BS) and Bayesian posterior probabilities (BPP) are indicated on branches (BS/BPP). Branches without values correspond to BS/BPP = 100/100.
Figure 2.
Second model tree (MT2) representing mitogenomic relationships among mammalian families with eight extra polytomies added to MT1 to obtain a tree compatible with recent molecular studies. Bootstrap values (BS) and Bayesian posterior probabilities (BPP) are indicated on branches (BS/BPP). Branches without values correspond to BS/BPP = 100/100.
Consensus and Supertree Methods
Individual datasets
For the consensus and supertree methods, the twelve individual mt genes were analyzed separately. Stationarity of base frequencies across taxa was tested on each of the twelve datasets using the chi-square test of homogeneity of base frequencies implemented in PAUP* 4.0, with a Bonferroni correction for multiple tests.83 Modeltest 3.7 was used to identify the best substitution model for each dataset. ML analyses were then performed on each dataset with PhyML 3.0, using the model suggested by the AIC criterion. Analytical parameters were identical to those described for the complete matrix analysis, except for the evolutionary model (listed in Table 1 for each gene). Individual gene trees are available upon request.
Table 1.
Statistical description of individual datasets (the twelve genes on the mitochondrial H-strand) and of concatenated datasets (AL). L (bp): length of the gene in base pairs. No cst: number of constant sites in the alignment. No info: number of informative sites in the alignment. AIC: Model selected according to AIC criterion in Modeltest, which always included parameters G (a gamma distribution of substitution rates) and I (a proportion of invariable sites). χ2 (1, 2): chi square test for homogeneity of base frequencies across species on datasets with third codon position removed (P = 1.0 in every case). χ2 (1, 2, 3): chi square test on datasets with codon positions 1, 2 and 3 included.
| Datasets | L (bp) | No cst (%) | No info (%) | AIC | χ2 (1, 2) | χ2 (1, 2, 3) |
|---|---|---|---|---|---|---|
| ATP6 | 452 | 200 (44.2) | 204 (45.1) | GTRa | 78.25 | 419.99* |
| ATP8 | 60 | 10 (16.7) | 47 (78.3) | TrNb | 145.87 | 198.83 |
| COX1 | 1022 | 780 (76.3) | 157 (15.4) | GTR | 15.85 | 573.75* |
| COX2 | 440 | 247 (56.1) | 150 (34.1) | TVMc | 28.26 | 290.95 |
| COX3 | 522 | 339 (64.9) | 137 (26.2) | TVM | 33.42 | 320.19 |
| CYTB | 754 | 402 (53.3) | 276 (36.6) | TIMd | 80.04 | 525.82* |
| NAD1 | 618 | 312 (50.5) | 239 (38.7) | GTR | 69.18 | 484.12* |
| NAD2 | 690 | 173 (25.1) | 463 (67.1) | GTR | 129.20 | 623.00* |
| NAD3 | 230 | 109 (47.4) | 100 (43.5) | TrN | 78.92 | 251.49 |
| NAD4 | 918 | 380 (41.4) | 458 (49.9) | GTR | 96.29 | 681.88* |
| NAD4 L | 192 | 69 (35.9) | 105 (54.7) | TVM | 65.08 | 251.49 |
| NAD5 | 1202 | 463 (38.5) | 619 (51.5) | GTR | 136.64 | 919.59* |
| ALL | 7100 | 3484 (49.1) | 2955 (41.6) | GTR | 267.78 | 4107.63* |
Given that all twelve datasets included an identical number of taxa (n = 102), the comparison of consensus and supertree methods was performed in a consensus setting.32 Therefore, even though we will maintain the use of “supertree” for methods that have been developed in a supertree context, all methods can be considered as consensus methods and can be divided into three categories: (1) consensus techniques based on topological relationships (topological consensus methods), (2) supertree techniques based on topological relationships (topological supertree methods), and (3) supertree techniques that take into account branch lengths (branch-length supertree methods).
Topological consensus methods
Four consensus methods were applied to combine the twelve independent gene trees in PAUP* 4.0: (1) strict, (2) majority rule (MR), (3) majority rule with compatible groupings (MRC), and (4) Adams consensus. The strict consensus only retains groups that are identical among all input trees.84,85 The majority rule consensus (MR) contains groups that are present in more than 50% of input trees,11,86 such that groups found in seven or more trees were kept. The second type of majority rule consensus (MRC) retains all compatible groupings below 50% of occurrence in addition to those above 50%. The Adams consensus presents groups that are nested within another without necessarily including identical taxa.87,88 Therefore, Adams consensus does not only propose monophyletic groups. A more complete description of these methods can be found in Swofford.11
Topological supertree methods
Three different optimality criteria were used to construct supertrees (consensus) from the twelve independent gene trees in CLANN 3.0.2:89 (1) matrix representation with parsimony (MRP), (2) most similar supertree (MSS), and (3) maximum splits fit (SFIT). In MRP, nodes present in each tree are coded into a binary matrix using the Baum and Ragan method.28–30 The binary matrix is then analyzed with a parsimony algorithm90 using ten TBR searches and a random starting tree. The MSS method calculates the symmetric differences between each gene tree and the supertree and sums these differences to obtain a supertree score.91 The optimal supertree is the one with the best score (smallest distance). For the SFIT method, the splits present in each gene tree and a candidate supertree are recorded and the supertree with the maximum split fit (sharing the greatest number of splits) is selected.89 For MSS and SFIT, a SPR heuristic search using ten repetitions each starting with a different NJ tree was selected to search among all possible supertree topologies (default parameters in CLANN). For all of these methods, a strict consensus was used to combine equally optimal supertrees, if any.
