Abstract
Poxvirus DNA replication generates linear concatemers containing many copies of the viral genome with inverted repeat sequences at the junctions between monomers. The inverted repeats refold to generate Holliday junctions, which are cleaved by the virus-encoded resolvase enzyme to form unit-length genomes. Here we report studies of the influence of metal cofactors on the activity and structure of the resolvase of fowlpox virus (FPV), which provides a tractable model for in vitro studies. Small molecule inhibitors of related enzymes bind simultaneously to metal cofactors and nearby surface amino-acid residues, so understanding enzyme-cofactor interactions is important for the design of antiviral agents. Analysis of inferred active site residues (D7, E60, K102, D132, D135) by mutagenesis and metal rescue experiments specified residues that contribute to binding metal ions, and that multiple binding sites are probably involved. Differential electrophoretic analysis was used to map the conformation of the DNA junction when bound by resolvase. For the wild-type complex in the presence of EDTA or Ca2+, migration was consistent with the DNA arms arranged in near tetrahedral geometry. However, the D7N active site mutant resolvase held the arms in a more planar arrangement in EDTA, Ca2+ or Mg2+ conditions, implicating metal-dependent contacts at the active site in the larger architecture of the complex. These data show how divalent metals dictate the conformation of FPV resolvase/ DNA complexes and subsequent DNA cleavage.
Keywords: poxvirus, resolvase, DNA, Holliday junction, fowlpox virus
INTRODUCTION
Poxviruses encode a Holliday junction resolving enzyme (designated A22 in vaccinia and fowlpox1) that is a potential target for antiviral agents. During poxvirus DNA replication, a concatemeric form of the viral DNA is initially produced. The junctions between monomer units within the concatemer form inverted repeat sequences1. These sequences can be extruded from the DNA double helix to form a cruciform, which at its base contains a Holliday junction2. The poxvirus resolvase can cleave such structures in vitro3–6 and inactivation of resolvase during infection results in accumulation of uncleaved concatemers, indicating that resolvase is required for concatemer resolution in vivo7. After cleavage, ligation of the nicked DNA ends forms the covalently closed hairpin termini characteristic of poxviruses.
The poxvirus resolvase enzymes are members of the RNase H superfamily of DNA modifying enzymes3. Each enzyme in the superfamily shares a similar protein-fold in the catalytic domain, containing a central five-stranded mixed beta-sheet sandwiched between alpha-helices8. Each active site is thought to be comprised of three or four acidic residues. For those family members that have been purified and studied in vitro, each was found to require divalent metal-ions for activity. Structural and functional studies have suggested that multiple metal atoms may bind the conserved acidic resid ues to form the catalytic center, and specific models have been proposed based on two metal mechanisms9, 10.
Members of the RNase H superfamily catalyze nucleophilic substitution at a nucleic acid phosphodiester bond, where a hydroxyl oxygen acts as the nucleophile, attacking the phosphorus atom, and the 3' oxyanion serves as the leaving group. For RNase H, this involves attack of water on the RNA strand of an RNA/ DNA hybrid. The RNase H superfamily also includes Holliday junction resolving enzymes, the most thoroughly studied of which is E. coli RuvC. For these enzymes, hydrolysis takes place at a DNA branch point, freeing the individual DNA duplexes from one another11, 12. The retroviral integrases, also members of the superfamily, catalyze the attack of a hydroxyl at the 3' end of a donor DNA chain on target DNA strand, thus achieving strand joining by a single step transesterification13.
The HIV integrase enzyme is the target of the FDA-approved drug raltegravir14,raising the hope that related drugs might be devised that target poxvirus resolvase and inhibit poxvirus replication. Raltegravir is believed to bind to integrase in part by chelating two metal atoms at the enzyme active site. Pendant groups on raltegravir make additional contacts to nearby positions on the integrase surface which confer specificity15.
Because we are interested in devising poxvirus resolvase inhibitors, we carried out studies to investigate metal atom binding at the active site by the poxvirus resolvase, the likely binding site for raltegravir-like molecules. Previous work with poxvirus resolvases has focused on the vaccinia virus enzyme and has been limited by the fact that vaccinia resolvase is relatively insoluble and thus difficult to study in vitro 3–5, 16. Here we present studies of the role of metal atoms in catalysis and the overall geometry of the protein-DNA complex using purified fowlpox resolvase, which is more soluble and therefore a more tractable model 17.
We first examined metal ion cofactor specificity for catalysis by fowlpox resolvase by testing seven different divalent metal ions in HJ resolution reactions. Next, we use fourteen mutant fowlpox resolvase proteins, each with a single amino acid substitution at one of five putative active site residues, to probe for residues required for efficient catalysis. By comparing the catalytic activity of Ala, Cys, and amide substituted proteins under different divalent ion metal conditions, we were also able to make inferences as to which active site residues may be involved in direct metal chelation. We then studied the HJ binding properties of each mutant protein to ensure that the substituted proteins contained no gross defects in DNA binding activity as compared to the wild type protein.
Using the mutants and assembly conditions developed in the above studies, we carried out comparative electrophoresis analysis to determine the geometry of the HJ DNA arms in resolvase complexes 18. HJ DNAs containing each of the six possible pairs of shortened DNA arms were synthesized, and the mobilities of the free DNA and resolvase complexes were compared using polyacrylamide gel electrophoresis. In the only two previous studies where predicted complex structures deduced from electrophoretic data could later be compared to X-ray crystallographic data, the predicted structures matched the X-ray data 18–23. The electrophoretic approach has the advantage that complexes are not subject to possible distortion as a result of crystal packing forces, though the resolution of the electrophoretic method is much lower. We found that the orientation of the arms differed between in the FPV resolvase/ DNA complex from that of free DNA, and also differed in the presence of divalent metal ions. The overall complex conformation, although similar to other family enzyme members studied previously, contained a measurable degree of asymmetry, a phenomenon not previously observed for a resolvase-HJ complex.
Taken together, these data show how metal atoms interact at the FPV resolvase active site and further how metals dictate the conformation of the complex. This study provides background and methods for design of small molecule inhibitors for the poxvirus resolvase potentially useful as antiviral therapy.
RESULTS
Analysis of the divalent metal requirements for catalysis by FPV resolvase
Previous work has established that the vaccinia virus resolvase requires either Mg2+ or Mn2 for its DNA cleavage activity3, 4. To investigate the requirements of fowlpox virus resolvase, we incubated the enzyme with a fluorescein-tagged DNA Holliday junction (HJFS) substrate under various metal ion conditions, and then analyzed the products by polyacrylamide gel electrophoresis (data not shown). We observed product formation in the presence of either Mg2+ or Mn2+, but not in the presence of EDTA. Previously we estimated the first order rate of Holliday junction cleavage in 15 mM Mg2+ under single turnover conditions to be 0.23s−1 at 25°C 6. We also tested Ca2+, Cu2+, Fe2+, Ni2+, and Zn2+ at concentrations ranging from 0.4–30 mM. Slight cleavage was seen in Cu2+, Fe2+, Ni2+, and Zn2+. No cleavage was observed in Ca2+. Thus only Mg2+ and Mn2+ act efficiently as cofactors, paralleling studies of other RNase H superfamily members 8, 26–30.
We also noticed that the titration curve for Mn2+ showed a sharp drop in activity between 1.1 and 3.3 mM. This suggested that higher concentrations of Mn2+ may be inhibitory, and tests with mixtures of Mn2+ and Mg2+ showed indeed that concentrations of Mn2+ above 3.3 mM inhibited cleavage (data not shown).
