Abstract
The constant accumulation of sequence data poses new computational and methodological challenges for phylogenetic inference, since multiple sequence alignments grow both in the horizontal (number of base pairs, phylogenomic alignments) as well as vertical (number of taxa) dimension. Put aside the ongoing controversial discussion about appropriate models, partitioning schemes, and assembly methods for phylogenomic alignments, coupled with the high computational cost to infer these, for many organismic groups, a sufficient number of taxa is often exclusively available from one or just a few genes (e.g., rbcL, matK, rDNA). In this paper we address scalability of Maximum-Likelihood-based phylogeny reconstruction with respect to the number of taxa by example of several large nested single-gene rbcL alignments comprising 400 up to 3,491 taxa. In order to test the effect of taxon sampling, we employ an appropriately adapted taxon jackknifing approach. In contrast to standard jackknifing, this taxon subsampling procedure is not conducted entirely at random, but based on drawing subsamples from empirical taxon-groups which can either be user-defined or determined by using taxonomic information from databases. Our results indicate that, despite an unfavorable number of sequences to number of base pairs ratio, i.e., many relatively short sequences, Maximum Likelihood tree searches and bootstrap analyses scale well on single-gene rbcL alignments with a dense taxon sampling up to several thousand sequences. Moreover, the newly implemented taxon subsampling procedure can be beneficial for inferring higher level relationships and interpreting bootstrap support from comprehensive analysis.
Keywords: RAxML, phylogenetic inference, many taxon analyses, taxon jackknifing
Introduction
At present phylogenetic inference using statistical models of evolution has come of age and several novel, fast, and accurate likelihood-based phylogenetic inference programs such as GARLI,1 RAxML,2–4 IQPNNI,5 PHYML,6,7 Tree-Finder,8 using maximum likelihood (ML), or Bayesian programs like for instance MrBayes9,10 have become available. One key question that arises is up to which number of taxa they scale on single-gene alignments with respect to accuracy because of the comparatively weak phylogenetic signal.
Despite the increasing popularity of phylogenomic analyses comprising up to 150 genes11 (see also refs.12–16 for recent examples of such studies), which has stimulated a growing controversy about their assembly17–19 and the choice of appropriate models as well as partitioning schemes,20,21 the inference of trees based on large single-gene alignments, e.g.,22 still remains an important issue for two reasons: Firstly, for many organismic groups comprehensive sequence data that provide a sufficiently dense taxon sampling are only available for commonly used gene markers such as rbcL for green plants, a small number of mitochondrial genes for animals, and the large subunit and small subunit ribosomal RNA genes for various unicellular organisms. Secondly, the addition of genes (increased gene sampling) leads to extremely “gappy” alignments that typically contain more than 70% of gaps due to unsampled genes. For instance, the datasets used by McMahon and Sanderson12 exhibit a gappyness of 90%. Thus, only little supplementary signal is provided by addition of more genes18 at the expense of significantly larger data matrices and a scarce taxon sampling. The information content of the data does not increase linearly with the alignment length, and more importantly, the computational cost. The high computational cost is generated by the extremely large inference times and memory footprints of the alignments under the widely used23 General Time Reversible (GTR) model24 of nucleotide substitution combined with the gamma (Γ) model of rate heterogeneity. 25 Analyses of such datasets typically require supercomputing resources like the IBM BlueGene/L or the SGI Altix26,27 which are difficult to exploit for most taxonomists (biologists). Current phylogenomic analysis projects with RAxML required 89 GB of main memory and 2.25 million CPU hours on an IBM BlueGene/L supercomputer. In addition, there is a realistic chance that such large-scale analyses on supercomputers might be limited by high energy costs in the near future. On the other hand, the recent introduction of a rapid Bootstrapping (BS)28 algorithm in RAxML version 7.0.429 allows for full, i.e., more than 100 BS replicates and a thorough search for the best-scoring ML tree, large-scale phylogenetic analyses on single-gene datasets of more than 1,000 taxa within a couple of days on a modern desktop computer. While the accuracy of phylogenetic reconstruction depends on the sequence length of the alignment30,31 and ML is consistent if the sequence length goes to infinity,32 it still remains crucial to explore the scalability limits for current single-gene alignments because of the aforementioned reasons. Due to the immense computational resource requirements in terms of random access memory and number of CPUs there is a clear trade-off: One can either compute trees with many taxa, e.g.,22,33 or with many genes, e.g.,14 but not both, i.e., one needs to choose between dense taxon sampling and dense gene sampling.
Finally, the discussion on the impact of appropriate taxon sampling34 on results of phylogenetic analyses tends to be neglected, despite recent findings that phylogeny reconstruction is more susceptible to incomplete taxon than to incomplete gene sampling (see ref.35 for a review).
Another problem that can potentially be resolved by increasing the number of terminal accessions is the selection (and taxon density) of outgroup(s) used in phylogenetic studies. Outgroup selection may bias subtree topologies by placing a false root, e.g.,36 which can be prevented by using all available (and “alignable”) sequence data to assemble a dense outgroup that contains multiple organisms.
Due to the undertaken systematic efforts by studies of single- or few-gene genealogies based on dense taxon samples as well as on multigene phylogenies, many (terminal) clades in contemporary systematics can be considered to be well established. For instance, this is the case for the currently accepted families and orders of angiosperms,37–44 among others. However, some interclade relationships remain unresolved.14,38,42,43 Thus, although one is not necessarily interested in obtaining additional support to merely confirm well-established clades, the usage of placeholder taxa for such clades can potentially bias results because of insufficient taxon sampling and hence a lack of sufficient phylogenetic signal from these clades. The impact of taxon sampling on the inferred topology has been demonstrated in several cases, in particular for angiosperms.34,45,46
We therefore address the important issue of scalability of ML to large single-gene alignments via an empirical study of the widely used rbcL gene for which currently over 26,000 sequences are available (NCBI GenBank query, 08/17/2008). We assembled several large alignments for eudicots (excluding euasterids, represented by c. 3,500 additional rbcL sequences), rosids, eurosids I, and eurosids II, containing 3,490, 2,259, 1,590, and 436 taxa respectively. The fact, that the rosids are a subset of the eudicots and that eurosids I and II are nested within the rosids, allowed us to assess the scalability of ML on these datasets. In addition to these comprehensive large-scale analyses, we also examined an alternative approach that uses Group-based Randomized Taxon Subsampling (GRTS) and allows for comparisons of trees with varying taxon numbers while maintaining the relative taxon composition. In contrast, the commonly known taxon jackknifing approach as introduced by Lanyon47 applies agnostic taxon subsampling; this leads to distinct sets of leaves in each jackknife replicate, which hinders computation of consensus trees and bipartition frequencies using currently available software. Also, Lanyon47 only removed a single leaf per replicate, which rather corresponds to a leave-one-out experiment. Hence, his approach is not well-suited for improving scalability. We here compare topologies as well as bipartition frequencies obtained via GRTS, which is primarily used as a vehicle to assess scalability of ML, to those obtained by straightforward comprehensive analyses of the aforementioned datasets.
The main objective of this paper is to assess how good comprehensive ML analyses (combined tree inference and bootstrapping analyses) using RAxML scale up to alignments that contain several thousand taxa by example of single rbcL genes. We use RAxML as a typical representative of modern ML-based algorithms, that, similarly to GARLI and the most recent version of PHYML, deploys an implementation of lazy SPR (Subtree Prunung and Re-grafting) moves. While RAxML, GARLI, and PHYML typically yield trees that are not significantly different from each other, based on the standard statistical significance tests, RAxML on average returns trees with the best-known likelihood values on datasets of more than 1,000 to 2,000 taxa. Nonetheless, the results obtained here with RAxML, are qualitatively very similar to those that could be obtained via GARLI or PHYML, because all programs deploy comparable search mechanisms.