Branch-length supertree methods
Three other optimality criteria, which take into account branch lengths of input trees, were also employed: (4) average consensus (AC), (5) unweighted super-distance matrix (SDM), and (6) weighted super-distance matrix (SDMw). These methods are implemented in the SDM program.92 The AC criterion optimizes the sum-of-squared distances between each source tree and the consensus tree, by averaging the path-length distance matrices computed from each gene tree and then applying a least-squares algorithm to this average matrix.93 SDM applies the same criterion, except that pathlength distance matrices are first transformed so as to minimize the sum-of-squared distances among them.92 The weighted version (SDMw) assigns a weight to each tree prior to computing the average matrix, based on the sequence length of the corresponding gene. All supertrees (consensus) were estimated using an ordinary least squares algorithm94 in PHYLIP 3.6895 with the FITCH program, using the jumble option (J = 10) which randomizes the input order of species for each run, and with global rearrangements allowed (SPR algorithm).
Distance metrics
Two dissimilarity measures were computed in PAUP* 4.0 to quantify the congruence between model trees (MT1 and MT2) versus consensus and supertrees. The symmetric-difference or partition metric (PM) counts the number of different splits in the trees being compared.96,97 PM was normalized by dividing each value by its maximal possible value (2n−6), where n is 102 taxa. The maximum agreement subtree index (D1) calculates the number of taxa that need to be pruned from the trees to obtain a congruent topology.98–100 Here again, normalized D1 are obtained by dividing each value by its maximum possible value (n−3), where n is 102 taxa. Rohlf’s consensus information index (CII)101 was also calculated on the consensus trees to measure their relative resolution (this index ranges from 0 when the consensus is a bush to 1 when the tree is fully resolved).
Results
Individual datasets
The length (L) of each of the twelve aligned mitochondrial genes varied from 90 to 1803bp, when all three codon positions were included. The homogeneity test of base frequencies indicated that seven out of the twelve datasets were heterogeneous (Table 1). However, when only the two first codon positions were considered, all datasets were homogeneous. Consequently, subsequent analyses were performed using alignments with only the first and second codon positions. The two optimality criteria (hLRTs and AIC) implemented in Modeltest suggested different models for some datasets. Indeed, whereas the hLRTs criterion proposed a GTR model for all datasets, AIC suggested varying models depending on the dataset, as listed in Table 1. The congruence among distance matrix test (CADM) suggested that all twelve datasets were congruent (Friedman’s χ2 = 44341.5, Kendall’s W = 0.7175, P = 0.0001).
Topological consensus methods
Important differences were observed between PM and D1 and among topological consensus methods (Table 2). These results may be explained by the fact that some consensus methods were poorly resolved, i.e. CII = 0.02 for strict consensus, 0.10 for MR and 0.18 for Adams, compared to CII ranging from 0.53 to 1.0 for all other methods. The resolution level affects the congruence indices. When comparing a fully resolved tree to a bush, PM will be of 0.5 because only the clades in the fully resolved tree are contributing to the distance. On the other hand, D1, which calculates the number of taxa that have to be pruned from both trees to obtain identical topologies, will exhibit a very big value given that n−2 taxa need to be deleted for both topologies to be compatible. Because the majority rule consensus that included all compatible groupings (MRC) is more resolved than other classical consensus methods (CII = 0.94), it provided the best results and was the closest to model tree topologies (PM = 0.22–0.23 for MT1–MT2). The majority rule consensus (MR) was the second most congruent consensus method (PM = 0.30–0.25 for MT1–MT2), although much less resolved (CII = 0.10), and thus D1 was considerably increased (0.79–0.73 for MT1–MT2, compared to 0.39–0.46 for MRC).
Table 2.
Congruence of phylogenetic trees inferred from consensus and supertree methods (that ignore or consider branch lengths). CI: Rohlf’s consensus information index (ranges from 0 to 1; 0 being a bush and 1 a fully resolved tree), PM: partition metric, D1: maximum agreement subtrees. Indices range from 0 to 1. MR: majority rule consensus, MRC: majority rule consensus with compatible groupings, MRP: matrix representation with parsimony, MSS: most similar supertree, SFIT: maximum splits fit, AC: average consensus, SDM: unweighted super-distance matrix, SDMw: weighted super-distance matrix.
| CII |
Model tree 1 (MT1) |
Model tree 1 (MT2) |
||||
|---|---|---|---|---|---|---|
| PM | D1 | PM | D1 | |||
| Topological consensus methods | Strict | 0.02 | 0.46 | 0.94 | 0.39 | 0.94 |
| MR | 0.10 | 0.30 | 0.79 | 0.25 | 0.73 | |
| MRC | 0.94 | 0.22 | 0.39 | 0.23 | 0.46 | |
| Adams | 0.18 | 0.42 | 0.78 | 0.36 | 0.75 | |
| Topological supertree methods | MRPa | 0.98 | 0.23 | 0.47 | 0.23 | 0.43 |
| MSSb | 0.91 | 0.54 | 0.62 | 0.56 | 0.60 | |
| SFITc | 0.53 | 0.24 | 0.51 | 0.22 | 0.50 | |
| Branch-length supertree methods | AC | 1.00 | 0.25 | 0.43 | 0.22 | 0.49 |
| SDM | 1.00 | 0.27 | 0.45 | 0.24 | 0.50 | |
| SDMw | 1.00 | 0.30 | 0.45 | 0.27 | 0.50 | |
Strict consensus of five most parsimonious trees.
Strict consensus of two equally optimal supertrees.
Strict consensus of 184 equally optimal supertrees.