Catalytic activity of fowlpox resolvase mutants
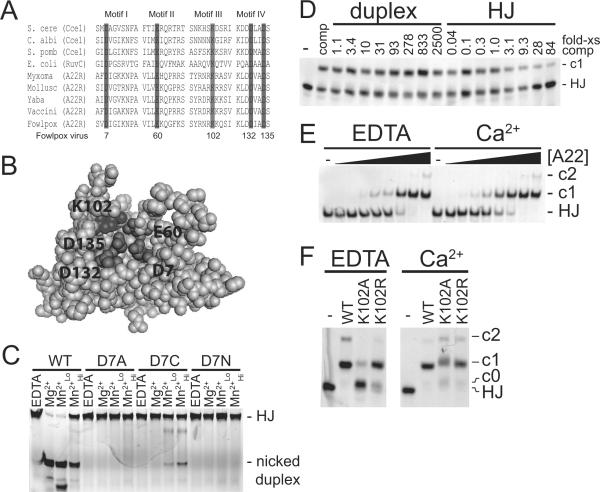
We wished to identify and analyze the amino-acid side chains comprising the fowlpox resolvase active site. Figure 1A shows an alignment of conserved residues from known resolvases of the RNase H superfamily, and Figure 1B shows their positions on a model of the structure inferred from the crystal structure of E. coli RuvC. Five conserved residues can be seen--D7, E60, K102, D132, D135 (fowlpox resolvase numbering scheme). Each of these residues has been implicated as important for function in vaccinia resolvase 5.
Figure 1.
DNA binding and cleavage by FPV resolvase and active site mutants. A) Protein sequence alignment of four conserved motifs of various RNase H-like family Holliday junction resolvases (adapted from Lilley and White12). Shading indicates residues that are 100% conserved within the RNase H-like resolvases. Amino acid numbering refers to fowlpox virus. S. cere, Saccharomyces cerevisiae; C. albi, Candida albicans; S. pomb, Schizosaccharomyces pombe; E. coli, Escherichia coli; Myxoma, myxoma virus; Mollusc, molluscum contagiosum virus; Yaba, Yaba monkey tumor virus; Vaccini, vaccinia virus; Fowlpox, fowlpox virus. B) Model of fowlpox resolvase tertiary structure showing the inferred active site, constructed with SWISS-MODEL37 using the RuvC crystal structure as a template. C) Native PAGE analysis of a representative metal rescue experiments. DNA cleavage reactions for the indicated amino acid substituted proteins were carried out in the various metal conditions indicated above the gel. EDTA, 1 mM EDTA; Mg2+, 15 mM MgCl2; Mn2+Lo, 0.5 mM MnCl2; Mn2+Hi, 5 mM MnCl2. Reaction products are labeled to the right of the gel. HJ, Holliday junction substrate. D) Native PAGE analysis of DNA binding by wild type protein in the presence of duplex or Holliday junction competitor DNA. DNA binding reactions were performed with 48 ng of wild type protein (2.4 pmol, 120 nM) and 2 nM Holliday junction probe DNA in the presence of 1 mM EDTA. Numbers above the gel lanes indicate the fold-excess of competitor DNA to probe DNA (wt/ wt). E) Binding of wild-type FPV resolvase to DNA in the presence of EDTA or Ca2+. DNA binding reactions were performed with various amounts of wild type protein and 2 nM Holliday junction DNA in the presence of either 1 mM EDTA (EDTA) or 1 mM CaCl2 (Ca2+). -, no protein; solid triangle indicate increasing amounts of protein from left to right: 0.3, 0.9, 2.6, 7.9, 24, 71, 213, 640 ng (the 640 ng amount corresponds to 1.6 μM). Resolvase-DNA complexes are labeled to the right of the gel. F) Effect of Ca2+ on DNA binding by amino acid substituted resolvases. DNA binding reactions were performed with 600 ng of the indicated protein (1.5 μM) and 2 nM of the Holliday junction probe DNA in the presence of either 1 mM EDTA (EDTA) or 1 mM CaCl2 (Ca2+).
In order to address whether or not the conserved acidic residues contribute to catalysis in FPV resolvase by binding directly to metal atoms, we compared substitutions that remove the side chain functional group (Ala) with those that preserve some potential for metal binding (Asn/ Gln or Cys). The Cys substitution is of particular interest because thiolate binds more tightly to Mn2+ than to Mg2+. Thus if Cys substitutions retain catalytic activity in Mn2+ but not Mg2+, it suggests that the side chain binds directly to the divalent metal-ion cofactor31–34.
Each resolvase derivative was incubated with HJ substrate in the presence of 1 mM EDTA, 15 mM Mg2+, 0.5 mM Mn2+, or 5 mM Mn2+. Data for all the amino acid substitution derivatives of FPV resolvase studied are presented in Table 1. A four hour incubation time was used to allow comparison of product accumulation for low activity mutant proteins. Table 2 presents kinetic data comparing estimated initial rates for the metal rescue studies in which FPV resolvase/ Holliday junction complexes were preformed, then reactions initiated by addition of metal cofactor. Reaction progression curves are shown in the Supplementary Report. Results for the different amino-acid substitutions are presented below, starting from the N-terminus.
Table 1.
Cleavage and DNA binding by FPV resolvase with active site substitutions.
Cleavage activity: Substrate and product bands from native-PAGE analysis of DNA cleavage reactions were quantified and percent product was calculated. The mean and standard error from n independent reactions are reported. Binding activity: Free probe and shifted bands from native-PAGE analysis of DNA binding reactions over a range of protein concentrations were quantified and the fraction bound was calculated. To compare binding affinities between enzymes, we estimated Kd values using nonlinear regression, using a resolvase dimer as the binding species. The normalized data were plotted as `fraction bound' versus `log[protein]' and fit with a sigmoidal dose-response model. Between 3 and 29 independent measurements went into each Kd measurement using 3 to 6 different protein concentrations over a 9 to 243-fold concentration range.
| Enzyme | Cleavage Activity (% Product ± SE) | Kd (nM) | 95% Confidence Interval | ||
|---|---|---|---|---|---|
| 15 mM Mg2+ | 0.5 mM Mn2+ | 5 mM Mn2+ | |||
| WT | 95.5 ± 2.2, n=13 | 98.1 ± 3.9, n=9 | 24.9 ± 2.8, n=9 | 53 | 39–73 |
| D7A | 0.0 ± 0.3, n=3 | 0.0 ± 0.0, n=2 | 0.0 ± 0.3, n=3 | 128 | 12–1419 |
| D7C | 0.1 ± 0.4, n=2 | 6.8 ± 0.7, n=2 | 14.1 ± 1.4, n=3 | <22 | NA |
| D7N | 0.2 ± 1.1, n=3 | 0.1 ± 0.5, n=2 | 0.1 ± 1.2, n=3 | 36 | 14–94 |
| E33A | 95.9 ± 3.2, n=3 | 58.1 ± 2.2, n=2 | 5.0 ± 1.3, n=3 | 1413 | 39–5246 |
| D55A | 98.5 ± 3.3, n=3 | 57.6 ± 3.4, n=2 | 4.6 ± 2.7, n=3 | 760 | 254–2270 |
| E60A | 0.8 ± 0.7, n=3 | 1.0 ± 0.2, n=2 | 0.6 ± 1.2, n=2 | 38 | 7–220 |
| E60C | 0.5 ± 1.4, n=3 | 8.3 ± 1.7, n=2 | 11.5 ± 2.4, n=2 | 5 | 0–254 |
| E60N | 1.1 ± 0.9, n=3 | 38.1 ± 13.6, n=2 | 57.5 ± 7.1, n=3 | 21 | 4–114 |
| E60Q | 0.8 ± 1.2, n=3 | 1.3 ± 0.7, n=2 | 0.6 ± 2.6, n=3 | 421 | 147–1205 |
| K102A | 0.9 ± 0.2, n=3 | 0.6 ± 0.7, n=2 | 1.1 ± 1.0, n=3 | 384 | 187–789 |
| K102R | 93.8 ± 4.9, n=3 | 88.0 ± 2.5, n=2 | 2.9 ± 1.5, n=3 | 77 | 37–162 |
| D132A | 0.2 ± 0.4, n=3 | 0.9 ± 0.2, n=2 | 0.4 ± 3.5, n=2 | 25 | 16–40 |
| D132C | 0.1 ± 1.0, n=3 | 7.1 ± 1.1, n=2 | 12.0 ± 3.6, n=2 | 144 | 103–201 |
| D135A | 0.1 ± 1.4, n=3 | 94.3 ± 4.9, n=2 | 12.5 ± 8.5, n=2 | 14 | 11–17 |
| D135C | 1.7 ± 1.2, n=3 | 24.3 ± 9.4, n=2 | 40.2 ± 9.8, n=3 | 195 | 76–504 |
| D135N | 1.6 ± 1.0, n=3 | 4.9 ± 1.2, n=2 | 31.1 ± 6.1, n=2 | 144 | 92–226 |
Table 2.