The results of the rbcL analyses are compared to previous results from the literature in the on-line supplements. We demonstrate that: (i) ML scales well on large single-gene matrices. (ii) GRTS using well-established groups as TU deserves further investigation to assess its potential for resolving higher level phylogenetic relationships. (iii) Analyses of densely sampled rbcL data are informative and in good agreement with “multigene analyses”.
Material and Methods
RbcL alignment assembly
Gene bank data was accessed in spring 2007 via the NCBI GenBank taxonomy portal. Searches and downloads of rbcL data were conducted at the ordinal level as provided by the NCBI taxonomy. Comprehensive alignments were assembled as follows: Initially, subalignments for each order (or several small orders as well as taxa not included in current orders) were constructed using the Clustal V algorithm as implemented in MegAlign (DNA Star Software package, LaserGene, Madison, WI, USA). These subalignments were visually inspected for apparent sequence artifacts: the rbcL is highly length-conserved among angiosperms, hence, any gap (or additional base) can be considered to be an artifact or to represent a pseudogene. Sequences containing a high degree of artifacts were eliminated. For the use with GRTS, the sequence name labels were transformed into a 5-digit code, followed by the gene bank accession number. The first three letters of the 5-digit code indicate the family or genus, to accommodate taxa that have not been assigned to a family, sensu APG II;39 the last two letters designate the order (or family or genus) sensu APG II, amended by information retrieved from the Angiosperm Phylogeny Website38 (APW) and additional web and print resources on taxonomy provided via APW. (Labeling was not automated since the current NCBI taxonomy contains several inconsistencies at the family and order level compared to taxonomic-systematic resources such as APG II and APW. A respective list of such inconsistencies can be found in the online appendix.) Note that, the coding reflects the systematic affinity of the organism and not the sequence, hence, we did not correct for misnamed/mislabeled sequences (see Results) at this stage of the analysis process. In a second step, the subalignments were successively merged (nested) into more comprehensive alignments, which were then used to conduct the phylogenetic analyses. The alignments contain the entire rbcL data for eurosids I (EURO1 matrix), eurosids II (EURO2), rosids (ROSID), and eudicots except euasterids (EUDIS). All respective NEXUS and PHYLIP alignment files are available for download at http://wwwkramer.in.tum.de/exelixis/rbcl.tar.bz2.
Comprehensive ML analyses
Comprehensive analyses of the full datasets were conducted with RAxML-VI-HPC version 2.2.3. (The most recent version 7.2.6 was not available at the time the phylogenetic inferences for this paper were conducted.) For each alignment we inferred 100 BS trees and conducted 20 ML searches to determine the best-scoring ML tree on distinct randomized stepwise addition MP starting trees using GTR and the CAT approximation of rate heterogeneity.48 This approximation serves as an efficient computational workaround for the significantly more memory- and floating point-intensive standard Γ model of rate heterogeneity. The CAT approximation simply provides a means to rapidly navigate into portions of the topological search space where tree topologies score well under GTR + Γ. The computations were conducted on the CIPRES (Cyberinfrastructure for Phylogenetic Research project, http://www.phylo.org) project cluster located at the San Diego Supercomputer Center (SDSC) that is equipped with 16 8-way 2.4 Ghz AMD Opteron nodes and on the infiniband cluster located at the Technische Universität München (TUM) that comprises 36 4-way 2.4 GHz AMD Opteron nodes. We denote the BS support values obtained from the comprehensive analyses as CA-BS.
Finally, we used the rapid Bootstrapping algorithm implemented in RAxML version 7.0.429 in combination with a Perl-script to assess the effect and applicability of the double Bootstrap procedure49 on the large 3,490 sequence alignment. Here we used the naïve approach, i.e., we computed 100 second-level BS analyses on 100 first-level BS replicates which amount to a total of 10,000 BS analyses. This analysis was carried out on 128 CPUs located at the Technische Universität München over a weekend. This first assessment of a double bootstrap procedure on a large single-gene dataset was included as an alternative to GRTS to evaluate whether it can be used to improve support values for such large analyses.
Group-based randomized taxon subsampling (GRTS) analyses
The group-based randomized taxon subsampling (GRTS) procedure was implemented via appropriate Perl-scripts. As mentioned above, taxon names in the alignments were assigned in a way such that grouping information, a taxonomic unit (TU), is encoded by certain characters in the taxon name. This grouping information was then used to reduce alignment sizes via directed taxon jackknifing with various alignment size reduction factors ranging from 1/2, 1/4, 1/8, down to 1/64 (where applicable) of the original number of taxa (procedure illustrated in Fig. 1). For instance, an alignment of 1,024 taxa will successively be reduced via taxon jackknifing to sizes of 512, 256, 128, ..., 16 taxa. However, since we conduct group-based taxon jackknifing (based on the meta-information contained in the sequence names) in order to maintain the taxon diversity and composition during the reduction process, every taxonomic group will be reduced proportionally to the number of representatives in the original alignment. For example, if a group has 256 members in an original 1,024 sequence alignment, it will successively be reduced to 128, 64, 32, ..., 4 members in the GRTS samples. This means that the jackknifing process is not completely conducted at random, but maintains the structure of the original alignment in terms of its taxonomic breadth and composition. In addition, the reduction factor was limited in such a way that at least two sequences were sampled per predefined group. The assignment of orders in addition to families as TU (Taxonomic Unit) ensured that the majority of TU comprised enough members to allow for application of large reduction factors such as 1/32 or 1/64. For each alignment and each applicable reduction factor we computed 100 replicates. For each of those 100 replicates we inferred 10 ML trees using the GTR + CAT approximation and determined the respective best-scoring tree under GTR + Γ. Thus, for every reduction factor and dataset, we computed 100 best-scoring ML trees for distinct randomized group-based subsamples. Finally, in order to investigate the effect of the GRTS approach on bipartition support values, we also conducted 100 standard BS replicates for only 10 out of 100 GRTS replicates per reduction factor (1,000 trees per dataset and reduction factor), in order to keep the computational requirements within acceptable limits. However, for a reduction factor of 1/32 we also computed 100 BS replicates on all 100 GRTS replicates (a total of 10,000 trees per dataset) to assess the effect of using all 100 GRTS replicates instead of only 10. This effect was, however, negligible. We carried out BS analyses of GRTS replicates for the three smaller alignments (EURO1, EURO2, ROSID) with reduction factors of 1/4, 1/8, ..., 1/64 (were applicable). The rationale behind the GRTS approach is to reduce those large alignments to subalignments with a significantly lower and hence, more favorable number of taxa for ML-based analyses with respect to the signal in the data and the NP-hard optimization problem, while maintaining the relative taxon composition. This can be regarded as zooming into the alignment while maintaining the relative taxon composition. This allows us to better assess the scalability of the tree search algorithms and conduct topological comparisons with respect to the placement of well-established groups. At the same time this provides a mechanism to assess the change (decrease/increase) of BS support values with increasing number of taxa. Henceforth, we denote GRTS support values obtained from the ML-searches on replicates (100 trees for 100 GRTS replicates) as GRTS-ML and GRTS support values obtained from 100 BS replicates on 10 GRTS replicates as GRTS-BS.