Supertree methods
The topological supertree techniques suggested more than one optimal supertree: 184 (SFIT), five (MRP), and two (MSS) optimal supertrees. These supertrees were combined using a strict consensus supertree, and thus, were not fully resolved (CII = 0.53 to 0.98, compared to a value of 1.0 for the branch-length supertree methods). The least congruent method was MSS (PM = 0.54–0.56 for MT1–MT2; D1 = 0.62–0.60 for MT1–MT2). MRP and SFIT performed well (for MRP: PM = 0.23 for MT1 and MT2 and D1 = 0.47–0.43; and for SFIT: PM = 0.24–0.22 and D1 = 0.51–0.50).
Weighted supertree methods that take branch lengths into account performed relatively well (PM range from 0.22 to 0.30, and D1 ranging from 0.43 to 0.50) and proposed one optimal, fully resolved supertree. AC was slightly more accurate than both SDM and SDMw. The weighted version of SDM (i.e. SDMw) did not improve phylogenetic performance (PM increased slightly while D1 remained the same).
Discussion
A method commonly used in large-scale studies is the construction of supertrees from individual source trees.14 Supertree methods combine trees that have overlapping taxon sets, whereas consensus methods summarize trees with identical taxon set. Both approaches have been extensively studied.6,18,32,92,102,103 They can be compared and tested in a consensus setting, where identical taxon sets are used.32,103–108
Among the consensus methods, the majority rule with compatible groupings (MRC) was the most congruent to MT1 (and second most congruent to MT2), when compared to all other consensus and supertree methods tested. Criticisms of consensus methods emphasized the poor resolution of consensus trees.9,18,36 However, MRC was well resolved (CII = 0.94), which may explain its performance relative to other topological consensus methods. Through simulations, Bininda-Emonds32 has also observed that MRC provided the highest accuracy amongst consensus methods.
In line with previous studies, we confirmed that supertree techniques based on topological relationships did not offer a fully resolved consensus tree. In general, most supertree methods gave similar results (except for MSS, see below). This result was surprising, given that numerous studies have proposed that methods accounting for branch lengths should provide more accurate supertrees.106–108 However, Criscuolo et al92 have shown that MRP (that do not account for branch lengths) and SDM (that do account for branch lengths) were equally accurate at low levels of missing data (i.e. 25% of deleted taxa), and that the benefit of accounting for branch lengths was only revealed at higher levels of missing data (e.g. 75% of deleted taxa). The consensus setting used in this study did not allow the investigation of the effect of missing data and therefore the difference in performance could have been seen in a supertree setting.
MSS was the least accurate of all supertree methods. Creevey and McInerney89 have compared their MSS approach to the AC technique, but without branch lengths (i.e. by setting all branch lengths equal to one). The better results obtained with AC (and SDM) with respect to MSS, might suggest that branch lengths contain information different from topological relationships, when supertree methods are used. A similar result was also observed by Criscuolo et al.92 As for the other supertree methods (SFIT and MRP), they provided similar results to techniques that use branch lengths. This result is consistent with studies that have shown that MRP is accurate under certain conditions.92,102,109,110 Through simulations, Bininda-Emonds and Sanderson102 have observed that MRP provided accuracy values comparable to those obtained from a supermatrix analysis (and that accuracy was slightly increased when a weighted MRP was used). Among the supertree methods that account for branch lengths, SDM outperformed slightly SDMw. The distance matrices are weighted according to sequence lengths in SDMw, with trees inferred from longer sequences contributing more to the “super” distance values. Thus, biases will be amplified if they are associated with longer sequence datasets. This might explain why SDMw might not always provide the optimal solution. Also, AC was slightly more accurate than SDM, in contrast with Criscuolo et al92 who showed the opposite under all conditions tested. Fitzpatrick et al109 in a fungal study comparing AC, MRP and the supermatrix approach, reported that AC might be prone to long-branch attraction, but this was not the case here. Supertree methods that account for branch lengths may thus provide additional information, which could help resolve some least-resolved clades.
The results from this study demonstrate that most of the supertree methods tested were highly congruent with both model trees. Interestingly, the majority rule consensus with compatible clades was also highly congruent, which suggest that it represents an accurate and fast approach to summarize information obtained in separate analyses.
Acknowledgments
For the computational resources, we would like to thank Marie-Hélène Duplain for granting access to the Laboratoire Interfacultaires de Micro-Informatique de l’Université de Montréal, outside opening hours. We would also like to thank Antoine Lapointe and Daniel Stubbs for their programming skills and the RQCHP (Réseau Québécois de Calcul de Haute Performance) for granting access to its HPC facilities for the Bayesian analyses. Also, we greatly appreciated the help of Stéphane Guindon for running some of the ML analyses. This study was supported by a FESP (Faculté des Études Supérieures de l’Université de Montréal) scholarship to VC and by NSERC grant OGP0155251 to FJL.
Supplementary data
Appendix 1.