Rate analysis for the metal rescue experiment.
Reactions were started by addition of metal cofactor. Substrate and product bands from native-PAGE analysis of DNA cleavage reactions were quantified and percent product was calculated. Rates were calculated using a single exponential decay model. From 15–18 points were used to determine each rate.
| Enzyme | Rate of Cleavage (pM/sec) | Ratio of rates 0.5 mM Mn/ 15 mM Mg | |
|---|---|---|---|
| 0.5 mM Mn | 15 mM Mg | ||
| D7C | 0.7543±0.0453 | 0.0428±0.0192 | 17.6 |
| E60C | 0.2072±0.0301 | 0.0990±0.0276 | 2.1 |
| D132C | 0.5117±0.0853 | 0.2135±0.0354 | 2.4 |
| D135C | 5.9166±0.3227 | 0.2541±0.0510 | 23.2 |
Figure 1C shows a representative gel image for reactions with the WT, D7A, D7C, and D7N enzymes. Reactions containing wild type protein showed no cleavage in the presence of EDTA (Figure 1C, lane marked “EDTA”). Reactions with Mg2+ or the lower concentration of Mn2+ showed near complete cleavage of the substrate, whereas reactions with the higher concentration of Mn2+ showed detectable cleavage, but only ~25% of the substrate was consumed. We also examined cleavage of a different branched DNA substrate and obtained similar results (data not shown). Substitution of Ala for the conserved Asp at position-7 (D7A) eliminated detectable activity in all the metal conditions. The substitution mutant D7N also was inactive, emphasizing the importance of the acidic side chain. However, D7C showed low but detectable activity in the presence of Mn2+ and barely detectable activity in Mg2+, with a ratio of rates of at least 18-fold (Table 2). Thus the observed Mn2+ rescue is as expected if metal binding is important for activity and the D7 carboxylate is involved in binding metal, though the enzyme bearing the Cys-tethered Mn atom was much less active than wild type.
Substitution of the non-conserved control positions E33 and D55 with Ala had little effect on activity, indicating that these side chains are not important for catalysis. E60A and E60Q were inactive under all metal conditions. E60C showed slight but detectable activity in Mn2+ but lower activity in Mg2+, again a metal rescue phenotype indicating that the E60 side chain likely binds metal to promote catalysis (Table 2).
Unexpectedly, the E60N substitution was also active in Mn2+ but much less active in Mg2+ (Table 1). One explanation of this observation is that the local environment of the N60-amide, being one C-C bond length closer to the protein backbone than the Q60-amide, is sufficiently more basic (perhaps due to closer proximity to K102) to deprotonate the amide and permit divalent metal-ion chelation and catalysis. Another model holds that the E60N mutant yields a less stable enzyme-substrate complex than E60Q, and this assists product formation.
Removing the positive charge at the conserved K102 residue via Ala substitution eliminated detectable cleavage activity under all metal conditions. However, preserving the positive charge by substitution of Arg at this position returned activity to wild-type levels (Table 1).
Substitution of Ala at D132 eliminated detectable cleavage activity under all metal conditions. The D132C substituted enzyme showed weak activity in Mn2+ and lower activity in Mg2+, consistent with a role for the side chain in binding metal during catalysis.
For D135, Asn and Cys substitutions showed slight activity in the presence of Mn2+ but much less in Mg2+ (Table 1). Substitution with Cys showed a 23-fold higher rate in Mn2+, suggesting a role for metal binding (Table 2). Substitution with Ala, a side chain not expected to participate in metal coordination, resulted in loss of activity in Mg2+, but unexpectedly retained activity in Mn2+, with low Mn2+ being more active than high Mn2+ as observed for the wild type enzyme. This complicates the interpretation and suggests that D135 plays a more specialized role in catalysis (we return to this point in the Discussion).
In summary, the metal rescue study implicated D7, E60, D132, and D135 in metal binding during catalysis. Some caution is warranted in interpreting these data because i) the cleavage rates of the Mn2+ rescued Cys substitutions were not restored to wild-type rates, and ii) the rate limiting steps in the catalytic cycle were not determined for each mutant and the wild type enzyme, so it is possible the rate limiting steps differed. However, one simple interpretation is that that the four acidic residues show Mn2+rescue of Cys substitutions because these residues promote catalysis by binding the metal cofactor.
DNA binding by mutant and wild type fowlpox resolvases
An advantage of the fowlpox resolvase model system is that, unlike vaccinia resolvase, simple band shift assays of DNA binding are feasible due to better protein solubility6. We previously reported that FPV bound to Holliday junction DNA with a Kd of <2 X 10−8 M in the absence of competitor DNA6. All of the amino acid substituted resolvases studied above were analyzed by band shift assays using the Holliday junction probe in the presence of 300-fold excess of sonicated salmon sperm competitor DNA (Table 1). To prevent cleavage by resolvase during the reaction, binding studies were carried out in the presence of 1 mM EDTA to chelate divalent metals. Several DNA complexes were formed by fowlpox resolvase and are designated as c0, c1, and c2 beside the gels in Figure 1 D–F.
Resolvase binding is specific for branched DNA6. To demonstrate the specificity on our fluorescein-tagged HJ substrate, fowlpox resolvase was add ed to mixtures of the labeled Holliday junction probe and an unlabeled duplex competitor DNA (Figure 1D). We found that a >2500-fold excess duplex linear competitor DNA was needed to compete for Holliday junction binding (Figure 1D, duplex). In contrast, only a 3-fold excess Holliday junction competitor DNA was required to observe a reduction in binding (Figure 1D, lanes marked “HJ”).
We tested each of the fowlpox resolvase mutants for DNA binding activity using the Holliday junction substrate in the presence of EDTA (Table 1). Examples of the quantitative binding data are presented in the Supplementary Report. All mutants bound to DNA detectably, but there was considerable variation among them. Errors for some measurements w ere large because only a narrow concentration range of protein could be interrogated. Two mutant proteins bound more tightly than wild-type (D7C and D135A). These mutants may be useful in efforts to form resolvase-DNA complexes for biophysical and structural analysis. Several showed impaired binding (E33A, D55A, E60Q, K102A, D132C, D135C, and D135N). In summary, all of the FPV resolvase proteins with amino acid substitutions at the four conserved acidic residues showed detectable formation of c1, though some showed altered affinity. Several proteins showed primarily formation of c0 only (E33A, D55A, and K102A), so for these we investigated binding in the presence of metal (instead of EDTA as was used above).
When Ca2+ was includ ed in binding assays, affinity of wild-type resolvase was increased approximately two-fold (Figure 1E). We then studied the effect of Ca2+on binding by K102A and K102R (Figure 1F), and the controls E33A and D55A (data not shown). All showed increased binding in the presence of Ca2+, suggesting that the impairment in DNA binding for these proteins in the presence of EDTA was partially overcome by addition of metal. This helps to explain why E33A and D55A showed little impairment in the cleavage reactions. We also observed some retention of DNA in the gel wells (< 5% of total DNA) after electrophoresis in binding experiments with E33A, D55A, and K102A, suggesting these proteins may be more prone to aggregation than the others studied.
Oriented cleavage on Holliday Junction 1 by FPV resolvase
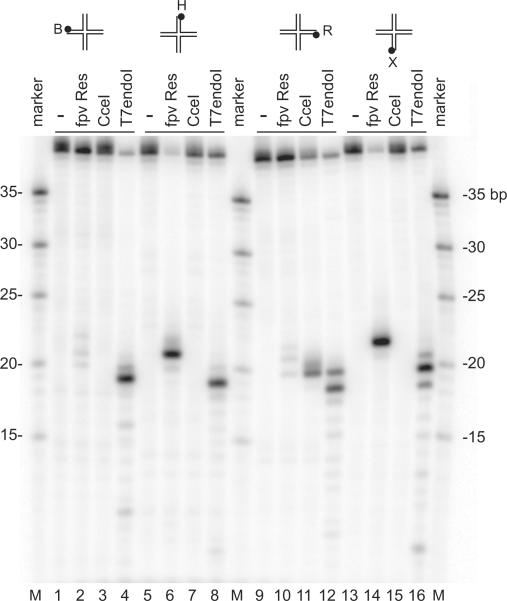
In the course of a comparative analysis of FPV resolvase interactions with Holliday Junction DN As, we noticed that Junction 1 from Lilley and coworkers18 was cleaved in a highly biased fashion, so that cleavage on one pair of strands was strongly favored over the other (Figure 2). The reason for working with Junction 1 was that it was designed to allow probing of resolvase/ HJDNA complexes by comparative electrophoresis--Junction 1 is designed to contain a unique restriction enzyme recognition site in each DNA arm, so that pairs of arms can be conveniently shorted by restriction enzyme cleavage. This makes possible analysis of the geometry of the DNA arms by comparative electrophoresis, as is described below. The finding of oriented cleavage by FPV resolvase provided a means of assembling oriented FPV resolvase/ HJDNA complexes for such studies.