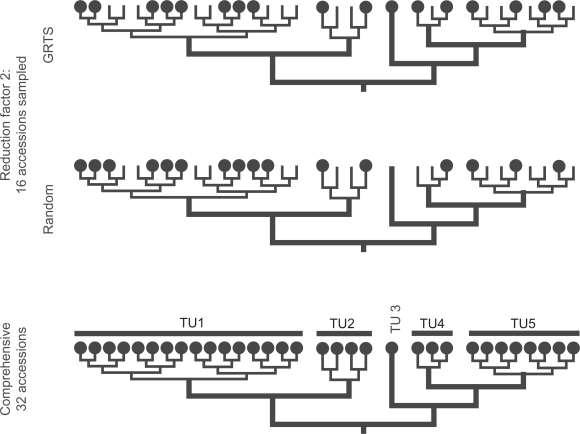
Figure 1.
Scheme illustrating the GRTS procedure (this study) in comparison to random taxon jackknifing.47 In contrast to random jackknifing, GRTS assures that each replicate includes always members of all pre-defined TU (in the given example TU 3 is missing in the random jackknife replicate) and that each TU is sampled proportionally to its original size. For instance, TU 1 includes 16 accessions. Thus, using a reduction factor of 2 each GRTS replicate will include exactly 8 members of TU 1. The number in random jackknife replicates may vary, resulting in an over- (TU 1 in given example) or underrepresentation of TUs (TUs 4 and 5).
Result analyses: comparing trees with distinct sets of leaves
We implemented three distinct tree comparison methods to extract comparable bipartition support values from the respective collections of trees:
a comparison (CA-BS with CA-BS) of the nested trees obtained via the comprehensive analyses
a comparison of GRTS-ML with GRTS-BS replicates among each other
a comparison of GRTS-ML/GRTS-BS replicates with CA-BS replicates
In order to compare CA-BS values from the comprehensive analyses with different nested sets of leaves, e.g., a eudicot tree with a rosid tree, leaves not present in the less comprehensive rosid tree were pruned from the larger eudicot phylogeny in all CA-BS replicates. “Nested” means in this context that all taxa in the smaller trees and alignments are also contained in the respective larger and more comprehensive trees. With respect to the taxon label sets (TLS) induced by the trees and alignments we have:
To prune the trees we used our own script newick.tcl that is freely available at http://www.goeker.org/mg/distance/. This script is equivalent to the DELETE/PRUNE command in PAUP*.50
The comparison of trees inferred via GRTS-BS and GRTS-ML is slightly more complicated because each taxonomic unit (TU) that is used for sampling is represented by a distinct set of sequence name labels in every GRTS replicate. Moreover, the representatives of a TU, which reflect an accepted angiosperm order or family, are not necessarily recognized as being monophyletic based on rbcL data alone. To reduce each TU to a single leaf representing this TU in every tree, that is, to extract the “big picture”, we used the following algorithm: Each homogeneous subtree that only comprises members of a single TU is reduced to a single leaf and the number of leaves in this homogeneous subtree is stored. If there is more than a single subtree per TU, i.e., if the TU is not monophyletic within the tree, all homogeneous subtrees for this TU except the largest one are pruned. The rationale for this is that the largest homogeneous subtree of a TU is most likely the best representative for the specific TU and will most probably contain true (e.g., non-mislabeled) members of this TU. On the other hand small and deviating clades, most likely comprise sequences from misidentified specimens or from wetlab artifacts. For comparison of GRTS-BS/GRTS-ML replicates with trees from comprehensive CA-BS analyses, the CA-BS trees as well as GRTS-BS and GRTS-ML were reduced to TU trees using the same topological reduction algorithm.
If several equally large (maximum size) homogeneous subtrees exist for a TU, one of them is chosen at random. The rationale for this random selection is that GRTS, like bootstrapping, includes random decisions and that the deviations induced by applying the above algorithm will average out if a sufficiently large number of GRTS samples is used for comparison.
This topology reduction algorithm is also implemented in the newick.tcl script, which includes a flexible mechanism to recognize the TU assignments of sequences that are encoded in their labels.
After application of these transformations for the three types of comparisons (CA-BS versus CA-BS, GRTS-BS versus GRTS-ML, GRTS-ML/GRTS-BS versus CA-BS) to the respective replicates in order to obtain collections of trees with consistent leave sets of equal size, we computed the Pearson correlation coefficient ρ between all bipartition frequencies, induced by the respective replicate sets (via the respective RAxML command line switch −f m). In addition, we computed the Pearson correlation coefficient, the slope, and the offset using an appropriate Perl script and the respective RAxML option (−f b) on the respective best-scoring ML trees with support values. This allowed us to compare scalability of support values induced by CA-BS analyses on the best-scoring ML trees of the respective smaller nested datasets. Finally, we computed the extended majority rule consensus tree (a bifurcating tree) for CA-BS replicates using consense from the PHYLIP package.
Visualization
We used the following tools to visualize the results of the comprehensive ML and BS, as well as GRTS-BS and GRTS-ML trees, analyses: Dendroscope51 (version 0.22) was used to draw and color the comprehensive ML trees; modules implemented in SplitsTree52 (versions 4.8 and 4.10) were used to analyze the bipartition patterns observed in the CA-BS and GRTS-BS/GRTS-ML analyses: The consensus network approach53 allowed us to visualize the distinct sets of tree replicates: CA-BS and GRTS-BS/GRTS-ML replicates were used to reconstruct splits graphs in which the length of each edge reflected the frequency of the respective bipartition in a sample of input trees (“bipartition network”, edge weight set to count);54 or in which the length of each edge corresponded to the mean branch length over all replicates (“confidence network”, edge weight set to mean).55 In the latter case, only those edges are represented in the splits graph occurring in more than a defined percentage of the replicate trees. For example, a 25% confidence network represents all splits that occur in more than 25% of the (BS/GRTS) replicates.
Results and Discussion
Scalability of nested comprehensive ML analyses
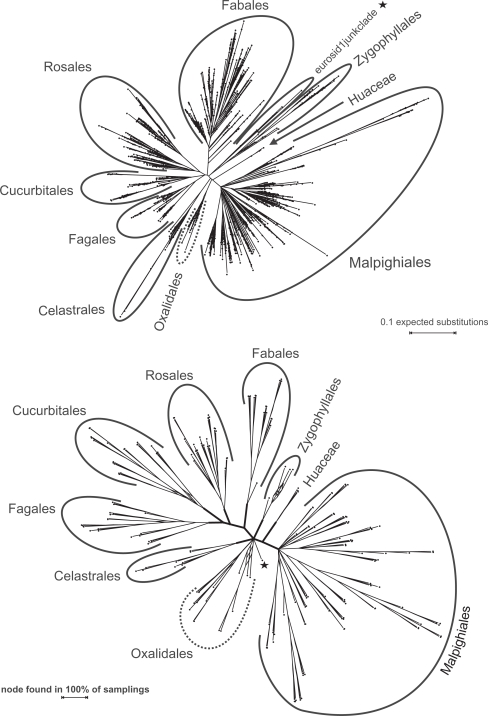
Figure 2 shows a condensed representation (family-level) of the ML tree inferred from the EUDIS matrix; the full tree as well as trees inferred from the other three matrices can be accessed via http://www.kramer.in.tum.de/rbcl.html. (This link provides also the complete results from the CA-BS, GRTS-BS, and GRTS-ML analyses.) Most orders and families represented by more than one rbcL accession (sensu APG II; in total 143 taxa) formed clades in the comprehensive ML trees and received moderate to high BS support by CA-BS, which highlights the systematic value of rbcL sequences for angiosperm systematics at the ordinal and subordinal level.37,44,56–59 Exceptions were mainly due to mislabeled sequences or sequences representing organisms of controversial systematic affiliation. (Details are provided in Online Supplement [OS] 1.) Our rbcL data does not support the monophyly (CA-BS ≥ 50; misplaced or controversial sequences not considered) of 20 families and one genus (Nelumbo; monotypic Nelumbaceae) that have been defined as TUs (putatively monophyletic according to APG II and APW; details provided in Online Supplement [OS] 2). However, members of such TUs occasionally formed clades in the best-known ML trees as well as in the majority of GRTS-based ML trees (see below). Of the 31 currently accepted order-level TUs (orders and unplaced families) covered by our data, 25 received moderate to high CA-BS support (Table S1 in OS 2).