GenBank accession numbers of complete mitochondrial DNA sequences from 102 species representing 93 mammalian families. Family and species taxonomy based on Wilson and Reeder.1
| Order | Family | Species | Complete |
|---|---|---|---|
| MONOTREMATA | Tachyglossidae | Tachyglossus aculeatus | NC_003321 |
| Ornithorhynchidae | Ornithorhynchus anatinus | NC_000891 | |
| DIDELPHIMORPHIA | Didelphidae | Didelphis virginiana | NC_001610 |
| PAUCITUBERCULATA | Caenolestidae | Caenolestes fuliginosus | NC_005828 |
| MICROBIOTHERIA | Microbiotheriidae | Dromiciops gliroides | NC_005826 |
| DASYUROMORPHIA | Thylacinidae | Thylacinus cynocephalus | NC_011944 |
| Myrmecobiidae | Myrmecobius fasciatus | NC_011949 | |
| Dasyuridae | Phascogale tapoatafa | NC_006523 | |
| PERAMELEMORPHIA | Thylacomyidae | Macrotis lagotis | NC_006520 |
| Peramelidae | Isoodon macrourus | NC_002746 | |
| NOTORYCTEMORPHIA | Notoryctidae | Notoryctes typhlops | NC_006522 |
| DIPROTODONTIA | Phascolarctidae | Phascolarctos cinereus | NC_008133 |
| Vombatidae | Vombatus ursinus | NC_003322 | |
| Phalangeridae | Trichosurus vulpecula | NC_003039 | |
| Potoroidae | Potorous tridactylus | NC_006524 | |
| Macropodidae | Macropus robustus | NC_001794 | |
| Pseudocheiridae | Pseudocheirus peregrinus | NC_006519 | |
| Petauridae | Petaurus breviceps | NC_008135 | |
| Tarsipedidae | Tarsipes rostratus | NC_006518 | |
| Acrobatidae | Distoechurus pennatus | NC_008145 | |
| XENARTHRA | Dasypodidae | Dasypus novemcinctus | NC_001821 |
| Bradypodidae | Bradypus tridactylus | NC_006923 | |
| Megalonychidae | Choloepus didactylus | NC_006924 | |
| Myrmecophagidae | Tamandua tetradactyla | NC_004032 | |
| PROBOSCIDEA | Elephantidae | Elephas maximus | NC_005129 |
| Loxodonta africana | NC_000934 | ||
| SIRENIA | Dugongidae | Dugong dugon | NC_003314 |
| Trichechidae | Trichechus manatux | NC_010302 | |
| HYRACOIDEA | Procaviidae | Procavia capensis | NC_004919 |
| Dendrohyrax dorsalis | NC_010301 | ||
| TUBULIDENTATA | Orycteropodidae | Orycteropus afer | NC_002078 |
| MACROSCELIDEA | Macroscelididae | Macroscelides proboscideus | NC_004026 |
| Elephantulus sp. | NC_004921 | ||
| AFROSORICIDA | Tenrecidae | Echinops telfairi | NC_002631 |
| Chrysochloridae | Chrysochloris asiatica | NC_004920 | |
| Eremitalpa granti | NC_010304 | ||
| CETACARTIODACTYLA | Balaenidae | Balaena mysticetus | NC_005268 |
| Balaenopteridae | Megaptera novaeangliae | NC_006927 | |
| Eschrichtiidae | Eschrichtius robustus | NC_005270 | |
| Neobalaenidae | Caperea marginata | NC-005269 | |
| Delphinidae | Lagenorhynchus albirostris | NC_005278 | |
| Monodontidae | Monodon monoceros | NC_005279 | |
| Phocoenidae | Phocoena phocoena | NC_005280 | |
| Physeteridae | Physeter catodon | NC_002503 | |
| Iniidae | Inia geoffrensis | NC_005276 | |
| Platanistidae | Platanista minor | NC_005275 | |
| Ziphiidae | Berardius bairdii | NC_005274 | |
| Suidae | Sus scrofa | NC_000845 | |
| Tayassuidae | Pecari tajacu | NC_012103 | |
| Hippopotamidae | Hippopotamus amphibius | NC_000889 | |
| Camelidae | Lama pacos | NC_002504 | |
| Giraffidae | Giraffa camelopardalis | NC_012100 | |
| Cervidae | Cervus elaphus | NC_007704 | |
| Bovidae | Bos taurus | NC_001567 | |
| PERISSODACTYLA | Equidae | Equus caballus | NC_001640 |
| Tapiridae | Tapirus terrestris | NC_005130 | |
| Rhinocerotidae | Ceratotherium simum | NC_001808 | |
| CARNIVORA | Ailuridae | Ailurus fulgens | NC_011124 |
| Ursidae | Ursus americanus | NC_003426 | |
| Canidae | Vulpes vulpes | NC_008434 | |
| Felidae | Felis catus | NC_001700 | |
| Herpestidae | Herpestes javanicus | NC_006835 | |
| Mustelidae | Gulo gulo | NC_009685 | |
| Otariidae | Eumetopias jubatus | NC_001050 | |
| Odobenidae | Odobenus rosmarus | NC_004029 | |
| Phocidae | Phoca vitulina | NC_001325 | |
| Procyonidae | Procyon lotor | NC_009126 | |
| Mephitidae | Mephitis mephitis | AY598529–AY598539, X94927 | |
| EULIPOTYPHLA | Soricidae | Crocidura russula | NC_006893 |
| Sorex unguiculatus | NC_005435 | ||
| Episoriculus fumidus | NC_003040 | ||
| Talpidae | Talpa europaea | NC_002391 | |
| Galemys pyrenaicus | NC_008156 | ||
| Mogera wogura | NC_005035 | ||
| Urotrichus talpoides | NC_005034 | ||
| CHIROPTERA | Pteropodidae | Pteropus dasymallus | NC_002612 |
| Vespertilionidae | Chalinolobus tuberculatus | NC_002626 | |
| Mystacinidae | Mystacina tuberculata | NC_006925 | |
| Rhinolophidae | Rhinolophus monoceros | NC_005433 | |
| Phyllostomidae | Artibeus jamaicensis | NC_002009 | |
| RODENTIA | Thryonomyidae | Thryonomys swinderianus | NC_002658 |
| Caviidae | Cavia porcellus | NC_000884 | |
| Gliridae | Myoxus glis | NC_001892 | |
| Sciuridae | Sciurus vulgaris | NC_002369 | |
| Dipodidae | Jaculus jaculus | NC_005314 | |
| Spalacidae | Nannospalax ehrenbergi | NC_005315 | |
| Cricetidae | Cricetulus griseus | NC_007936 | |
| Muridae | Mus musculus | NC_005089 | |
| LAGOMORPHA | Ochotonidae | Ochotona princeps | NC_005358 |
| Leporidae | Oryctolagus cuniculus | NC_001913 | |
| PRIMATES | Lemuridae | Lemur catta | NC_004025 |