Figure 2.
Preferential orientation for DNA cleavage by FPV resolvase on Junction 1S. DNA oligonucleotides were 5'-end labeled with 32P on the strands indicated above the gels. Resolvase enzymes studied are marked above the gel lanes. Fowlpox resolvase, Cce I protein or T7 endonuclease I (New England Biolabs, Beverley, MA) were added to a solution containing HJ1S DNA (final concentrations; 10 nM resolvase, 10 nM Cce I, 2U of T7 endonuclease and 2nM HJ1S DNA substrate). Reaction mixtures were incubated at 37 °C for 30 min. Products of the reactions were separated on urea-containing 10% polyacrylamide gels by electrophoresis in 1X TBE buffer and visualized by autoradiography.
Figure 2 documents the oriented cleavage using Junction 1S,which is a version of Junction 1 synthesized with shorter DNA arms, which we used to improve electrophoretic separation on DNA sequencing-type gels. Incubation of Junction 1S with FPV resolvase resulted in cleavage to near completion on strands H and X (Figure 2, lanes 6 and 14), whereas only barely detectable cleavage was seen on strands B and R (Figure 2, lanes 2 and 10). For comparison, the yeast Cce1 enzyme showed weak cleavage on strand R only (Figure 2 lane 11), indicating that Junction 1S lacked sequence features required for efficient cleavage by Cce1. In contrast, T7 endonuclease I cleaved efficiently on all four strands (Figure 2, lanes 4, 8, 12, and 16). Kinetic analysis showed that cleavage with FPV resolvase was slightly faster on the X strand than on the H strand, consistent with the asymmetry in binding described below, though the difference was slight (data not shown).
We next used order of addition experiments to investigate the role of metal atoms in formation of the oriented cleavage complex. We compared cleavage in reactions where i) DNA was mixed with metal, then started with enzyme, or ii) DNA was mixed with enzyme, then the reaction started with metal. In both cases, oriented cleavage was seen (data not shown). This is consistent with the idea that formation of the oriented complex does not depend on the presence of metal, though we cannot rule out an alternative explanation that the complex is in fast equilibrium, so that the enzyme releases, binds metal, then rebinds in an oriented fashion.
Comparative gel electrophoresis of FPV resolvase/Holliday Junction 1 complexes
We took advantage of the ability to assemble oriented FPV resolvase/ DNA complexes to analyze the geometrical positions of the DNA arms using comparative electrophoresis. In previous studies, this method successfully predicted the global orientation of the DNA arms in a variety of resolvase/ HJ complexes, and the predictions have been confirmed by X-ray crystallography for T7 endonuclease I and T4 endonuclease VII bound to HJDNA18–23.
To carry out the analysis, six different HJDN As were synthesized, all of which contain the same numbers of base pairs, but each of which had one of the six possible combinations of two long DNA arms and two short arms 18. For any given HJ species, the angle between the two long arms of the junction mainly dictates the mobility of the junction through a polyacrylamide gel matrix, with angles nearer 180° resulting in faster mobility and relatively smaller angles resulting in slower mobility. Assuming that the geometries of the arms are consistent for resolvase/ DNA complexes containing all six variants, then electrophoresis of the six species side-by-side allows determination of the relative angles between all pairs of arms of the junction. In this study, we used Junction 1, a HJ previously analyzed by this method to allow for comparison to previous work18.
We prepared FPV resolvase/ DNA complexes with Junction 1 using both the wild-type enzyme and the active-site mutant enzyme D7N. As a control, we also studied the six species in the absence of protein, and obtained data identical to published results18. To analyze the effects of metal on the conformation of the DNA, we compared 2 mM EDTA, 200 uM CaCl2, or 200 uM MgCl2. In each case, the complex was incubated in the indicated metal or in EDTA prior to electrophoresis, and the metal or EDTA was added to the electrophoretic buffer as well. Our finding that Ca2+does not support DNA cleavage allowed us to study the wild-type enzyme under divalent metal ion conditions without DNA cleavage. Use of the active site mutant D7N allowed us to study FPV resolvase in the presence of Mg2+ without concurrent DNA cleavage. DNA and FPV resolvase/ DNA complexes were analyzed by electrophoresis on 8% native polyacrylamide gels.
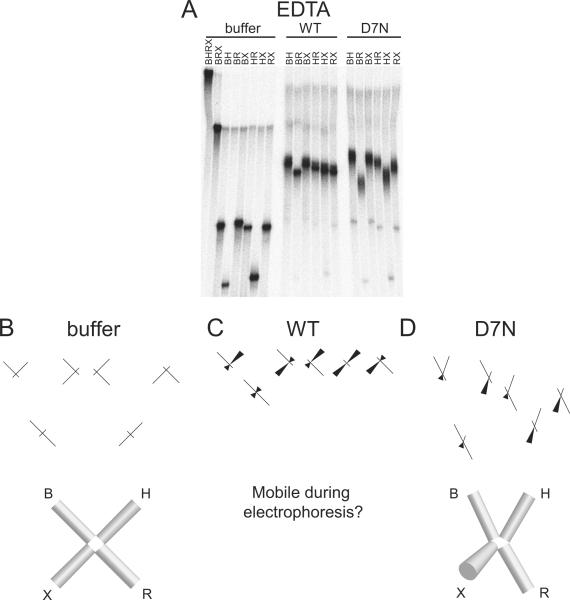
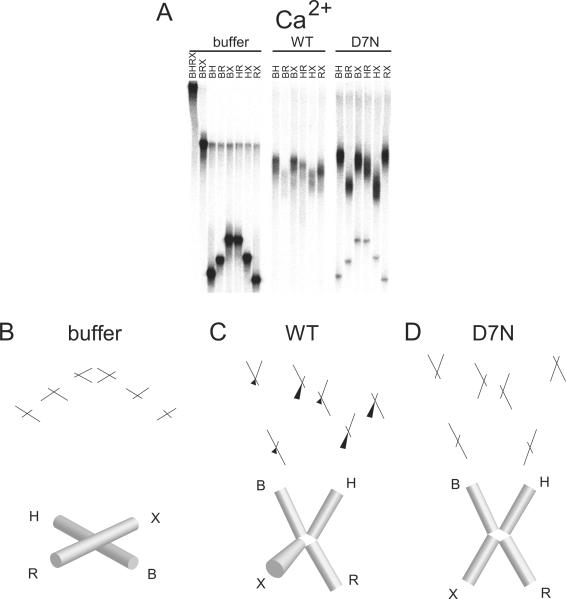
In the absence of divalent metal ion and resolvase protein (Figure 3A), we obtained an electrophoretic pattern for free Junction 1 of slow-fast-slow-slow-fast-slow. This pattern matches previous results and corresponds to an open squar e planar conformation of Junction 1 in which all four arms of the junction lie in the same plane and the angle between each set of adjacent arms is ~90° (Figure 3B). In the comparative electrophoresis experiment with wild-type or D7N resolvase in EDTA, we observed discrete slower migrating bands, as expected for formation of FPV resolvase/ DNA complexes. In each case, we observed a sharp change in the relative electrophoretic pattern of the six species between the unbound and bound conditions, indicating that HJ binding by FPV resolvase altered the global conformation of the DNA junction.
Figure 3.