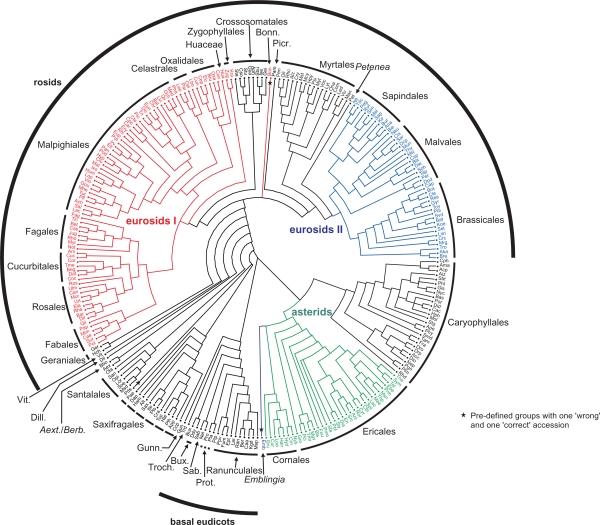
Figure 2.
A family-level representation of the best-known ML tree inferred from the EUDIS matrix. The basic tree includes more than 3,500 leaves and has here been reduced to family-level TU (see Material and Methods); the latter have also been used for the GRTS analyses (following chapter).
The Pearson correlation coefficient ρ between CA-BS values from the distinct nested datasets, calculated after pruning leaves down to the leave set of the smaller tree from the respective larger and more comprehensive tree via newick.tcl, are shown in Table 1. For the CA-BS analyses we do not prune down trees to TUs, but just prune down the respective larger trees to the taxon set of the nested smaller trees. Column # bipartitions in Table 1 provides the number of bipartitions induced by the respective (pruned-down) tree collections. Column ρ-best provides the correlations of the support values on the respective smaller, best-scoring tree, e.g., the correlation between CA-BS values of the pruned-down eudicot replicates and the rosid replicates, drawn on the best-scoring rosid ML tree. The computed slope and the offset for the comparisons of support values on the best-scoring trees showed only insignificant variations. The slope lies between 1.0019 and 0.9861, while the offset varies between −0.4087 and 1.8442. The average support on the respective best-scoring ML trees for the four datasets (original support values and pruned-down support values) is highly stable regardless of the number of taxa, and varies between 59.90 and 61.29. This also holds for the average support (80.98 and 81.66) on the respective extended majority rule (binary) consensus trees extracted from the replicates. The higher average support on CA-BS consensus trees compared to the best-scoring ML trees is not surprising, since the extended majority rule consensus algorithm in essence just maximizes the average support for a given set of bipartitions. In general, correlations appear to slightly decrease with increasing differences in the number of leaves, but are nonetheless very high (minimum: 0.987 for all bipartitions; 0.982 for bipartitions on best-scoring tree). In addition, the number of bipartitions induced by the pruned-down trees from the respective large datasets is slightly higher (2.5%) than for the nonpruned datasets.
Table 1.
Correlation between bootstrap support values for comprehensive analyses and number of bipartitions in the respective tree collections of nested rbcL datasets.
| Data sets | # taxa | ρ-all | # bipartitions | ρ-best | WRF |
|---|---|---|---|---|---|
| Eudicots ⊇ | 3,490 | 0.989 | Eudicots: 31,124 | 0.986 | 0.021 |
| rosids | 2,259 | Rosids: 30,338 | |||
| Eudicots ⊇ | 3,490 | 0.987 | Eudicots: 22,060 | 0.982 | 0.032 |
| eurosids I | 1,590 | Eurosids I: 21,688 | |||
| Eudicots ⊇ | 3,490 | 0.988 | Eudicots: 6,110 | 0.983 | 0.048 |
| eurosids II | 436 | Eurosids II: 5,894 | |||
| Rosids ⊇ | 2,259 | 0.993 | Rosids: 22,060 | 0.993 | 0.019 |
| eurosids I | 1,590 | Eurosids I: 21,983 | |||
| Rosids ⊇ | 2,259 | 0.992 | Rosids: 6,110 | 0.990 | 0.053 |
| eurosids II | 436 | Eurosids II: 6,019 |
The overall high correlation coefficients show that the number of taxa in the alignments, which reflects the taxonomic breadth of the data set, has little effect on the bipartitions induced by the BS analyses. In other words, the support of a node defining any eurosid I clade X, is not influenced by inclusion of eurosids II, other rosids, or other eudicots in the alignment. This is in agreement with the observation that, with respect to branch support, a large and dense outgroup provides a better subtree rooting than a sparse outgroup.36 However, there is a slightly higher correlation among the rosids, eurosids I, and eurosids II data sets than within the eudicots data set. The inclusion of most eudicots (outgroups from the perspective of eurosids I, eurosids II, and rosids) has some, although small, effect on the BS support of nodes within the rosids, eurosids I, and eurosids II subclades (Tables S1–S3 in OS 2). There are three possible explanations: First, it might be that the effect of the less favorable number of taxa to number of base pair ratio becomes more prevalent.30,31 As a consequence, ‘correct’ relationships, according to the rbcL genealogy, are less resolved. Second, exactly the reverse phenomenon might occur: the eurosids I, eurosids II, and rosids analyses might have yielded support for ‘incorrect’ bipartitions, which were correctly resolved in the eudicots analyses due to the more comprehensive taxon sampling. Third, the support of a few isolated nodes may vary significantly depending on the underlying data. A case-by-case investigation of changing support with reference to the well established phylogenetic framework for Angiosperms shows that all potential explanations apply (OS 2).
The general agreement between the inferred topologies and current knowledge (outlined in Figs. 2, 3), the comparably high CA-BS support of commonly accepted nodes (OS 2), and the general high correlation (Table 1) indicates that ML scales well in the studied case of large rbcL data sets. However, deeper (interordinal or backbone) and well-supported relationships based on several to many genes, e.g.,14,43,44 generally lack CA-BS support from rbcL alone (Table S3 in OS 2). Only the nitrogen fixing clade, received moderate BS support (Fig. 3; CA-BSAll matrices = 54).
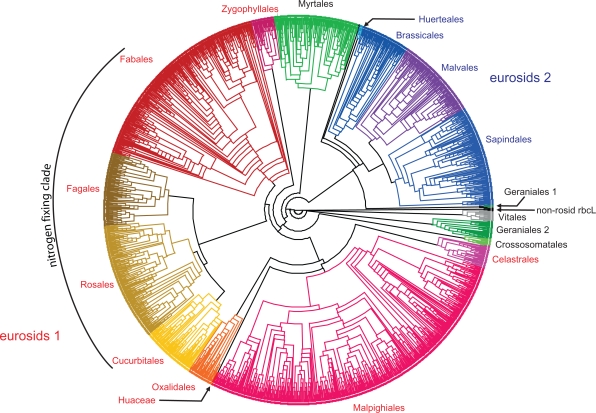
Figure 3.
A circle cladogram of the best-known ML tree inferred from the ROSI D matrix. By far the most sequences are placed according to well-known clades (occasional mislabeled sequences not addressed). Note that the CA-BS support of these higher order clades is often low (<50; Tables S2, S3 in OS 2).