| Indriidae | Propithecus coquereli | NC_011053 | |
| Daubentoniidae | Daubentonia madagascariensis | NC_010299 | |
| Lorisidae | Nycticebus coucang | NC_002765 | |
| Tarsiidae | Tarsius bancanus | NC_002811 | |
| Cebidae | Cebus albifrons | NC_002763 | |
| Aotidae | Aotus trivirgatus | AY250707 | |
| Cercopithecidae | Macaca mulatta | NC_005943 | |
| Hylobatidae | Hylobates lar | NC_002082 | |
| Hominidae | Pan troglodytes | NC_001643 | |
| DERMOPTERA | Cynocephalidae | Cynocephalus variegatus | NC_004031 |
| SCANDENTIA | Tupaiidae | Tupaia belangeri | NC_002521 |
Wilson DE, Reeder DM. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd ed. Baltimore, Maryland: The Johns Hopkins University Press; 2005.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425(6960):798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- 2.Driskell AC, Ané C, Burleigh JG, McMahon MM, O’Meara BC, Sanderson MJ. Prospects for building the tree of life from large sequence databases. Science. 2004;306(5699):1172–4. doi: 10.1126/science.1102036. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Loffelhardt W, et al. Monophyly of primary photosynthetic eukaryotes: Green plants, red algae, and glaucophytes. Curr Biol. 2005;15(14):1325–30. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 4.Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–9. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 5.Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6(5):361–75. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 6.de Queiroz A. For consensus (sometimes) Syst Biol. 1993;42(3):368–72. [Google Scholar]
- 7.Gatesy J, Baker RH, Hayashi C. Inconsistencies in arguments for the supertree approach: Supermatrices versus supertrees of Crocodylia. Syst Biol. 2004;53(2):342–55. doi: 10.1080/10635150490423971. [DOI] [PubMed] [Google Scholar]
- 8.Eernisse DJ, Kluge AG. Taxonomic congruence versus total evidence, and amniote phylogeny inferred from fossils, molecules, and morphology. Mol Biol Evol. 1993;10(6):1170–95. doi: 10.1093/oxfordjournals.molbev.a040071. [DOI] [PubMed] [Google Scholar]
- 9.Kluge AG, Wolf AJ. Cladistics: What’s in a word? Cladistics. 1993;9(2):183–99. doi: 10.1111/j.1096-0031.1993.tb00217.x. [DOI] [PubMed] [Google Scholar]
- 10.de Queiroz A, Gatesy J. The supermatrix approach to systematics. Trends Ecol Evol. 2007;22(1):34–41. doi: 10.1016/j.tree.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Swofford DL. When are Phylogeny Estimates from Molecular and Morphological Data Incongruent? In: Miyamoto MM, Cracraft J, editors. Phylogenetic Analyses of DNA Sequences. Oxford: Oxford University Press; 1991. pp. 295–333. [Google Scholar]
- 12.Farris JS, Källersjö M, Kluge AG, Bult C. Constructing a significance test for incongruence. Syst Biol. 1995;44(4):570–2. [Google Scholar]
- 13.Huelsenbeck JP, Bull JJ. A likelihood ratio test to detect conflicting phylogenetic signal. Syst Biol. 1996;45(1):92–8. [Google Scholar]
- 14.Sanderson MJ, Purvis A, Henze C. Phylogenetic supertrees: Assembling the trees of life. Trends Ecol Evol. 1998;13(3):105–9. doi: 10.1016/S0169-5347(97)01242-1. [DOI] [PubMed] [Google Scholar]
- 15.Bininda-Emonds ORP, Gittleman JL, Steel MA. The (Super)tree of life: Procedures, problems, and prospects. Annu Rev Ecol Syst. 2002;33:265–89. [Google Scholar]
- 16.Bininda-Emonds ORP. Trees versus characters and the supertree/supermatrix “paradox”. Syst Biol. 2004;53(2):356–9. doi: 10.1080/10635150490440396. [DOI] [PubMed] [Google Scholar]
- 17.Gadagkar SR, Rosenberg MS, Kumar S. Inferring species phylogenies from multiple genes: Concatenated sequence tree versus consensus gene tree. J Exp Zool Part B. 2005;304B(1):64–74. doi: 10.1002/jez.b.21026. [DOI] [PubMed] [Google Scholar]
- 18.de Queiroz A, Donoghue MJ, Kim J. Separate versus combined analysis of phylogenetic evidence. Annu Rev Ecol Syst. 1995;26:657–81. [Google Scholar]
- 19.Huelsenbeck JP, Bull JJ, Cunningham CW. Combining data in phylogenetic analysis: Reply. Trends Ecol Evol. 1996;11(8):335. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 20.Huelsenbeck JP, Bull JJ, Cunningham CW. Combining data in phylogenetic analysis. Trends Ecol Evol. 1996;11(4):152–8. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 21.Wiens JJ. Combining data sets with different phylogenetic histories. Syst Biol. 1998;47(4):568–81. doi: 10.1080/106351598260581. [DOI] [PubMed] [Google Scholar]
- 22.Bininda-Emonds ORP. The evolution of supertrees. Trends Ecol Evol. 2004;19(6):315–22. doi: 10.1016/j.tree.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Crandall KA, Buhay JE. Genomic databases and the tree of life. Science. 2004;306(5699):1144–5. doi: 10.1126/science.1106198. [DOI] [PubMed] [Google Scholar]