Conformation of FPV resolvase-HJ complexes analyzed by comparative electrophoresis in the presence of 2 mM EDTA. A) Comparative gel electrophoresis analysis of fowlpox resolvase-DNA complexes. The long arms of the junctions are designated above the gel lanes by the letters B, H, R, or X. The protein used in each binding reaction is indicated above the gel. buffer: no protein, WT: wild-type protein, D7N: D7N mutant protein. B) Structural interpretation of comparative electrophoresis data for the buffer only control. Each cylinder represents a double helical arm of the HJ and its identity is labeled by the letters B, H, R, or X. The lack of continuity between adjacent cylinders indicates that the DNA arms are unstacked. More conical shaped cylinders indicate angling of that DNA arm out of the plane of the paper. C) Structural interpretation of the data for complex with wild-type FPV. The DNA arms of the complex may be mobile. D) Structural interpretation of the data for complex with D7N resolvase. More conical shaped cylinders indicate angling of that DNA arm out of the plane of the paper.
For the wild-type samples, we observed a relative electrophoretic pattern in which the difference among the six species showed only slight differences (Figure 3A, “WT”). To the extent that they differed, the samples showed slow-fast-slow-intermediate-intermediate-intermediate mobilities (Figure 3A). The extreme case of all species having the same mobility would imply either that i) all pairs of arms have identical angles between them, which can only be satisfied by a tetrahedral geometry, or ii) the junction is mobile so that the geometry changes during electrophoresis, resulting in mobilities that reflect multiple conformations. An example of rapid exchange in HJ conformation has been reported previously18,22. If we assume a single geometry, we interpret the observed pattern to arise from a conformation of the DNA lying somewhere between the tetrahedral conformation (all arm-arm angles ~109.5°) and a conformation in which ∠BR is ~180° and the other five arm-arm angles are ~90° , though it may be more likely that the junction is mobile during electrophoresis.
For the D7N active-site mutant samples analyzed in the absence of divalent metalion (Figure 3A and D), we observed an electrophoretic pattern of slow-fast-slow-intermediate-fast-intermediate. We interpret this pattern to arise from a structure close to a two-fold symmetric open-X conformation, where all four arms lie in the same plane and ∠BH and ∠RX are <90° and ∠HR and ∠BX are >90°. However, a true two-fold symmetric open-X structure would have a pattern of slow-fast-intermediate-intermediate-fast-slow , as is approximated more closely in Figure 4C (Mg2+/ D7N) and Figure 5D (Ca2+/ D7N). We suggest the deviation from this pattern observed in Figure 3A and D may be due to the X arm coming slightly out of plane under these conditions, thus deviating from two-fold symmetry. It is also possible that the other arms have at least modest out of plane bending toward a tetrahedral arrangement, and that the ∠XH angle is smaller than the others.
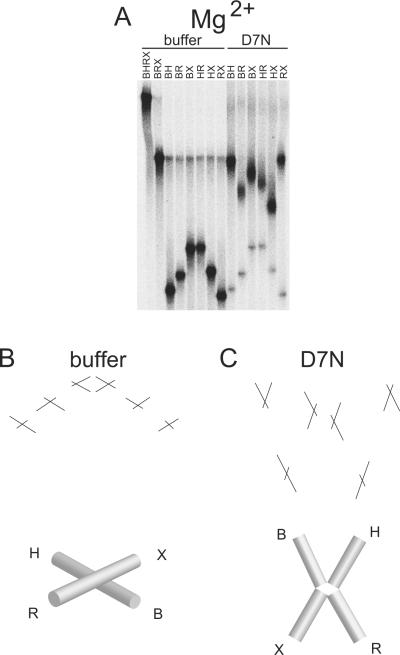
Figure 4.
Conformation of FPV resolvase-HJ complexes analyzed by comparative electrophoresis in the presence of 200 uM MgCl2. A) Comparative gel electrophoresis analysis of fowlpox resolvase-DNA complexes. Markings as in Figure 3. B) Structural interpretation of comparative electrophoresis data for buffer only or C) D7N resolvase. The continuity of adjacent cylinders seen in the buffer only condition indicates that the adjacent arms of the HJ are coaxially stacked.
Figure 5.
Conformation of FPV resolvase-HJ complexes analyzed by comparative electrophoresis in the presence of 200 uM CaCl2. A) Comparative gel electrophoresis analysis of fowlpox resolvase-DNA complexes. Markings as in Figure 3. B) Structural interpretation of comparative electrophoresis data for buffer only, C) wild-type FPV resolvase, or D) D7N. Markings as in Figure 3. The continuity of adjacent cylinders seen in the buffer only condition indicates that the adjacent arms of the HJ are coaxially stacked.
In the presence of divalent metal ion (Figures 4 shows data for Mg2+ and Figure 5 shows data for Ca2+), we obtained an electrophoretic pattern for free Junction 1 of fast-intermediate-slow-slow-intermediate-fast (Figure 4 and 5, buffer only). This pattern matches previous analysis of this junction and corresponds to the anti-parallel stacked-X conformation in which the arms of the junction are coaxially stacked upon one another with B stacked on H and R stacked on X.
For the samples containing wild-type or active-site mutant FPV resolvase, we observed discrete slower migrating bands, as expected for formation of protein-DNA complexes. In each case, we observed a sharp change in the relative electrophoretic pattern of the six species between the unbound DNA and the bound DNA, signifying that HJ binding by FPV resolvase also altered the global conformation of the DNA junction in the presence of divalent metal ion.
For the active site mutant D7N in the presence of divalent metal ion (Ca2+ or Mg2+), we observed an electrophoretic pattern of slow-fast-intermediate-intermediate-fast-slow (Figure 4A and C, Mg2+/ D7N and Figure 5A and D, Ca2+/ D7N). We interpret this pattern to arise from an approximately two-fold symmetric open-X conformation, where all four arms lie in the same plane and ∠BH and ∠RX are <90° and ∠HR and ∠BX are >90°.
The wild type FPV could only be studied under metal conditions in Ca2+, because in Mg2+ the enzyme cleaves DNA. For wild-type FPV resolvase in the presence of Ca2+, we observed an electrophoretic pattern of slow-fast-slow-intermediate-fast-intermediate, close to the pattern observed for the active-site mutant under EDTA conditions (Figure 5B, compare EDTA/ D7N to Ca2+/ WT). We interpret this pattern again to arise from an open-X structure with a slightly out of plane X arm (see above).
DISCUSSION
In this paper we present a study of the role of divalent metal ions in the structure and activity of FPV resolvase. Analysis of metal dependence and amino acid substitutions delineated the requirements for efficient catalysis. The active site mapping yielded results closely paralleling previous studies of vaccinia virus resolvase and other RNaseH superfamily enzymes. Our use of metal rescue experiments helped establish the roles for some of these active site constituents in metal binding. In the course of these studies, we found that we could assemble FPV protein onto the Junction 1 substrate in a defined orientation. This provided the tools necessary to analyze the geometry of the DNA arms by comparative electrophoresis in oriented complexes, which yielded results that paralleled but were not identical to previous studies of the RNaseH superfamily resolvase RuvC. Some of the implications of these studies are discussed below.
Residues that were likely components of the FPV resolvase active site could be identified by homology to other RNase H superfamily members. All five of the conserved charged residues in vaccinia resolvase have been substituted with neutral residues, and for each cleavage activity was abolished. However, for the vaccinia protein, due to the poor solubility of the purified proteins it was not possible to assess whether DNA binding was affected by the substitutions, leaving the functions of the residues uncertain 5. For fowlpox resolvase, substitution of the five conserved charged residues with neutral residues abolished the catalytic activity in Mg2+, and all proteins could be shown to retain DNA binding activity. As controls, two non-conserved acidic residues were substituted with Ala, and both of these proteins retained catalytic activity.