The bootstrap of the bootstrap on 3,491 eudicots rbcL sequences
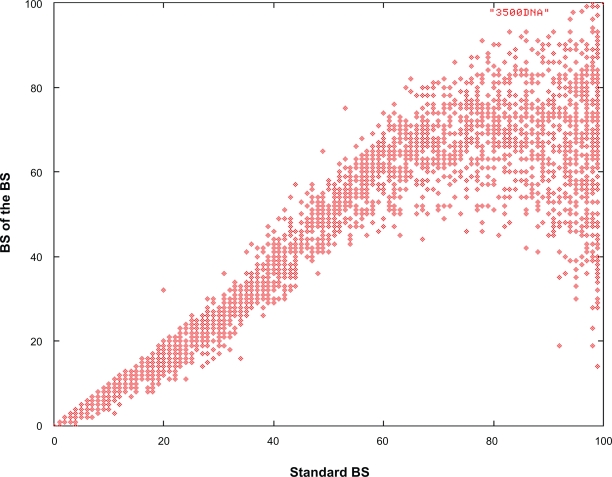
We mainly assessed the effects of applying the double BS procedure to large single-gene datasets since it had become computationally feasible due to the recent implementation of a rapid bootstrapping algorithm in RAxML29 and had never been tested in practice on large datasets before. We therefore conducted an empirical assessment of this approach to determine if it can be deployed as an alternative to improve the quality of support values on large trees. Despite the theoretically favorable statistical properties of second-level bootstrap procedures, in practice, and in particular on large single-gene alignments, a double bootstrap procedure does not appear to be applicable. The main reason is the significant reduction of the number of distinct (unique) alignment column patterns with respect to the original alignment and hence, phylogenetic signal in the BS replicates and to an even larger extent in the second-level BS replicates. In our experiments with the large eudicot dataset, the original alignment has 1,370 distinct column patterns, while the 100 first level bootstrap alignments have an average of 868 patterns per replicate and the 10,000 second level replicates only contain 641 patterns per replicate, less than half the length of the original alignment. In addition, first-level as well as second-level BS replicates contain a relatively high number of sequences that are exactly identical under the ML model, while the original alignment does not contain duplicate sequences. For first-level replicates there are on average 67 identical, thus essentially indistinguishable, sequences per replicate while for second-level BS replicates this number increases to 140 on average. Therefore, the phylogenetic signal contained in second-level BS replicates is reduced significantly and thus does not represent a good solution to infer support values for large single-gene analyses. This is depicted in Figure 4, where we plot the support values on the best-scoring ML tree of first-level BS replicates against support values from second-level BS analyses. The second-level BS values are significantly lower than first-level BS above a threshold of 75%. Thus, second-level BS does not scale as well as first-level BS (see results of CA-BS above) in the case of the herein analyzed data sets.
Figure 4.
Support values of second-level BS values over first-level BS support values on the best-scoring ML tree for the eudicots.
CA-BS versus GRTS-BS/GRTS-ML
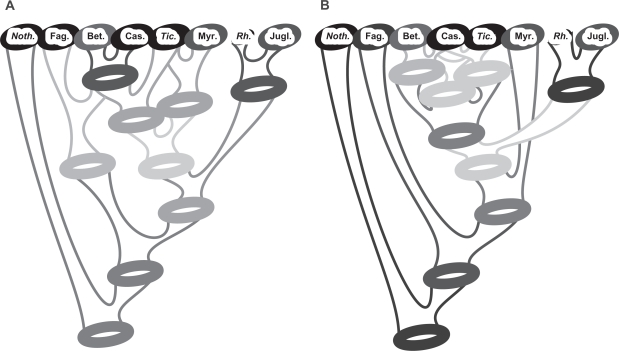
With a reduction factor of 1/4, the EUDIS matrix was reduced to 440 terminal taxa by GRTS. In the case of our focus groups rosids (matrix ROSID) and eurosids I (matrix EUROS 1; Fig. 5), datasets were reduced down to 282 and 199 taxa respectively. Nodes that received high BS support (CA-BS ≥ 70) based on the comprehensive data matrices were recovered in most (>50%) to all best-scoring GRTS-ML trees (Tables S1, S2 in OS 2). Representatives (subsamples) of the same TU clustered together when the same group was supported by moderate (>50) to high CA-BS values (OS 2). The consensus networks of the GRTS-ML trees indicated further relationships between predefined TUs that received low CA-BS (<50; example given in Fig. 6). For instance, large-scale multigene analyses43,44 supported an eurosid I subclade including the Celastrales, Malpighiales, Huaceae, and Oxalidales. CA-BS support for this subclade is low, even when mislabeled sequences are not considered, but the representatives of these TU consistently group using GRTS-ML (Table S3 in OS 2). The high overall similarity between CA-BS and GRTS-ML based topologies, irrespective of the actual support values, can be visualized via consensus networks (bipartition networks) of the 100 GRTS-ML trees, which depict the same relationships (Fig. 5), or alternative relationships, as indicated by CA-BS analyses of the comprehensive matrices (example shown in Fig. 6; see also OS 2): Equally CA-BS supported topological alternatives could be found with a certain frequency also in the GRTS-ML replicates.
Figure 5.
Potential of GRTS-ML to recover ‘correct’ relationships. Top, ML phylogram based on EURO1 matrix. Bottom, bipartition network based on 100 GRTS-ML replicates (family-level TU, reduction factor 1/4).
Figure 6.
Competing topological alternatives in GRTS-ML and CA-BS replicates. Unlabeled circles represent ‘candidate’ common ancestors (topological alternatives); support indicated by intensity of gray tones.
Pearson correlation coefficients between all bipartition support values obtained via CA-BS and GRTS-ML (reduction factor 1/4), all pruned-down to family-level TU trees, are shown in Table 2. Column # bipartitions indicates the total number of bipartitions induced by the pruned-down CA-BS trees and the GRTS trees. Correlations vary between 0.90 and 0.93 and are relatively high, but lower than for the CA-BS support values among the nested comprehensive datasets shown above. In Table 2 there is no prevalent tendency for the correlation to increase or decrease with increasing number of leaves in the GRTS-ML replicates. The number of bipartitions induced by the GRTS-ML replicates is significantly smaller than the respective number of bipartitions induced by the comprehensive trees, which has a direct effect on the average support on the pruned-down best-scoring ML trees from the comprehensive analyses: GRTS-ML support values (average: 61.16) are significantly higher than support values obtained via CA-BS (average: 53.78). This is also reflected by larger offsets between 9.48 and 11.05 compared to the offsets among support values induced by comparison of CA-BS between each other. Likewise, average support values as calculated from the majority rule consensus trees range between 82.15 and 87.69 for GRTS-ML, but between 77.65 and 83.10 for CA-BS. The reason for this is that GRTS-ML tends to be more decisive than CA-BS and to favor topological alternatives to different degrees (an example is given in Fig. 6). Such decisiveness could be positive or negative: Positive would mean that GRTS-ML has a higher chance than CA-BS to find support for ‘correct’ nodes; put in a negative context, the higher decisiveness could indicate that GRTS-ML exhibits a higher risk of yielding too high support for ‘wrong’ nodes. In our case, the effect appears to be positive rather than negative: The relationships indicated by GRTS-ML are in good agreement with the inferred comprehensive ML trees (e.g., example shown in Fig. 5) and earlier multigene analysis (Tables S1, S2 in OS 2). Using higher reduction factors (only feasible if order-level TUs are used; see below) seem to even increase the positive effect considering the recovered relationships.
Table 2.