- 24.Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol Biol Evol. 2005;22(5):1246–53. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- 25.Nishihara H, Okada N, Hasegawa M. Rooting the eutherian tree: The power and pitfalls of phylogenomics. Genome Biol. 2007;8(9):R199. doi: 10.1186/gb-2007-8-9-r199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiens JJ. Missing data and the design of phylogenetic analyses. J Biomed Inform. 2006;39(1):34–42. doi: 10.1016/j.jbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Telford MJ. Resolving animal phylogeny: A sledgehammer for a tough nut? Dev Cell. 2008;14(4):457–9. doi: 10.1016/j.devcel.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Baum BR. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41(1):3–10. [Google Scholar]
- 29.Ragan MA. Matrix representation in reconstructing phylogenetic relationships among the Eukaryotes. Biosystems. 1992;28(1–3):47–55. doi: 10.1016/0303-2647(92)90007-l. [DOI] [PubMed] [Google Scholar]
- 30.Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol Phyl Evol. 1992;1(1):53–8. doi: 10.1016/1055-7903(92)90035-f. [DOI] [PubMed] [Google Scholar]
- 31.Miyamoto MM, Fitch WM. Testing species phylogenies and phylogenetic methods with congruence. Syst Biol. 1995;44(1):64–76. [Google Scholar]
- 32.Bininda-Emonds ORP. MRP Supertree Construction in the Consensus Setting. In: Janowitz M, Lapointe FJ, McMorris FR, Mirkin B, Roberts FS, editors. Bioconsensus. Providence: American Mathematical Society; 2003. pp. 231–42. [Google Scholar]
- 33.Wilkinson M, Cotton JA.Supertree Methods for Building the Tree of Life: Divide-and-Conquer Approaches to Large Phylogenetic Problems Hodkinson T, Parnell J, Waldren S.Towards the Tree of Life: Taxonomy and Systematics of Large and Species Rich Taxa CRC Press; Systematic Association special volume; 200661–75. [Google Scholar]
- 34.Bininda-Emonds ORP. Phylogenetic Supertrees: Combining Information to Reveal the Tree of Life. Dordrecht, the Nethelands: Kluwer Academic Publishers; 2004. [Google Scholar]
- 35.Chippindale PT, Wiens JJ. Weighting, partitioning, and combining characters in phylogenetic analysis. Syst Biol. 1994;43(2):278–87. [Google Scholar]
- 36.Barrett M, Donoghue MJ, Sober E. Against Consensus. Syst Zool. 1991;40(4):486–93. [Google Scholar]
- 37.Wiens JJ. Does adding characters with missing data increase or decrease phylogenetic accuracy? Syst Biol. 1998;47(4):625–40. doi: 10.1080/106351598260635. [DOI] [PubMed] [Google Scholar]
- 38.Bininda-Emonds ORP. Supertree construction in the genomic age. Method Enzymol. 2005;395:745–57. doi: 10.1016/S0076-6879(05)95038-6. [DOI] [PubMed] [Google Scholar]
- 39.Gatesy J, Matthee C, DeSalle R, Hayashi C. Resolution of a supertree/supermatrix paradox. Syst Biol. 2002;51(4):652–64. doi: 10.1080/10635150290102311. [DOI] [PubMed] [Google Scholar]
- 40.Springer MS, de Jong WW. Which mammalian supertree to bark up? Science. 2001;291(5509):1709–11. doi: 10.1126/science.1059434. [DOI] [PubMed] [Google Scholar]
- 41.Springer MS, Murphy WJ. Mammalian evolution and biomedicine: New views from phylogeny. Biol Rev. 2007;82(3):375–92. doi: 10.1111/j.1469-185X.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- 42.Springer MS, Burk-Herrick A, Meredith R, et al. The adequacy of morphology for reconstructing the early history of placental mammals. Syst Biol. 2007;56:673–84. doi: 10.1080/10635150701491149. [DOI] [PubMed] [Google Scholar]
- 43.Wildman DE, Uddin M, Opazo JC, Liu G, Lefort V, Guindon S, et al. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci (U S A) 2007;104:14395–400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes A, Gissi C, Catzeflis F, Nevo E, Pesole G, Saccone C. Congruent mammalian trees from mitochondrial and nuclear genes using Bayesian methods. Mol Biol Evol. 2004;21:397–403. doi: 10.1093/molbev/msh033. [DOI] [PubMed] [Google Scholar]
- 45.Phillips MJ, McLenachan PA, Down C, Gibb GC, Penny D. Combined mitochondrial and nuclear DNA sequences resolve the interrelations of the major Australasian marsupial radiations. Syst Biol. 2006;55:122–37. doi: 10.1080/10635150500481614. [DOI] [PubMed] [Google Scholar]
- 46.Arnason U, Adegoke JA, Gullberg A, Harley EH, Janke A, Kullberg M. Mitogenomic relationships of placental mammals and molecular estimates of their divergences. Gene. 2008;421(1–2):37–51. doi: 10.1016/j.gene.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 47.Gibson A, Gowri-Shankar V, Higgs PG, Rattray M. A comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol Biol Evol. 2005;22(2):251–264. doi: 10.1093/molbev/msi012. [DOI] [PubMed] [Google Scholar]
- 48.Higgins DG, Sharp PM. Clustal: A package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73(1):237–44. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 49.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony and Other Methods. Sunderland, MA: Sinauer Associates Inc; 1998. [Google Scholar]
- 50.Legendre P, Lapointe FJ. Assessing congruence among distance matrices: Single-malt Scotch whiskies revisited. Aust Nz J Stat. 2004;46(4):615–29. [Google Scholar]
- 51.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 52.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: 2009. [Google Scholar]
- 53.Paradis E. Analyses of Phylogenetics and Evolution with R. New York: Springer; 2006. [Google Scholar]