Thus studies of the amino acid substitutions established the importance of these residues for function, but not whether they act by binding metal. Studies of Cys substitutions at the four conserved acidic residues showed detectable restoration of activity in the presence of Mn2+ compared to the Ala substitutions. Because Cys binds Mn2+ more tightly than Mg2+, these findings indicate that residues at this position likely promote catalysis by binding metal. Some caution is warranted for this conclusion, since the rate-limiting step was not determined for wild-type and mutants, and this may differ, but differential metal binding offers a reasonable model. We also studied substitutions of Asn and/ or Gln at three of the conserved acidic residues and some of these proteins also displayed detectable activity in Mn2+. Our studies of the conserved residue D135 revealed an unexpected result that differed from the other three conserved acidic residues. Ala substitution at D135 resulted in loss of detectable activity in Mg2+, but preservation of wild type activity in Mn2+, whereas Ala substitution at the other catalytic acidic residues resulted in loss of detectable activity under all metal conditions. Since Ala is not expected to chelate metal, these data indicate that metal chelation at D135 is not a requirement for catalysis in Mn2+, however the D135-carboxylate is a requirement for catalysis in Mg2+. Given that Mg2+ has stricter coordination geometry than Mn2+ and is believed to be the physiologic cofactor for this superfamily of enzymes, we speculate the role of D135 is to stabilize a geometry of Mg2+ at the enzyme active-site that is necessary for catalysis. Also, our finding that the D135A enzyme retains inhibition with high Mn2+, suggests D135 does not directly participate in high Mn2+ inhibition.
For K102, Ala substitution at the homologous residue in the E. coli RuvC protein suggested this conserved basic residue played a role in DNA binding 35. However, our finding that Ala or Arg substitution at K102 have similar DNA binding properties, but only the Ala substitution is inactive for cleavage, suggests the function of this residue may be to stabilize the catalytic orientation of the scissile phosphate or nucleophile within the active site and/ or to participate directly in the catalytic mechanism as a general base.
The results from our metal rescue experiments are consistent with a cleavage mechanism involving multiple metal atoms bound at the active site, and this suggests approaches to inhibitor development. Using our model (Figure 1B), we measured the distances between the side chain carboxylates of D7, E60, and D132, and found the distance between E60 and D132 to be 11.2 Å, between E60 and D7 to be 5.9 Å, and between D7 and D132 to be 7.4 Å. The distance between E60 and D132 is too large to have both residues coordinate a common Mg2+ ion simultaneously, and the same conclusion can be mad by inspection of the homologous residues in the RuvC crystal structure. Our metal rescue data suggest that both of these residues do in fact contact metal, however, thus providing biochemical evidence supporting the idea of multiple metal-ion binding sites. This finding has implications for targeting of this enzyme with small molecule inhibitors. HIV-1 integrase inhibitors of the raltegravir class are proposed to chelate two metal atoms at the integrase active site 15, 36, and so could potentially bind and inhibit poxvirus resolvases. Inhibitors of this class have been identified in a high throughput screen for molecules active against fowlpox resolvase and are presently under study (Culyba, Hwang and Bushman, unpublished data).
The diagrams of the geometry of Junction 1 in complex with FPV resolvase in Figure 3–5 summarize our interpretation of the comparative electrophoresis data. Binding of FPV resolvase to Junction 1 alters the global conformation of the HJ in both the absence and presence of divalent metal ions. In the presence of divalent metal ions, the protein-DNA complexes adopt structures that allow adjacent pairs of arms to more closely approach one another. For example, for the WT resolvase/ HJ complexes, addition of divalent metal switches the conformation of the arms from a more tetrahedral-like configuration to a configuration where three of the arms of the junction lie approximately within the same plane and the fourth arm is slightly deviated out of this plane. Similarly, for the D7N resolvase-HJ complexes, addition of divalent metal ion switches the conformation of the arms from one arm being slightly out of plane to all four arms residing in approximately the same plane. These effects are likely due in part to binding of the divalent metal ions neutralizing charge repulsion between negatively charged phosphates on the adjacent DNA arms. We also observed a similar effect with the active site mutant--replacing a negatively charged residue at the active site with a neutral residue in D7N also seemed to help the arms of the junction to more closely approach one another. Since we observed this phenomenon under EDTA conditions, it appears to be a largely divalent metal ion independent effect. One explanation may be that a neutral residue can accommodate the negatively charged scissile phosphodiester at the active site, whereas the wild -type acidic residue cannot due to charge repulsion, and this leads to a tighter mobility constraint on the arms of the HJ by the active-site mutant protein.
We also observed relative electrophoretic patterns of resolvase/ HJ complexes consistent with deviation from two-fold symmetry of the HJ arm positions. This was unexpected, since no significant asymmetry was noted in the patterns obtained in the absence of protein, and FPV resolvase is expected to bind the HJ as a dimer with two-fold symmetry. Although the asymmetry was detectable in all the FPV resolvase/ DNA electrophoretic patterns, it was most prominent for the D7N protein in the presence of EDTA, where we interpreted the pattern to indicate that three of the arms lie approximately in the same plane and the fourth arm deviating out of plane. We speculate that the observed asymmetry in the patterns may be due to an inherent sequence preference of the enzyme, which is evident also in the highly oriented choice of cleavage sites on Junction 1.
Comparative electrophoresis studies have been carried out for bacterial RuvC, yeast Cce1, and the phage enzymes T4 endonuclease VII, T7 endonuclease I, and RusA18–23, allowing comparison to FPV resolvase. Each of these five protein -HJ complexes showed a different electrophoretic pattern, likely representing a unique overall structure for each. The Cce1 complex adopted a square planar structure both in the presence and absence of divalent metal ion. The RuvC complex showed a pattern consistent with a 2-fold symmetric X-like structure where all four arms of the HJ lie in the same plane both in the presence and absence of divalent metal ion. The T4 endonuclease VII complex showed a pattern consistent with a structure similar to that of RuvC, except that two diametrically opposed arms were deviated out of a plane and towards each other. This pattern also did not change in the presence or absence of divalent metal ion. The T7 endonuclease I complex showed a pattern consistent with the arms of the junction remaining coaxially stacked both in the presence and absence of divalent metal ion. Lastly, the RusA structure showed a pattern suggesting a tetrahedral positioning of the junction arms whether in the presence or absence of divalent metal ion.
In comparison to the above patterns, our data are most similar to the electrophoretic pattern of the RuvC complex, but with three notable differences. First, the fowlpox resolvase pattern had the asymmetries described above, second, the pattern was sensitive to divalent metal ion conditions, and third, the pattern was sensitive to an amino acid substitution at the enzyme active site. These three differences have not been observed in other published comparative electrophoresis experiments of HJ/ resolvase complexes. This suggests that the FPV resolvase/ HJ complex studied here has unique structural features.
In their analysis of the crystal structure of T4 endonuclease VII bound to a HJ, Biertumpfel, et al. observed that the S-shaped positive electrostatic surface potential of endonuclease VII explains much of the overall binding mode to the HJ studied 23. They proposed that other HJ resolving enzymes with similar S-shaped positive electrostatic surface potentials, such as RuvC and Cce1, may also be expected to have binding modes similar to that of endonuclease VII. In the endonuclease VII/ HJ crystal structure, the protein dimer contacts the HJ predominantly on the minor groove face of the HJ and the S-shape positive surface facilitates protein-DNA contacts on all four arms of the junction. Of the known HJ resolving enzymes, poxvirus resolvases share the most sequence similarity with that of RuvC and Cce1 and vaccinia virus resolvase has similarly been shown to bind the HJ as a dimer. Our cleavage site data indicate that the active sites of the FPV resolvase contact the minor-groove surface, since the cleavage sites are 3' to the branch point of the junction. Assuming a similar binding mode for FPV resolvase as with T4 endonuclease VII, we infer that FPV resolvase interacts predominantly with the minor-groove face of the HJ.
In summary, our structural models for the fowlpox resolvase-HJ complex presented here are most similar to those proposed for the RuvC, Cce1, and T4 endonuclease VII complexes. In each of these models, the arms of the HJ are unstacked, the branch point is open, and the complex has two-fold rotational symmetry about the dyad axis of the protein dimer, though for FPV resolvase bound to Junction 1, our data indicated slight departures from perfect two-fold symmetry, perhaps related to an inherent sequence preference. Finally, our investigation of the role of divalent metal ion cofactors in fowlpox resolvase catalysis is consistent with a catalytic mechanism involving multiple metal atoms, similar to that proposed for HIV integrase and other RNase H family enzymes. This implies that raltegravir-like inhibitors may also be active against poxvirus resolvase and useful for treating poxvirus infections.