Correlation between family-level TU CA-BS and family-level TU GRTS-ML, using a reduction factor of 1/4, support values for the distinct datasets.
| Datatset | # taxa | ρ-all | # bipartitions | ρ-best | WRF |
|---|---|---|---|---|---|
| Eudicots | 256 | 0.925 | CA-BS: 4,427 | 0.883 | 0.079 |
| GRTS-ML: 2,987 | |||||
| Rosids | 153 | 0.915 | CA-BS: 2,587 | 0.889 | 0.092 |
| GRTS-ML: 1,390 | |||||
| Eurosids I | 80 | 0.908 | CA-BS: 1,683 | 0.899 | 0.094 |
| GRTS-ML: 771 | |||||
| Eurosids II | 43 | 0.924 | CA-BS: 322 | 0.824 | 0.229 |
| GRTS-ML: 139 |
Effects of different reduction factors
As mentioned in Material and Methods, numerous family-level TUs are represented by a few accessions only. Therefore, we used the order-level TUs to be able to apply larger reduction factors. For the ROSID matrix, order-level GRTS was based on 18 TUs (15 rosid orders, 2 families, one outgroup order/family), the respective subsets (matrices EURO1 and EURO2) contained nine and five TUs. The usage of these more coarse-grained TUs is feasible because, according to APG II as well as APW, the according clades are generally well supported (see also Table S1 in OS 2 and cited literature; for a review see Soltis and Soltis).42
The effect of varying reduction factors on the correlation between CA-BS and GRTS-ML as well as GRTS-BS support values is shown in Tables 3 and 4, respectively, which also allows for a comparison between the GRTS-BS and GRTS-ML approach. Here the trees were pruned to order-level TU topologies instead of family-level TUs. This more coarse-grained reduction allows for assessing the effect of high reduction factors (see Material and Methods). Except for GRTS-ML values for rosids (Table 4), the correlation with CA-BS values generally decreases with increasing reduction factors. Overall, GRTS-BS values are more in agreement with the respective CA-BS values, than is the case for GRTS-ML (Tables 3 vs. 4). In combination with a moderate reduction factor of 1/4 or 1/8, the correlation between GRTS-BS and CA-BS can be as high as 0.999. These highest correlations are obtained for a data set (eurosid II) with well-sampled and well-supported TUs. The data set includes only five TUs that are mutually monophyletic. Four of these groups are extremely well represented by numerous rbcL sequences. Based on the overall good correlation, GRTS-BS may be a valid alternative to CA-BS to investigate support of backbone relationships of large datasets. In particular in the light of misplaced and mislabeled sequences: The subsampling of GRTS can handle a certain amount of misplaced and mislabeled representatives per TU if compensated by correctly placed and labeled representatives. For example, if the data includes 99 sequences of a TU that are correctly placed in the phylogenetic inference, the single misplaced (erroneous) sequence has only a probability of 0.01* samplingFactor to be sampled per replicate.
Table 3.
Correlation between order-level TU CA-BS and order-level TU GRTS-ML support values for the distinct datasets and reduction factors.
| Dataset | Reduction factor | # taxa | ρ-all | ρ-best | WRF |
|---|---|---|---|---|---|
| Rosids | 1/4 | 565 | 0.836 | 0.842 | 0.262 |
| 1/8 | 282 | 0.845 | 0.847 | 0.240 | |
| 1/16 | 141 | 0.882 | 0.869 | 0.213 | |
| 1/32 | 71 | 0.884 | 0.868 | 0.160 | |
| Eurosids I | 1/4 | 398 | 0.828 | 0.781 | 0.283 |
| 1/8 | 199 | 0.828 | 0.584 | 0.253 | |
| 1/16 | 99 | 0.693 | 0.390 | 0.206 | |
| 1/32 | 50 | 0.639 | 0.345 | 0.229 |
Table 4.
Correlation between order-level TU CA-BS and order-level TU GRTS-BS support values for the distinct datasets and reduction factors.
| Dataset | Reduction factor | # taxa | ρ-all | ρ-best | WRF |
|---|---|---|---|---|---|
| Rosids | 1/4 | 565 | 0.977 | 0.974 | 0.162 |
| 1/8 | 282 | 0.966 | 0.959 | 0.120 | |
| 1/16 | 141 | 0.946 | 0.940 | 0.119 | |
| 1/32 | 71 | 0.950 | 0.938 | 0.123 | |
| 1/64 | 35 | 0.891 | 0.875 | 0.055 | |
| Eurosids I | 1/4 | 398 | 0.977 | 0.878 | 0 |
| 1/8 | 199 | 0.932 | 0.714 | 0.136 | |
| 1/16 | 99 | 0.918 | 0.640 | 0.097 | |
| 1/32 | 50 | 0.865 | 0.500 | 0.053 | |
| 1/64 | 25 | 0.836 | 0.522 | 0.146 | |
| Eurosids II | 1/4 | 109 | 0.999 | ∼ | 0.517 |
| 1/8 | 55 | 0.999 | ∼ | 0.517 | |
| 1/16 | 28 | 0.999 | ∼ | 0.513 | |
| 1/32 | 14 | 0.993 | ∼ | 0.485 | |
| 1/64 | 7 | 0.970 | ∼ | 0.467 |
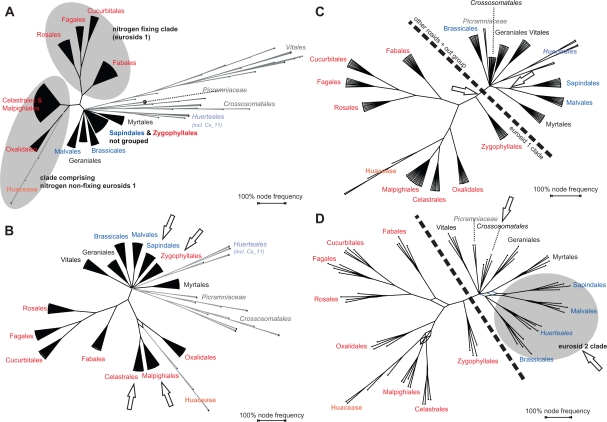
As mentioned before the decreased correlation between GRTS-ML, GRTS-BS (to a lesser degree) and CA-BS (Tables 3, 4) is coupled with the observation that GRTS-ML (and GRTS-BS) increasingly recover and support commonly accepted (inter-)ordinal relationships that received only low CA-BS support (OS 2). This is exemplarily illustrated in Figure 7 using the results of the GRTS-ML analyses based on the ROSID matrix. A reduction factor of 1/32 implied that from the 2,445 original accessions four are sampled per TU, plus the two rbcL accessions representing Picramminaceae (in total 70 terminal taxa per GRTS matrix). It has been demonstrated, especially for angiosperms, that increased taxon sampling is favorable.34,46 However, we obtain increased support for relationships, which were originally poorly supported, by reducing the number of leaves. Does this mean that fewer leaves are more prospective than many? Notably, only ‘correct’—or better, unchallenged—relationships gained support. Moreover, we have to keep in mind that the predefined TUs represented ‘good’ taxonomic units (well-supported clades based on multigene data). This is a major difference to ‘blind’ (unguided) random taxon jackknifing and/or using arbitrarily selected placeholders. An apparent effect of GRTS seems to be that a stable, ‘correct’ signal is maintained over the replicates while inconsistent, ‘wrong’ signal, is filtered out.
Figure 7.
GRTS-ML-based bipartition networks using different reduction factors and order-level TUs. Red, eurosid I clades, blue, eurosid II clades; important changes in the recognition of well-known groups are highlighted by arrows. The analyses were done based on the ROSID matrix. A) Reduction factor of ¼. The nitrogen fixing clade is recovered in most replicate trees. B) Reduction factor of 1/8. Sapindales are separated from Zygophyllales and Celastrales from Malpighiales. C) Reduction factor of 1/16. Zygophyllales are placed with other eurosids I. D) Reduction factor of 1/32. Eurosid 2 clades and Crossosomatales, respectively, are grouped.