- 54.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 55.Springer MS, Stanhope MJ, Madsen O, de Jong WW. Molecules consolidate the placental mammal tree. Trends Ecol Evol. 2004;19(8):430–8. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 56.Montgelard C, Forty E, Arnal V, Matthee CA. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol Biol. 2008;8:321. doi: 10.1186/1471-2148-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Penny D, Hasegawa M. The platypus put in its place. Nature. 1997;387(6633):549–50. doi: 10.1038/42352. [DOI] [PubMed] [Google Scholar]
- 58.Reyes A, Gissi C, Catzeflis F, Nevo E, Pesole G, Saccone C. Congruent mammalian trees from mitochondrial and nuclear genes using Bayesian methods. Mol Biol Evol. 2004;21(2):397–403. doi: 10.1093/molbev/msh033. [DOI] [PubMed] [Google Scholar]
- 59.Schmitz J, Ohme M, Zischler H. The complete mitochondrial sequence of Tarsius bancanus: Evidence for an extensive nucleotide compositional plasticity of primate mitochondrial DNA. Mol Biol Evol. 2002;19(4):544–53. doi: 10.1093/oxfordjournals.molbev.a004110. [DOI] [PubMed] [Google Scholar]
- 60.Huttley GA, Wakefield MJ, Easteal S. Rates of genome evolution and branching order from whole genome analysis. Mol Biol Evol. 2007;24(8):1722–30. doi: 10.1093/molbev/msm094. [DOI] [PubMed] [Google Scholar]
- 61.Delsuc F, Scally M, Madsen O, Stanhope MJ, de Jong WW, Catzeflis FM, et al. Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting. Mol Biol Evol. 2002;19(10):1656–71. doi: 10.1093/oxfordjournals.molbev.a003989. [DOI] [PubMed] [Google Scholar]
- 62.Douzery EJP, Delsuc F, Stanhope MJ, Huchon D. Local molecular clocks in three nuclear genes: Divergence times for rodents and other mammals and incompatibility among fossil calibrations. J Mol Evol. 2003;57:S201–13. doi: 10.1007/s00239-003-0028-x. [DOI] [PubMed] [Google Scholar]
- 63.Lin YH, McLenachan PA, Gore AR, Phillips MJ, Ota R, Hendy MD, et al. Four new mitochondrial genomes and the increased stability of evolutionary trees of mammals from improved taxon sampling. Mol Biol Evol. 2002;19(12):2060–70. doi: 10.1093/oxfordjournals.molbev.a004031. [DOI] [PubMed] [Google Scholar]
- 64.Lin YH, Waddell PJ, Penny D. Pika and vole mitochondrial genomes increase support for both rodent monophyly and glires. Gene. 2002;294(1–2):119–29. doi: 10.1016/s0378-1119(02)00695-9. [DOI] [PubMed] [Google Scholar]
- 65.Philippe H. Rodent monophyly: Pitfalls of molecular phylogenies. J Mol Evol. 1997;45(6):712–5. [PubMed] [Google Scholar]
- 66.Kjer KM, Honeycutt RL. Site specific rates of mitochondrial genomes and the phylogeny of eutheria. BMC Evol Biol. 2007;7:8. doi: 10.1186/1471-2148-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horner DS, Lefkimmiatis K, Reyes A, Gissi C, Saccone C, Pesole G. Phylogenetic analyses of complete mitochondrial genome sequences suggest a basal divergence of the enigmatic rodent Anomalurus. BMC Evol Biol. 2007;7:16. doi: 10.1186/1471-2148-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bergsten J. A review of long-branch attraction. Cladistics. 2005;21(2):163–93. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 69.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 70.Tavaré S. Some probabilistic and statistical problems on the analysis of DNA sequences. Lect Math Life Sci. 1986;17:57–86. [Google Scholar]
- 71.Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20(1):86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- 72.Rodriguez F, Oliver JL, Marin A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142(4):485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- 73.Yang ZH. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol Biol Evol. 1993;10(6):1396–401. doi: 10.1093/oxfordjournals.molbev.a040082. [DOI] [PubMed] [Google Scholar]
- 74.Felsenstein J. Maximum-likelihood estimation of evolutionary trees from continuous charaters. Am J Hum Genet. 1973;25:471–92. [PMC free article] [PubMed] [Google Scholar]
- 75.Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981;17(6):368–76. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 76.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 77.Rannala B, Yang ZH. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J Mol Evol. 1996;43(3):304–11. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 78.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 79.Phillips MJ, Penny D. The root of the mammalian tree inferred from whole mitochondrial genomes. Mol Phyl Evol. 2003;28(2):171–85. doi: 10.1016/s1055-7903(03)00057-5. [DOI] [PubMed] [Google Scholar]
- 80.Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, et al. The delayed rise of present-day mammals. Nature. 2007;446(7135):507–12. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 81.Meredith RW, Westerman M, Case JA, Springer MS. A Phylogeny and timescale for marsupial evolution based on sequences for five nuclear genes. J Mamm Evol. 2008;15(1):1–36. [Google Scholar]