EXPERIMENTAL PROCEDURES
Site-directed mutagenesis
The Holliday junction resolvase ORF was PCR amplified from fowlpox virus DNA, cloned into pET29a, expressed in E. coli, and purified as described 6. Expression plasmids conferring amino acid changes to the resolvase gene product were constructed using the QuikChange site directed mutagenesis kit (Stratagene) using the pET29a-resolvase vector as template with the appropriate oligonucleotide primer pairs. The desired DNA nucleotide changes were confirmed by DNA sequencing. Enzyme storage buffer was 300 mM NaCl, 20 mM HEPES (pH 8.0), 1 mM beta-mercaptoethanol, 0.1 mM EDTA, 40% glycerol. In some cases 50 mM arginine plus 50 mM glutamate was added as a solubilizing agent. Cce1 was purified as described 24, 25.
Construction of synthetic Holliday junctions
Sequences of the DNA oligonucleotides used to construct the synthetic Holliday junctions are compiled in Supplementary Table 1. HJFS was used for activity assays. Oligonucleotide 1 contains a 6-carboxyfluorescein (F) 3'-end label and was purified by high performance liquid chromatography (HPLC). Oligonucleotides 2–5 were purified by polyacrylamide gel electrophoresis (PAGE). DNA concentrations were determined by UV-spectrophotometry. The Holliday junction was assembled by annealing together oligonucleotides 1–4. The 40 bp duplex marker DNA was constructed by annealing together oligonucleotides 1 and 5. Duplex competitor DNA was constructed by annealing together an unlabeled version of oligonucleotide 1 with oligonucleotide 5. Annealing reactions contained 10 uM labeled DNA and 1.1-fold excess unlabeled DNA and were carried out in the presence of 100 mM NaCl by heating to 95 °C and allowing the solutions to cool slowly to room temperature over a period of 90 minutes. Junction 1 and Junction 1S were annealed as described under `Comparative electrophoresis analysis' below. The oligonucleotides which comprise these DNA junctions were purified by PAGE.
DNA cleavage assays
For tests of mutants in the metal rescue studies, except where noted, 240 nM enzyme was used with 2 nM HJFS (experiments in Table 1) or 20 nM HJFS (experiments in Table 2) in a final volume of 20 ul. Final solution conditions were 25 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM DTT, and 1% glycerol. Metal concentrations used for each reaction are specified in the Tables and Figure legends. Proteins were diluted in 10 mM HEPES pH 8.0, 1mM beta-mercaptoethanol, 0.1 mM EDTA, and 10% glycerol prior to addition to cleavage reactions. Reactions were initiated by addition of resolvase (Table 1) or metal cofactor (Table 2). Reaction mixtures were incubated at 37°C for the indicated times and then stopped with a 5x stop solution containing 100 mM EDTA, 5% SDS, 30% glycerol, and 0.1% bromophenol blue. Products of the reaction were separated on 10% polyacrylamide gels by electrophoresis under native conditions in 1X TBE buffer. The fluorescein-label was visualized using a Typhoon instrument (GE Healthcare). Gels were scanned for fluorescence (526 nm SP filter) after excitation with an Argon laser (488 nm). For kinetic assays, 15–18 points were used to define the reaction progression profile (see the Supplementary Report). Data was fit to a single exponential decay model using Prism software.
For the test of cleavage on the four strands of Junction 1S, wild type FPV resolvase protein was added to a solution containing 32P-labeled Junction 1S DNA to obtain a final volume of 20 ul (final concentrations 10 nM resolvase and 2 nM substrate). Final reaction conditions were 25 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM DTT, and 5% glycerol. Reaction mixtures were incubated at 37 °C for 30 min and then stopped with a formamide solution containing 100 mM EDTA, and, 0.1% bromophenol blue. Products of the reactions were separated on urea-containing 10% polyacrylamide gels by electrophoresis in 1X TBE buffer. Radioactivity was quantified using a Typhoon instrument (GE Healthcare). Cce1 was analyzed at 10 nM. T7 endo I was purchased from New England Biolabs (Beverley, MA).
Gel shift DNA binding assays
In binding reactions for gel shift analysis, except where noted, the indicated amount of fowlpox resolvase protein was added to a solution containing 2 nM of fluorescein-labeled Holliday junction DNA and 300-fold excess (wt/ wt) sonicated salmon sperm DNA to obtain a final volume of 20 ul. The final solution conditions were 25 mM Tris-HCl [pH 7.9], 100 mM NaCl, 1 mM EDTA, and 1–5% glycerol. Binding reactions were incubated at 37 °C for 15 min and then a 6x loading dye consisting of 30% glycerol and 0.1% bromophenol blue was added. 10 ul of the resultant solution was loaded on a 1.5 mm thick, 20 cm polyacrylamide gel (4%) and electrophoresed at ~85 V at 4 °C until the bromophenol blue reached the bottom of the gel. Products of the binding reaction were visualized as above. To compare binding affinities between enzymes, we estimated Kd values using nonlinear regression, taking a dimer as the binding species. The normalized data were plotted as 'fraction bound' versus 'log[protein]' and fit with a sigmoid dose-response model. Between 3 and 29 measurements went into each Kd measurement using 3 to 6 different protein concentrations over a 9 to 243-fold concentration range (we note that due to the narrow protein concentration range studied the error was high for a few samples). The Supplementary Report shows some examples of the quantitative binding data. Curves were fit using Prism software.
Comparative electrophoresis analysis
To study FPV resolvase binding during comparative electrophoresis, we first analyzed the stability of the wild-type enzyme/ DNA complex in the presence of EDTA. Competitor challenge experiments showed that dissociation was slow (>1hr for 50% dissociation, data not shown). In addition, FPV resolvase/ DNA complexes showed discrete mobilities in gels even during prolonged electrophoresis, further substantiating their stability for both D7N and wild-type in both the presence and absence of metal cofactors, all indicating that complexes were sufficiently stable for analysis.
The six long-arm/ short-arm versions of Junction 1 were prepared by first annealing stoichiometric proportions of the oligonucleotides which comprise Junction 1 together (see Table S1). DNA concentrations were determined by UV spectrophotometry. Before the annealing reaction, one of the DNA strands was 5'-end labeled with 32P using T4 polynucleotide kinase and gamma-32P-ATP, and the excess gamma-32P-ATP was removed using a G50 spin column (GE Healthcare). Annealing reactions contained 200 nM of each oligonucleotide, 100 mM NaCl, and 5 mM MgCl2. Annealing reactions were heated to 95 °C and then slow cooled to room temperature over five hours. After annealing, Junction 1 was digested with the six pairwise combinations of four restriction enzymes (BamHI [B], HindIII [H], EcoRI [R], and XbaI [X]) in reaction buffer containing 10 mM MgCl2, as recommended by the manufacturer (New England BioLabs). Each arm of Junction 1 contains a unique restriction site (B, H, R, or X, as above). The nomenclature of restriction digested Junction 1 products used in the text denotes the “long arms” (i.e. the non-digested arms), so that Junction “BR” refers to Junction 1 digested with HindIII and XbaI.
For the MgCl2 or EDTA binding reactions, the digested products were used directly without further purification. For binding reactions carried out in CaCl2, the residual MgCl2 in the DNA solution was first removed by dialysis against 1x TAE buffer (10 mM Tris-HCl [pH 7.9], 1 mM EDTA), so as to prevent DNA cleavage in binding reactions using wild-type fowlpox resolvase.