Conclusion and Outlook
Comprehensive and GRTS-based analyses were conducted on large rbcL datasets to investigate whether the broad sampling of a single genetic marker is useful for large-scale, in terms of number of taxa, phylogeny reconstruction. The good correlations among the CA-BS results (Table 1) of the nested large matrices show that comprehensive ML analyses with RAxML scale well in terms of accuracy and support values, despite the fact that the amount of signal available in the data is, in principle, less favorable. Furthermore, ML-based CA-BS and to a greater extent, GRTS are able to recover higher-level relationships obtained via multigene studies; relationships that received poor support based on single-gene rbcL analyses in previous studies (OS 2).
The overall high correlation between CA-BS and GRTS-ML (Tables 2, 3) and in particular, between CA-BS and GRTS-BS (Table 4) demonstrates that the predefined family-level TUs and order-level TUs were well chosen. One may expect significantly worse correlations between CA-BS and GRTS-ML/GRTS-BS if the selected TUs do not represent well-supported clades. GRTS-based analyses yield largely the same phylogenetic relationships as inferred via comprehensive large-scale analyses (Figs. 5–7; Tables S1–S3 in OS 2). However, despite these promising results (e.g., Fig. 7), the statistical properties of GRTS-ML in comparison to CA-BS need to be investigated in more detail via computational experiments with simulated and additional real-world data sets prior to using GRTS-ML as additional means in phylogenetic inference based on large datasets. On the other hand, the high correlation between GRTSBS and CA-BS values provide a useful tool for the interpretation of the CA-BS values. The execution times for CA-BS and GRTS-BS are roughly similar because the taxon subsampling approaches require, in total, a larger number of ML searches to be conducted. In addition even many-taxon single-gene alignments rarely require more than 500 bootstrap replicates to generate stable support values.60 The group-based taxon jackknifing in combination with bootstrapping (GRTS-BS) allows for comparing support for the relationship between the main clades, in terms of family- or order-level TUs, without investing too much effort (human and computational resources) on the optimization of intra-clade relationships. In addition, it can be assessed how, and which, BS values decrease with an increasing number of taxa, and thus, generally lower average support values on larger single-gene trees can be more easily interpreted. It may be of interest to apply the here introduced methods to multigene data: The basic concept of GRTS would allow combining gene data with only partly overlapping sets of species per predefined TU (e.g., well-supported families or orders) without the need to decide on a placeholder taxa and/or filtering of occasionally misplaced or misnamed accessions. Conversely, GRTS could be used to select placeholder sequences in conjunction with the new placement algorithm61,62 that has recently been implemented in RAxML and that is particularly suited for the identification of short sequence reads that range between 100–400 bp in length. It appears reasonable to choose those sequences as placeholders that resulted in those trees closest to the GRTS majority-rule tree; such a preselection would further accelerate sequence identification and could be applied as long as within-group placement of query sequences is not of interest.
Supplementary Data
Misplaced rbcL accessions and upgrade to NCBI taxonomy tree. http://wwwkramer.in.tum.de/exelixis/Stam_et_al_OS1.pdf
Details of the results of comprehensive ML analyses. http://wwwkramer.in.tum.de/exelixis/Stam_et_al_OS2.pdf
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Zwickl DJ. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. Austin: University of Texas at Austin; 2006. [Google Scholar]
- 2.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for Maximum Likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–63. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 3.Stamatakis A. RAxML-VI-HPC: Maximum-Likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 4.Stamatakis A, Blagojevic F, Antonopoulos CD, Nikolopoulos DS. Exploring new search algorithms and hardware for phylogenetics: RAxML meets the IBM Cell. Journal of VLSI Signal Processing Systems. 2007;48:271–86. [Google Scholar]
- 5.Minh BQ, Vinh LS, von Haeseler A, Schmidt HA. pIQPNNI: parallel reconstruction of large maximum likelihood phylogenies. Bioinformatics. 2005;21:3794–96. doi: 10.1093/bioinformatics/bti594. [DOI] [PubMed] [Google Scholar]
- 6.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 7.Hordijk W, Gascuel O. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics. 2005;21:4338–47. doi: 10.1093/bioinformatics/bti713. [DOI] [PubMed] [Google Scholar]
- 8.Jobb G, von Haeseler A, Strimmer K. Treefinder: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–55. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 10.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–74. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 11.Dunn CW, Hejnol A, Matus DQ, et al. (18 co-authors). Broad phylogenomic sampling improves resolution of the animal tree of life Nature 2008452745–49. [DOI] [PubMed] [Google Scholar]
- 12.McMahon MM, Sanderson MJ. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionoid legumes. Syst Biol. 2006;55:818–36. doi: 10.1080/10635150600999150. [DOI] [PubMed] [Google Scholar]
- 13.Gueidan C, Roux C, Lutzoni F. Using a multigene phylogenetic analysis to assess generic delineation and character evolution in Verrucariaceae (Verrucariales, Ascomycota) Mycol Res. 2007;111:1145–68. doi: 10.1016/j.mycres.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Jansen RK, Cai Z, Raubeson LA, et al. (16 co-authors). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns Proc Natl Acad Sci U S A 200710419369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackett SJ, Kimball RT, Reddy S, et al. (18 co-authors). A phylogenomic study of birds reveals their evolutionary history Science 20083201763–68. [DOI] [PubMed] [Google Scholar]
- 16.Yoon HS, Grant J, Tekle YI, et al. (10 co-authors). Broadly sampled multigene trees of eukaryotes BMC Evol Biol 2008814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gee H. Ending incongruence. Nature. 2003;425:782. doi: 10.1038/425782a. [DOI] [PubMed] [Google Scholar]
- 18.Philippe H, Snell EA, Bapteste E, Lopez P, Holland PWH, Casane D. Phylogenomics of Eukaryotes: Impact of missing data on large alignments. Mol Biol Evol. 2004;21:1740–52. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- 19.Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–31. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Mc Guire JA, Witt CC, Altshuler DL, Remsen JV., Jr Phylogenetic systematics and biogeography of humming birds:Bayesian and maximum likelihood analyses of partitioned data and selection of an appropriate partitioning strategy. Syst Biol. 2007;56:837–56. doi: 10.1080/10635150701656360. [DOI] [PubMed] [Google Scholar]
- 21.Kolokotronis SO. Molecular evolution of Elephantidae and their tuberculosis pathogens. New York: Columbia University; 2008. [Google Scholar]
- 22.Smith SA, Beaulieu JM, Donoghue MJ. Mega-phylogeny approach for comparative biology: an alternative to supertree and supermatrix approaches. BMC Evol Biol. 2009;9:37. doi: 10.1186/1471-2148-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ripplinger J, Sullivan J. Does choice in model selection affect maximum likelihood analysis? Syst Biol. 2008;57:76–85. doi: 10.1080/10635150801898920. [DOI] [PubMed] [Google Scholar]
- 24.Tavare S. Some probabilistic and statistical problems on the analysis of DNA sequences. Lect Math Life Sci. 1986;17:57–86. [Google Scholar]
- 25.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites. J Mol Evol. 1994;39:306–14. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 26.Ott M, Zola J, Aluru S, Stamatakis A.Large-scale Maximum Likelihood-based phylogenetic analysis on the IBM BlueGene/L On-Line Procceedings of IEEE/ACM Supercomputing Conference 2007. Available at: http://www.sc07.supercomputing.org/schedule/pdf/pap271.pdf
- 27.Stamatakis A, Ott M. Exploiting fine-grained parallelism in the phylogenetic likelihood function with MPI, Pthreads, and OpenMP: A performance study. Proceedings of third IAPR International Conference on Pattern Recognition in Bioinformatics (PRIB 2008) 2008:424–35. [Google Scholar]
- 28.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 29.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–71. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 30.Bininda-Emonds ORP, Brady SG, King J, Sanderson MJ. Scaling of accuracy in extremely large phylogenetic trees. Procceedings of Pacific Symposium on Biocomputing. 2001;6:547–58. doi: 10.1142/9789814447362_0053. [DOI] [PubMed] [Google Scholar]
- 31.Moret BME, Roshan U, Warnow T.2002Sequence-length requirements for phylogenetic methods Goos G, Hartmanis J, van Leeuwen J.Procceedings of Algorithms in Bioinformatics: Second International Workshop, WABI 2002 Heidelberg, New York: Springer; 2002343–56.Lecture Notes in Computer Science; 2452 [Google Scholar]
- 32.Yang Z. Statistical properties of the Maximum Likelihood method of phylogenetic estimation and comparison with distance matrix methods. Syst Biol. 1994;43:329–42. [Google Scholar]
- 33.Goloboff PA, Catalano SA, Marcos Mirande J, et al. (7 co-authors). Phylogenetic analysis of 73 060 taxa corroborates major eukaryotic groups Cladistics 200925211–30. [DOI] [PubMed] [Google Scholar]
- 34.Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 2002;51:588–98. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
- 35.Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of live. Nat Rev Genet. 2005;6:361–75. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 36.Graham SW, Olmstead RG, Barrett SCH. Rooting phylogenetic trees with distant outgroups: A case study from the Commelinoid monocots. Mol Biol Evol. 2002;19:1769–81. doi: 10.1093/oxfordjournals.molbev.a003999. [DOI] [PubMed] [Google Scholar]
- 37.Savolainen V, Chase MW, Hoot SB, et al. (10 co-authors). Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences Syst Biol 200049306–62. [DOI] [PubMed] [Google Scholar]
- 38.Stevens PF.2001. onwards. Angiosperm Phylogeny Website Version 8, June 2007 (and more or less continuously updated since). Available at: http://www.mobot.org/MOBOT/research/APweb/welcome.html Accessed October 10 2009.