- 82.Meredith R, Westerman M, Springer M. A phylogeny of Diprotodontia (Marsupialia) based on five nuclear genes. Mol Phyl Evol. 2009 doi: 10.1016/j.ympev.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 83.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43(1):223–5. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 84.Sokal RR, Rohlf FJ. Taxonomic congruence in the Leptopodomorpha reexamined. Syst Zool. 1981;30(3):309–25. [Google Scholar]
- 85.Page RDM. Comments on component-compatibility in historical biogeography. Cladistics. 1989;5(2):167–82. doi: 10.1111/j.1096-0031.1989.tb00563.x. [DOI] [PubMed] [Google Scholar]
- 86.Margush T, McMorris FR. Consensus n-trees. B Math Biol. 1981;43(2):239–44. [Google Scholar]
- 87.Adams EN. Consensus techniques and the comparison of taxonomic trees. Syst Zool. 1972;21:390–7. [Google Scholar]
- 88.Adams EN. N-trees as nestings: Complexity, similarity, and consensus. J Class. 1986;3(2):299–317. [Google Scholar]
- 89.Creevey CJ, McInerney JO. Clann: Investigating phylogenetic information through supertree analyses. Bioinformatics. 2005;21(3):390–2. doi: 10.1093/bioinformatics/bti020. [DOI] [PubMed] [Google Scholar]
- 90.Edwards AWF, Cavalli-Sforza LL. The reconstruction of evolution. Ann Hum Genet. 1963;27:105–6. [Google Scholar]
- 91.Creevey CJ, Fitzpatrick DA, Philip GK, Kinsella RJ, O’Connell MJ, Pentony MM, et al. Does a tree-like phylogeny only exist at the tips in the prokaryotes? P Roy Soc B-Biol Sci. 2004;271(1557):2551–8. doi: 10.1098/rspb.2004.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Criscuolo A, Berry V, Douzery EJP, Gascuel O. SDM: A fast distancebased approach for (super) tree building in phylogenomics. Syst Biol. 2006;55(5):740–55. doi: 10.1080/10635150600969872. [DOI] [PubMed] [Google Scholar]
- 93.Lapointe FJ, Cucumel G. The average consensus procedure: Combination of weighted trees containing identical or overlapping sets of taxa. Syst Biol. 1997;46(2):306–12. [Google Scholar]
- 94.Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis: Models and estimation procedures. Evolution. 1967;32:550–70. doi: 10.1111/j.1558-5646.1967.tb03411.x. [DOI] [PubMed] [Google Scholar]
- 95.Felsenstein J. PHYLIP: Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–6. [Google Scholar]
- 96.Robinson DF, Foulds LR. Lecture Notes in Mathematics. Berlin: Springer-Verlag; 1979. Comparisons on weighted labelled trees; pp. 119–26. [Google Scholar]
- 97.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math Biosci. 1981;53(1–2):131–47. [Google Scholar]
- 98.Finden CR, Gordon AD. Obtaining common pruned trees. J Classif. 1985;2(2–3):255–76. [Google Scholar]
- 99.Goddard W, Kubicka E, Kubicki G, Mcmorris FR. The agreement metric for labeled binary trees. Math Biosci. 1994;123(2):215–26. doi: 10.1016/0025-5564(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 100.Gordon AD. On the Assessment and Comparison of Classifications. In: Tomassone R, editor. Analyse de Données et Informatique. Le Chesnay, France: I.N.R.I.A.; 1980. pp. 149–60. [Google Scholar]
- 101.Rohlf FJ. Consensus indices for comparing classifications. Math Biosci. 1982;59(1):131–44. [Google Scholar]
- 102.Bininda-Emonds ORP, Sanderson MJ. Assessment of the accuracy of matrix representation with parsimony analysis supertree construction. Syst Biol. 2001;50(4):565–79. [PubMed] [Google Scholar]
- 103.Wilkinson M, Cotton JA, Creevey C, Eulenstein O, Harris SR, Lapointe FJ, et al. The shape of supertrees to come: Tree shape related properties of fourteen supertree methods. Syst Biol. 2005;54(3):419–31. doi: 10.1080/10635150590949832. [DOI] [PubMed] [Google Scholar]
- 104.Wilkinson M, Cotton JA, Lapointe FJ, Pisani D. Properties of supertree methods in the consensus setting. Syst Biol. 2007;56(2):330–7. doi: 10.1080/10635150701245370. [DOI] [PubMed] [Google Scholar]
- 105.Lapointe FJ. For Consensus (with Branch Lengths) In: Rizzi A, Vichi M, Bock H-H, editors. Advances in Data Science and Classification. Berlin: Springer-Verlag; 1998. pp. 73–80. [Google Scholar]
- 106.Lapointe FJ, Kirsch JAW, Hutcheon JM. Total evidence, consensus, and bat phylogeny: A distance-based approach. Mol Phyl Evol. 1999;11(1):55–66. doi: 10.1006/mpev.1998.0561. [DOI] [PubMed] [Google Scholar]
- 107.Levasseur C, Lapointe FJ. War and peace in phylogenetics: A rejoinder on total evidence and consensus. Syst Biol. 2001;50(6):881–91. doi: 10.1080/106351501753462858. [DOI] [PubMed] [Google Scholar]
- 108.Levasseur C, Lapointe FJ. Total evidence, average consensus and matrix representation with parsimony: What a difference distances make. Evol Bioinform. 2006;2:1–5. [PMC free article] [PubMed] [Google Scholar]
- 109.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:1–15. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Higdon JW, Bininda-Emonds ORP, Beck RMD, Ferguson SH. Phylogeny and divergence of the pinnipeds (Carnivora:Mammalia) assessed using a multigene dataset. BMC Evol Biol. 2007;7:216. doi: 10.1186/1471-2148-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tamura K, Nei M. Estimation of the number of nucleotide sustitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]