Binding reactions were carried out by adding 10 pmol (0.2 ug, 1 uM) of fowlpox virus resolvase protein to a solution containing 10 fmol (1 nM) of 32P-labeled Junction 1 and 30 ug/ ml poly(dI-dC)·poly(dI-dC) competitor DNA in a final volume of 10 ul. Solution conditions were 10 mM Tris-HCl [pH 7.9], 100 mM NaCl, 1 mM DTT, 5% glycerol, and either 10 mM EDTA (10-fold excess to MgCl2), 1 mM MgCl2, or 1 mM CaCl2. Binding reactions were incubated at 37 °C for 10 min and then 1/ 6 volume of a loading dye consisting of 15% Ficoll-400 and 0.1% bromophenol blue was added. Binding reactions were loaded directly onto 8% native polyacrylamide gels (width × length × thickness = 20.0 × 45.0 × 0.15 cm or 32.5 × 37.5 × 0.15 cm, 6 mm teeth) running at a constant 500 V. Electrophoresis was carried out at room temperature and stopped after 16 hours. The electrophoresis buffer used for the EDTA binding reactions was 1x TBE buffer (89 mM Tris-borate [pH 8.4], 2 mM EDTA). The electrophoresis buffers used for the MgCl2 and CaCl2 binding reactions were the same except, instead of 2 mM EDTA, the buffers contained 200 uM MgCl2 and 200 uM CaCl2, respectively. After electrophoresis, the wet gel was exposed to a phosphor screen and the DNA was visualized using a Typhoon instrument (GE Healthcare).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to members of the Bushman laboratory for help and suggestions. This work was supported by a grant from the NIH NIAID Mid-Atlantic Regional Center of Excellence for Biodefense Research Grant 2 U54 AI057168. The training grant T32 AI 07324 supported M. J. C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
abbreviations: FPV: fowlpox virus; HJ: Holliday junction (DNA four-way junction)
REFERENCES
- 1.Moss B. Poxviridae: The viruses and their replication. In: Fields BN, editor. Virology. Lippincott-Raven; Philadelphia: 2001. pp. 2637–2672. [Google Scholar]
- 2.Dickie P, Morgan AR, McFadden G. Cruciform extrusion in plasmids bearing the replicative intermediate configuration of a poxvirus telomere. J Mol Biol. 1987;196:541–58. doi: 10.1016/0022-2836(87)90031-3. [DOI] [PubMed] [Google Scholar]
- 3.Garcia AD, Aravind L, Koonin EV, Moss B. Bacterial-type DNA holliday junction resolvases in eukaryotic viruses. Proc Natl Acad Sci U S A. 2000;97:8926–31. doi: 10.1073/pnas.150238697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Culyba MJ, Harrison JE, Hwang Y, Bushman FD. DNA cleavage by the A22R resolvase of vaccinia virus. Virology. 2006;352:466–76. doi: 10.1016/j.virol.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Culyba MJ, Minkah N, Hwang Y, Benhamou OM, Bushman FD. DNA branch nuclease activity of vaccinia A22 resolvase. J. Biol. Chem. 2007;282:34644–34652. doi: 10.1074/jbc.M705322200. 10.1074/ jbc.M705322200. [DOI] [PubMed] [Google Scholar]
- 6.Culyba MJ, Hwang Y, Minkah N, Bushman FD. DNA binding and cleavage by the fowlpox virus resolvase. J. Biol. Chem. 2009;284:1190–1201. doi: 10.1074/jbc.M807864200. 10.1074/ jbc.M807864200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia AD, Moss B. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J Virol. 2001;75:6460–71. doi: 10.1128/JVI.75.14.6460-6471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Steitz TA. Recombining the structures of HIV integrase, RuvC, and RNase H. Structure. 1995;3:131–134. doi: 10.1016/s0969-2126(01)00142-3. [DOI] [PubMed] [Google Scholar]
- 9.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/ DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Lovell S, Goryshin IY, Reznikoff WR, Rayment I. Two-metal active site binding of a Tn5 transposase synaptic complex. Nat Struct Biol. 2002;9:278–81. doi: 10.1038/nsb778. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. Embo J. 1991;10:4381–9. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lilley DM, White MF. Resolving the relationships of resolving enzymes. Proc Natl Acad Sci U S A. 2000;97:9351–3. doi: 10.1073/pnas.97.17.9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA Integration: Mechanism of Viral DNA Cleavage and DNA Strand Transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 14.Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, Jones P, Kinzel O, Laufer R, Monteagudo E, Muraglia E, Nizi E, Orvieto F, Pace P, Pescatore G, Scarpelli R, Stillmock K, Witmer MV, Rowley M. Discovery of Raltegravir, a Potent, Selective Orally Bioavailable HIV-Integrase Inhibitor for the Treatment of HIV-AIDS Infection. J. Med. Chem. 2008;25:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 15.Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, Wai JS, Young S, Vacca J, Hazuda DJ. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci U S A. 2002;99:6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia AD, Otero J, Lebowitz J, Schuck P, Moss B. Quaternary stru cture and cleavage specificity of a poxvirus holliday junction resolvase. J Biol Chem. 2006;281:11618–26. doi: 10.1074/jbc.M600182200. [DOI] [PubMed] [Google Scholar]
- 17.Culyba MJ, Hwang Y, Minkah N, Bushman FD. DNA binding and cleavage by the fowlpox virus resolvase. J. Biol. Chem. 2008;284:1190–1201. doi: 10.1074/jbc.M807864200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilley DM, White MF. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2001;2:433–443. doi: 10.1038/35073057. [DOI] [PubMed] [Google Scholar]
- 19.Hadden JM, Convery MA, Declais AC, Lilley DM, Phillips SE. Crystal structure of the Holliday junction resolving enzyme T7 en donuclease I. Nat Struct Biol. 2001;8:62–7. doi: 10.1038/83067. [DOI] [PubMed] [Google Scholar]
- 20.Hadden JM, Declais AC, Phillips SE, Lilley DM. Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I. Embo J. 2002;21:3505–15. doi: 10.1093/emboj/cdf337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadden JM, Declais AC, Carr SB, Lilley DM, Phillips SE. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature. 2007;449:621–624. doi: 10.1038/nature06158. 10.1038/ nature06158. [DOI] [PubMed] [Google Scholar]
- 22.Declais AC, Lilley DM. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr. Opin. Struct. Biol. 2008;18:86–95. doi: 10.1016/j.sbi.2007.11.001. 10.1016/ j.sbi.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Biertumpfel C, Yang W, Suck D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007;449:616–620. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- 24.White MF, Lilley DM. The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction. J. Mol. Biol. 1997;266:122–134. doi: 10.1006/jmbi.1996.0795. [DOI] [PubMed] [Google Scholar]
- 25.White MF, Lilley DM. The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J. Mol. Biol. 1996;257:330–341. doi: 10.1006/jmbi.1996.0166. [DOI] [PubMed] [Google Scholar]
- 26.Bujacz G, Alexandratos J, Wlodawer A, Merkel G, Andrake M, Katz RA, Skalka AM. Binding of Different Divalent Cations to the Active Site of Avain Sarcoma Virus Integrase and Their Effects on Enzymatic Activity. J. Biol. Chem. 1997;272:18161–18168. doi: 10.1074/jbc.272.29.18161. [DOI] [PubMed] [Google Scholar]
- 27.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka AM. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure. 1996;4:89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 28.Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 29.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 30.Katzman M, Katz RA, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao K, Wong S, Bushman F. Metal binding by the D,DX35E motif of human immunodeficiency virus type 1 integrase: selective rescue of Cys substitutions by Mn2+ in vitro. J. Virol. 2004;78:6715–22. doi: 10.1128/JVI.78.13.6715-6722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diamond TL, Bushman FD. Role of metal ions in catalysis by HIV integrase analyzed using a quantitative PCR disintegration assay. Nucleic Acids Res. 2006;34:6116–25. doi: 10.1093/nar/gkl862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon PM, Sontheimer EJ, Piccirilli JA. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: extending the parallels between the spliceosome and group II introns. RNA. 2000;6:199–205. doi: 10.1017/s1355838200992069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida A, Sun S, Piccirilli JA. A new metal ion interaction in the Tetrahymena ribozyme reaction revealed by double sulfur substitution. Nat Struct Biol. 1999;6:318–21. doi: 10.1038/7551. [DOI] [PubMed] [Google Scholar]
- 35.Yoshikawa M, Iwasaki H, Kinoshita K, Shinagawa H. Two basic residues, Lys-107 and Lys-118, of RuvC resolvase are involved in critical contacts with the Holliday junction for its resolution. Genes Cells. 2000;5:803–13. doi: 10.1046/j.1365-2443.2000.00371.x. [DOI] [PubMed] [Google Scholar]
- 36.Espeseth AS, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed JY, Young S, Hamill T, Cole JL, Hazuda DJ. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl. Acad. Sci. U S A. 2000;97:11244–9. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.