- 39.Angiosperm Phylogeny Group II An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- 40.Hilu KW, Borsch T, Müller K, et al. (16 co-authors). Angiosperm phylogeny based on matK sequence information Am J Bot 2003901758–76. [DOI] [PubMed] [Google Scholar]
- 41.Davies JT, Barradough TG, Chase MW, et al. (6 co-authors). Darwin’s abominable mystery: Insights from a supertree of the angiosperms Proc Natl Acad Sci U S A 20041011904–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soltis PS, Soltis DE. The origin and diversification of angiosperms. Am J Bot. 2004;91:1614–26. doi: 10.3732/ajb.91.10.1614. [DOI] [PubMed] [Google Scholar]
- 43.Qiu YL, Dombrovska O, Lee J, et al. (20 co-authors). Phylogenetic analyses of basal angiosperms based on nine plastid, mitochondrial, and nuclear genes Int J Plant Sci 2005166815–42. [Google Scholar]
- 44.Soltis DE, Gitzendanner MA, Soltis PS. A 567-taxon data set for angiosperms: The challenges posed by Bayesian analyses of large data sets. Int J Plant Sci. 2007;168:137–57. [Google Scholar]
- 45.Sanderson MJ, Wojciechowski MF, Hu J-M, Sher Khan T, Brady SG. Error, bias, and long-branch attraction in data of two chloroplast photosystem genes in seed plants. Mol Biol Evol. 2005;17:782–97. doi: 10.1093/oxfordjournals.molbev.a026357. [DOI] [PubMed] [Google Scholar]
- 46.Rydin C, Källersjö M. Taxon sampling and seed plant phylogeny. Cladistics. 2002;18:485–513. doi: 10.1111/j.1096-0031.2002.tb00288.x. [DOI] [PubMed] [Google Scholar]
- 47.Lanyon SM. Detecting internal inconsistencies in distance data. Syst Zool. 1985;34:397–403. [Google Scholar]
- 48.Stamatakis A.Phylogenetic Models of Rate Heterogeneity: A High Performance Computing Perspective Procceedings of 20th IEEE/ACM International Parallel and Distributed Processing Symposium (IPDPS 2006)2006; no page numbers [Proceedings on CD]. [Google Scholar]
- 49.Efron B, Halloran E, Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad Sci U S A. 1996;93:13429. doi: 10.1073/pnas.93.23.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods) 4 ed. Champaign, USA: Sinauer Associates; 2002. [Google Scholar]
- 51.Huson DH, Dezulian T, Rausch C, Richter D, Rupp R. Dendroscope 0.22: Visualization of large trees. Tübingen: University of Tübingen, ZBIT, Department Algorithms in Bioinformatics; 2007. [Google Scholar]
- 52.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 53.Holland B, Moulton V. Consensus Networks: A Method for Visualising Incompatibilities in Collections of Trees. In: Benson G, Page R, editors. Algorithms in Bioinformatics: Third International Workshop, WABI, Budapest, Hungary Proceedings. Berlin, Heidelberg, Stuttgart: Springer Verlag; 2003. pp. 165–76. [Google Scholar]
- 54.Grimm GW, Renner SS, Stamatakis A, Hemleben V. A nuclear ribosomal DNA phylogeny of Acer inferred with maximum likelihood, splits graphs, and motif analyses of 606 sequences. Evol Bioinform. 2:7–22. [PMC free article] [PubMed] [Google Scholar]
- 55.Holland B, Huber KT, Moulton V, Lockhart PJ. Using consensus networks to visualize contradictory evidence for species phylogeny. Mol Biol Evol. 2004;21:1459–61. doi: 10.1093/molbev/msh145. [DOI] [PubMed] [Google Scholar]
- 56.Nandi OI, Chase MW, Endress PK. A combined cladistic analysis of angiosperms using rbcL and non-molecular data sets. Ann MO Bot Gard. 1998;85:137–212. [Google Scholar]
- 57.Qiu YL, Lee JH, Bernasconi-Quadroni F, et al. (10 co-authors). The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes Nature 1999402404–07. [DOI] [PubMed] [Google Scholar]
- 58.Soltis DE, Senters AE, Zanis MJ, et al. (9 co-authors). Gunnerales are sister to other core eudicots: Implications for the evolution of pentamery Am J Bot 200390461–70. [DOI] [PubMed] [Google Scholar]
- 59.Kim S, Soltis DE, Soltis PS, Zanis MJ, Suh Y. Phylogenetic relationships among early-diverging eudicots based on four genes: were the eudicots ancestrally woody. Mol Phylogenet Evol. 2004;31:16–30. doi: 10.1016/j.ympev.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 60.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How Many Bootstrap Replicates are Necessary? Proceedings of RECOMB 2009, Springer Lecture Notes in Bioinformatics. 2009;5541:184–200. [Google Scholar]
- 61.Stamatakis A, Komornik Z, Berger SA. Evolutionary Placement of Short Sequence Reads on Multi-Core Architectures Accepted for publication at 8th ACS/IEEE International Conference on Computer Systems and Applications (AICCSA-10). PDF: http://wwwkramer.in.tum.de/exelixis/pubs/Exelixis-RRDR-2009-2.pdf
- 62.Berger SA, Stamatakis A. Evolutionary Placement of Short Sequence Reads, Exelixis Rapid Research Dissemination Report 2009–3, TU Munich, November 2009. PDF: http://wwwkramer.in.tum.de/exelixis/pubs/Exelixis-RRDR-2009-3.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Misplaced rbcL accessions and upgrade to NCBI taxonomy tree. http://wwwkramer.in.tum.de/exelixis/Stam_et_al_OS1.pdf
Details of the results of comprehensive ML analyses. http://wwwkramer.in.tum.de/exelixis/Stam_et_al_OS2.pdf